Abstract
During fasting, mammals maintain glucose homeostasis by stimulating hepatic gluconeogenesis1. Elevations in circulating glucagon (GLU) and epinephrine trigger the cAMP mediated phosphorylation of Creb and dephosphorylation of the Creb coactivator Crtc22. Although the underlying mechanism is unclear, hepatic gluconeogenesis is also regulated by the circadian clock, which coordinates glucose metabolism with changes in the external environment3–6. Here we show that Creb activity during fasting is modulated by Cryptochromes (Cry1 and Cry2), core components of the clock that are rhythmically expressed in the liver. Cry was elevated during the night/day transition, when it reduced fasting gluconeogenic gene expression by blocking GLU-mediated increases in intracellular cAMP concentrations and in the PKA-mediated phosphorylation of Creb. In biochemical reconstitution studies, we found that Cry inhibited accumulation of cAMP in response to G protein coupled receptor (GPCR) activation but not to forskolin, a direct activator of adenyl cyclase. Cry appeared to modulate GPCR activity directly through interaction with Gsα . As hepatic over-expression of Cry lowered blood glucose concentrations and improved insulin sensitivity in insulin resistant db/db mice, our results suggest that compounds which enhance Cry activity may provide therapeutic benefit to individuals with type II diabetes.
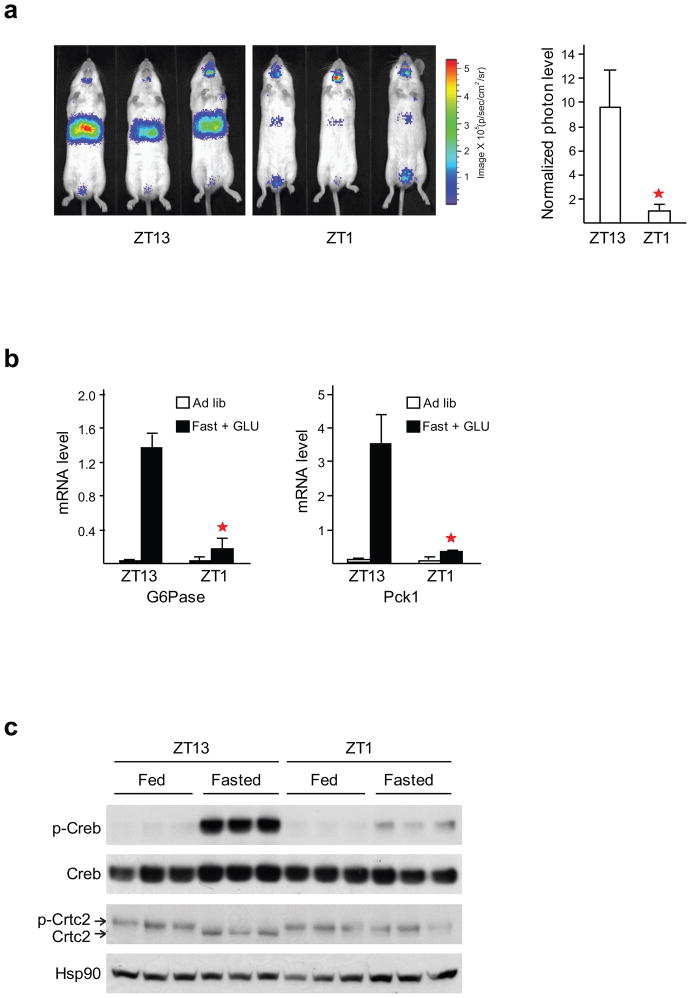
We used an adenovirally encoded CRE:Luciferase reporter to evaluate the rhythmicity of hepatic Creb activity under fasting conditions7. Intraperitoneal GLU administration (GLU-IP) increased Ad-CRE:Luc activity 40-fold better in mice fasted at Zeitgeber time 10–13 (ZT10–13, the day-to-night transition) than at ZT22–1, corresponding to the night-to-day transition (Fig. 1a and Supplementary Fig. 1). Consistent with a rhythmic oscillation in Creb activity, GLU re-injection to the same mice fasted at ZT6–9, near the day-to-night transition, caused a robust increase in hepatic CRE:Luciferase reporter activity. These results suggest that the amplitude of hepatic Creb activation by fasting is gated rhythmically.
Figure 1. Hepatic Creb:Crtc2 activity is modulated by the circadian clock.
(a) CRE:Luciferase reporter activity in mice fasted for 3 hours followed by intra-peritoneal (IP) GLU administration. Relative effect of fasting at ZT10–13 and ZT22–1 is indicated. Right, bar graph shows CRE:Luc activity from hepatic lysates normalized to beta galactosidase activity from co-infected Ad-RSV:β-gal adenovirus. Asterisks indicate P < 0.01, n = 5. (b) Q-PCR analysis of gluconeogenic gene expression (G6Pase, left; Pck1, right) in mice fasted at ZT10–13 or ZT22–1 and then injected IP with GLU. Asterisks indicate P < 0.01, n = 5. (c) Immunoblot of Creb and Crtc2 proteins in liver extracts from mice fasted ZT10–13 or ZT22–1 followed by IP with GLU. Relative effects ZT13 and ZT1 fasting on hepatic amounts of phospho-Crtc2 and phospho-Creb (Ser133) indicated.
Based on the changes in fasting CRE:Luc reporter activity, we hypothesized that the circadian clock modulates Creb and Crtc2 effects on the gluconeogenic program. Supporting this idea, GLU administration increased gluconeogenic gene expression (G6Pase, Pck1) to a greater extent in mice fasted at ZT10–13 (Fig. 1b). Consistently, GLU-IP administration increased hepatic Creb phosphorylation and Crtc2 de-phosphorylation robustly in ZT10–13 fasted mice, but less in ZT22–1 fasted mice (Fig. 1c). Together, these results indicate that the clock modulates fasting gluconeogenesis in part by regulating Creb-Crtc2 activities. These observations are consistent with recent reports showing that the phosphorylation of Creb is rhythmic in vivo8.
Owing to the rhythmic Creb phosphorylation, we considered that the liver clock may exert global effects on the expression of cAMP inducible genes. In line with this idea, we identified 194 Creb targets among a set of 652 clock-controlled genes (CCGs) in published microarray databases of mouse liver9–11. We plotted the sum of the Creb targeted CCGs, along with their ratio compared to the total CCGs at each time-point, versus circadian time (CT) based on their peak expression. Both the number and ratio curves show a clear rhythmic pattern with depletion of Creb targets at CT22 (Supplementary Fig. 2), suggesting that Creb targets are generally rhythmic in the liver with the trough phase coinciding with that of Per2.
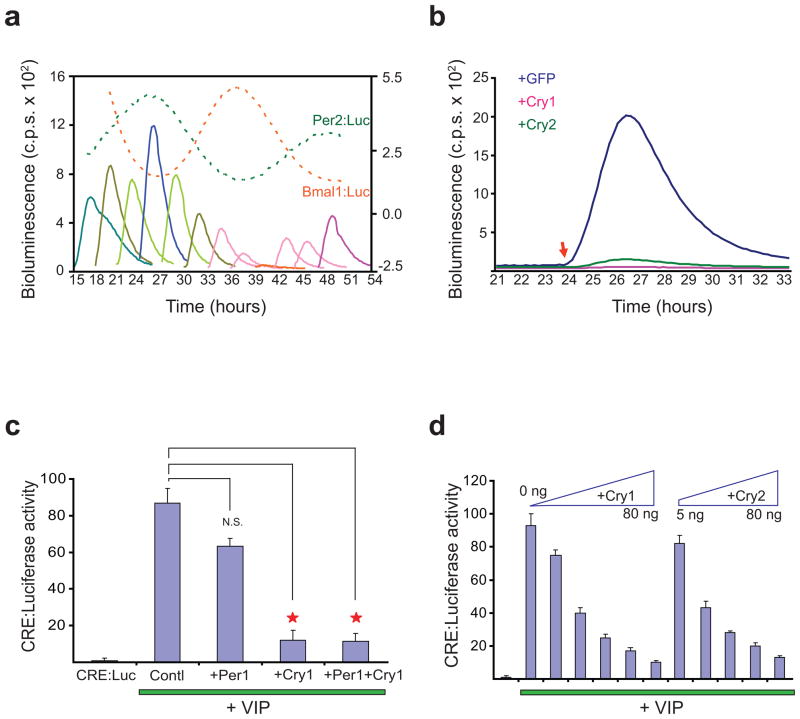
We evaluated the extent to which rhythmic gating of Creb activity is cell autonomous. Exposure to vasoactive intestinal peptide (VIP) increased CRE:Luc activity in mouse adult fibroblasts harboring the receptor Vipr212 (Fig. 2). After synchronizing the cells by serum shock, exposure to VIP at specific times resulted in CRE:Luc reporter activation that showed a rhythmic pattern, with peak (24, 48 h) and trough (36–39 h) activities coinciding with those for a Per2:Luciferase reporter (Fig. 2a). These results show the regulation of Creb pathway induction by cell autonomous circadian clock, and are reminiscent of the circadian gating seen in the light signaling induction of a PER2::LUC reporter in melanopsin-containing fibroblasts13.
Figure 2. Cry inhibits Creb activity.
(a) CRE:Luciferase reporter activity in Vipr2-expressing mouse fibroblasts synchronized by serum shock and then exposed to VIP at indicated times. Replicate samples (a total of 24) received VIP treatment every 3 h post-synchronization from 15–48 h. For clarity, one responsive curve per time point was shown in different colour. Mouse fibroblasts harbouring Per2:dLuc (green dashed lines) or Bmal1:dLuc reporters (red dashed lines) were used to report the two distinct phases of clock gene expression (scale at right). Data are representative of three experiments. (b) Effect of Cry over-expression on CRE:Luc reporter activity. Vipr2/CRE:Luc cells were infected with lentiviral particles expressing either GFP, Cry1 or Cry2. Cells were treated with VIP 24 h after medium change (red arrow). (c–d) Cry represses CRE:Luc induction in transfected cells expressing Vipr2. CRE:Luc (20 ng) and Vipr2 (40 ng) expression plasmids were co-transfected into HEK 293T cells along with Per1, Cry1 or Cry2 (40 ng) as indicated. 24 h post transfection, cells were treated with VIP (1 nM) for 8 h to induce CRE:Luc activity. N.S., not significance; Asterisks indicate P < 0.01 analyzed by student t-test.
Because CRE:Luc activity in liver and in synchronized fibroblasts is lowest when Per-Cry activity is highest (ZT22), we tested the potential role of Cry in regulating Creb signaling. Over-expression of Cry1 or Cry2 in Vipr2/CRE:Luc fibroblasts or HEK293T cells attenuated CRE:Luc activity in cells exposed to VIP (Fig. 2b–d), while Per1 did not show significant effect (Fig. 2c). The repression was dosage-dependent (Fig. 2d). Consistent with the absence of E-boxes within the CRE:Luc reporter, Clock-Bmal1 did not activate CRE:Luc expression (Supplementary Fig. 3)10. Furthermore, Cry1/2 overexpression reduced effects of urocortin-3 on CRE:Luc activity in cells expressing Corticotropin-releasing factor receptor 2β 14 (Supplementary Fig. 4). Thus, these data indicate that Cry may exert more general effects on GPCR-mediated increases in Creb-Crtc2 dependent transcription.
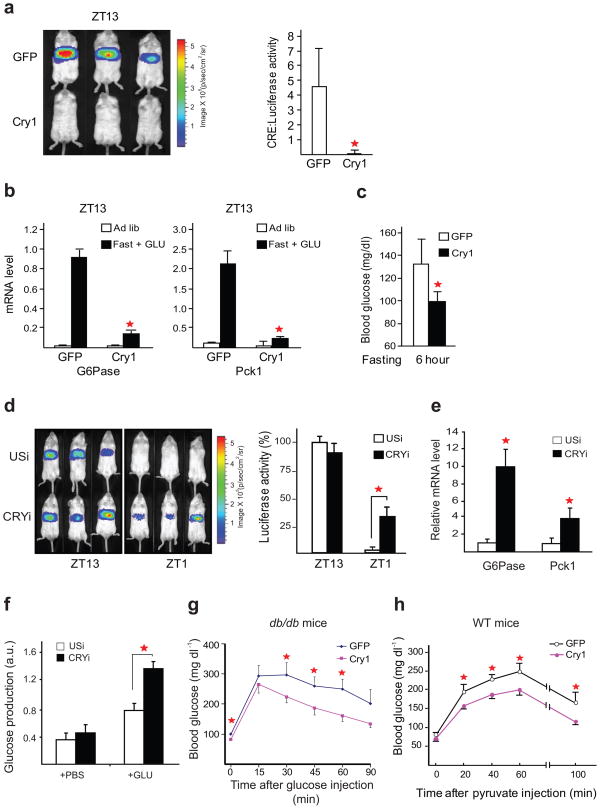
Based on its inhibitory effects on GPCR signaling in fibroblasts, Cry may correspondingly modulate GLU effects on liver gene expression. Supporting this idea, hepatic expression of adenovirally encoded Cry1 (Ad-Cry1) reduced GLU-induced CRE:Luc activity in liver of fasted mice at ZT10–13, when endogenous Cry1 expression is low (Fig. 3a). Moreover, Ad-Cry1 also down-regulated gluconeogenic gene expression (G6Pase, Pck1) in fasted, GLU-IP administered mice (Fig. 3b). Consequently, Ad-Cry1 expressing mice exhibited lower circulating blood glucose concentrations compared to Ad-GFP controls (Fig. 3c).
Figure 3. Cry blocks induction of the gluconeogenic program by Creb and Crtc2.
(a) Left, live imaging analysis of hepatic CRE:Luc reporter activity in control (Ad-GFP) and Ad-Cry1 expressing mice. CRE:Luc activity in fasted mice (ZT10–13) injected with GLU is shown. Right, quantitative analysis of CRE:Luc activity. Asterisks indicate P < 0.01, n = 5. (b) Q-PCR analysis of gluconeogenic gene expression (G6Pase, Pck1) in control and Cry1 expressing fasted and fed mice. Asterisks indicate P < 0.05, n = 5. (c) Effect of Cry1 over-expression on fasting blood glucose concentrations relative to control GFP expressing mice. P < 0.05, n = 3. (d) Left, live imaging analysis of CRE:Luc reporter activity in mice expressing adenovirally encoded Cry1/2 or unspecific RNAis (USi) in liver. Right, quantitative analysis of CRE:Luc activity. Asterisk indicates P < 0.01, n = 4. (e) Q-PCR analysis of hepatic G6Pase and Pck1 mRNA amounts in control (USi) and CRYi expressing mice. P < 0.001 for G6Pase and P < 0.01 for Pck1, n = 3. (f) Glucose production assay in primary hepatocytes infected with Ad-CRYi or control. Asterisks indicate P < 0.01, n = 3. (g) Glucose tolerance testing of db/db mice expressing adenovirus encoded Cry1 or GFP. Asterisks indicate P < 0.01, n = 5. (h) Pyruvate tolerance testing of WT mice expressing adenovirus-encoded Cry1 or GFP. Fasted mice were injected with pyruvate and blood glucose concentrations were measured at indicated times. Asterisks indicate P < 0.05, n = 5.
We tested whether depletion of Cry, either by targeted disruption of Cry1 and Cry2 genes15, or by RNAi-mediated knockdown in the liver, was sufficient to increase Creb activity. By contrast with the circadian oscillation of hepatic CRE:Luc induction in wild-type mice, GLU-stimulated CRE:Luc activity was constitutively elevated in Cry1−/−Cry2−/− animals (Supplementary Fig. 5). Similarly, RNAi-mediated depletion of hepatic Cry1/2 increased GLU-stimulated CRE:Luc activity at ZT1 relative to control animals expressing unspecific RNAi (USi, Fig. 3d). As a result, hepatic mRNA amounts for gluconeogenic genes (G6Pase, Pck1) were increased in Cry1/2-knockdown mice compared to controls (Fig. 3e). Circulating glucose concentrations were elevated following Cry depletion (Supplementary Fig. 6), suggesting that circadian changes in hepatic Cry expression are sufficient to modulate the gluconeogenic program in part through their inhibitory effects on the Creb-Crtc2 pathway. Indeed, knockdown of Cry1/2 also increased glucose production in primary hepatocytes following exposure to GLU, indicating that Cry effects on hepatic glucose metabolism are likely cell autonomous (Fig. 3f).
We wondered whether Cry would similarly modulate circulating glucose concentrations in the setting of insulin resistance. Supporting this idea, adenoviral expression of Cry1 reduced fasting blood glucose concentrations and improved whole body insulin sensitivity in insulin resistant db/db mice, as measured by glucose tolerance testing (Fig. 3g). Pointing to an effect on hepatic gluconeogenesis, Cry1 over-expression also lowered glucose excursions in pyruvate tolerance testing studies (Fig. 3h).
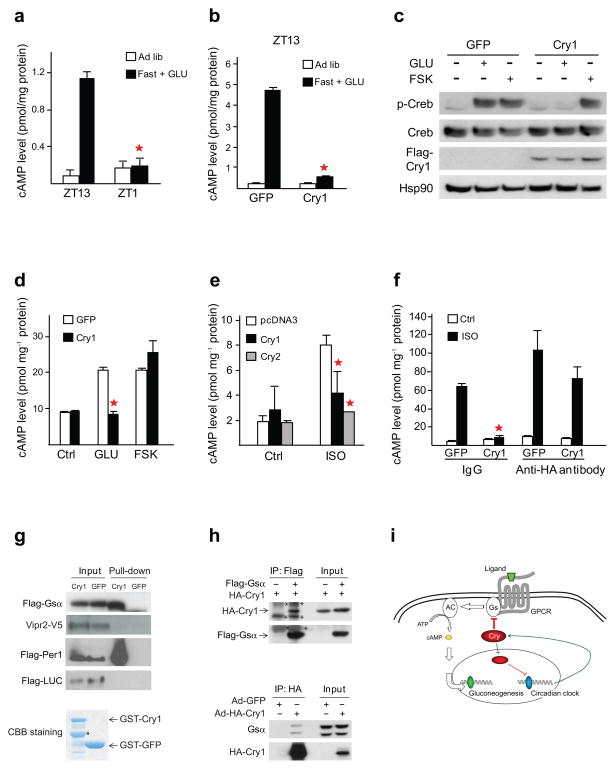
Based on the rhythmic changes in hepatic Creb phosphorylation, we suspected that Cry1 may disrupt effects of GLU on adenyl cyclase (AC) activation during fasting (Fig. 1c). Supporting this idea, GLU-IP administration increased hepatic cAMP concentrations at ZT10–13, when Cry1 expression is low, and to a lesser extent at ZT22–1, when Cry1 levels are elevated (Fig. 4a). Consistently, Ad-Cry1 expression reduced hepatic cAMP concentrations in ZT10–13 fasted mice exposed to GLU (Fig. 4b). Indeed, amounts of phosphorylated Creb were down-regulated in livers of fasted Ad-Cry1 mice compared to GFP control animals (Fig. 4c).
Figure 4. Cry inhibits GPCR-dependent increases in adenyl cyclase activity.
(a) Hepatic cAMP concentrations in fed or fasted mice following IP injection of GLU as indicated. (b) Effect of Cry1 over-expression on hepatic cAMP levels in fasted mice following IP GLU administration. (c) Immunoblot showing effect of Ad-Cry1 expression on hepatic amounts of phospho-Creb (Ser133) in fed and fasted mice. (d) Effect of Adenoviral Cry1 expression on intracellular cAMP concentrations in hepatocytes exposed to FSK or GLU. (e–f) Effect of Cry1 and Cry2 on cAMP accumulation. In vitro reconstitution studies showing effect of cytosolic fractions from control (GFP) and Cry1 expressing cells on cAMP accumulation in reactions containing plasma membrane fractions from HEK293T cells. (g) Immunoblot showing recovery of Gsα and Vipr2 from membrane fractions of transfected HEK293T cells incubated with recombinant GST- Cry1 or GST-GFP control. Upper panels: input and pull-down signals using 293T cells overexpressed with Flag-Gsα , Vipr2-V5, Flag-Per1, and Flag-Luciferase (LUC), respectively. Lower panel: GST-tagged proteins were purified and stained with Coomassie Brilliant Blue (CBB). Asterisk points to truncated GST-Cry1 polypeptide. (h) Co-immunoprecipitation of Gsα and Cry1. Upper panels: Immunoblot showing recovery of HA-Cry1 from immunoprecipitates of Flag-Gsα prepared from HEK293T cells. Asterisks point to non-specific signals. Lower panels: Immunoblot showing recovery of endogenous Gsα from immunoprecipitates of HA-Cry1 prepared from primary mouse hepatocytes. (i) Circadian regulation of cAMP signaling in liver is Cry-dependent. Blue oval represents E-box bound transcription factors CLOCK-BMAL1, and green oval represents CRE-mediated transcription activators or co-activators such as Creb, p300/Cbp and Crtc2.
We evaluated effects of Cry on Creb-Crtc2 signaling in cultures of primary hepatocytes. Similar to its effects in liver, Ad-Cry1 also down-regulated CRE:Luc reporter activity, as well as gluconeogenic gene expression in cells exposed to GLU (Supplementary Fig. 7a,b). By contrast with its effect in cells exposed to GLU, Ad-Cry1 did not reduce gluconeogenic gene expression or CRE:Luc reporter activity in cells treated with forskolin (FSK), a direct activator of AC. Consistent with these results, Ad-Cry1 expression reduced cAMP accumulation in cells exposed to GLU but not to FSK (Fig. 4d). In addition, RNAi-mediated knockdown of Cry1 and 2 increased cAMP content in primary hepatocytes exposed to GLU but not to FSK (Supplementary Fig. 7c). Pointing to a more general effect of this repressor, Cry1/2 over-expression also reduced cAMP accumulation in response to VIP treatment in fibroblasts expressed Vipr2 (Supplementary Fig. 8). Together, these results indicate that Cry1 acts upstream of AC to regulate hepatic Creb activity during fasting.
To evaluate further the mechanism, we performed in vitro reconstitution assays. Addition of isoproterenol (ISO) to plasma membranes from HEK293T cells, which express β-adrenergic receptors, increased cAMP accumulation in vitro (Fig. 4e,f). Co-incubation of plasma membrane fractions with cytoplasmic extracts from transfected HEK293T cells expressing Cry1 potently inhibited cAMP increases in response to ISO relative to control (GFP) extracts. We considered that Cry1 could disrupt cAMP accumulation either directly by binding to relevant signaling components, or indirectly by stimulating the expression of an intra-cellular inhibitor. Supporting the former hypothesis, immuno-depletion of HA-tagged Cry1 with HA-specific antiserum restored cAMP accumulation in response to ISO, whereas control IgG had no effect (Fig. 4f). Taken together, these results demonstrate that Cry1 inhibits activation of the Creb-Crtc2 pathway in liver by disrupting cAMP production in response to ligand-dependent activation of Gs coupled receptors.
Based on its ability to disrupt GPCR but not FSK-mediated increases in cAMP production, Cry would be predicted to associate with either GPCRs or with the Gsα subunit of heterotrimeric G protein. Although it did not interact detectably with the GPCR family member Vipr2, Cry associated with Gsα in pull-down assays of membrane fractions from transfected HEK293T cells incubated with recombinant glutathione-S-transferase (GST)-Cry protein (Fig. 4g). We also observed the Cry-Gsα interaction in reciprocal co-immunoprecipitation assays of HEK293T cells and primary hepatocytes (Fig. 4h). Together, these data demonstrate a role for extra-nuclear Cry in regulating cAMP production.
The circadian clock has been known to modulate hepatic gluconeogenesis for over 20 years, although the underlying mechanism has remained unclear. We found that the liver clock regulates the gluconeogenic program through Cry-mediated inhibition of Creb activity during fasting. Historically regarded as a transcriptional repressor, Cry appeared to inhibit gluconeogenic gene expression primarily by blocking GPCR mediated increases in cAMP and in the subsequent phosphorylation of Creb (Fig. 4i). These results are consistent with recent findings that hepatic Creb phosphorylation oscillates in a circadian fashion8. Similar to our observations in liver, cAMP concentrations have also been found to oscillate in the clock master organ, the suprachiasmatic nucleus, reaching a nadir when Cry expression is highest16. Future studies should provide further insight in to the detailed mechanism by which Cry modulates Gs effects on cAMP in liver and other tissues.
Methods
Adenoviruses and animals
We delivered adenoviruses (1 × 108 p.f.u.) Cry1, control GFP, Cry1i, Cry2i, or control USi into 8–10-week-old male C57BL/6J-Tyrc-2J/J (Jackson Laboratories) mice together with CRE:Luc (1 × 108 p.f.u.) and RSVβ-gal (5 × 107 p.f.u.) by tail- vein injection. Targeted sequences for knockdown were: 5’-GGAAATTGCTCTCAAGGAAGT-3’ (Cry1i) and 5’-GCTGAATTCGCGTCTGTTTGT-3’ (Cry2i). All mice were housed in colony cages with 12 h light-dark cycle for 4 weeks before study.
In vivo Imaging
We fasted mice beginning at ZT10 or ZT22 and injected intraperitoneally (IP) with GLU (100 mg kg−1; Sigma) at ZT13 or ZT1, respectively, and continued fasting for 1 h. At the end of fasting (ZT14 or ZT2), we injected IP with 50 mg kg−1 Nembutal (Abbott Laboratories) and 100 mg kg−1 sterile firefly D-luciferin (Biosynth AG), and imaged them on the IVIS-100 Imaging System within 15–45 min post-injection of luciferin, and analyzed with Living Image software (Xenogen) 7,17.
In vivo Analysis
We sonicated mouse tissues at 4 °C, centrifuged, and reserved supernatants for β-gal activity, protein determinations, and immunoblotting analysis. Blood glucose values were determined using a LifeScan automatic glucometer.
Cell Culture
We cultured cells in DMEM with 10% FBS and transfected with plasmid DNA by using lipofectamine2000. Primary hepatocytes were isolated and cultured as described7.
Luminometery and luciferase assay
We grew cells harboring Vipr2 and CRE:Luc reporter to confluence in 35 mm dishes, followed by a 2 h serum-shock and released to serum-free medium. We applied synthetic VIP (2 nM in saline, American Peptide) to different dishes 15–48 h post-serum shock at 3 h intervals. We collected data in a luminometer (Actimetrics). Luciferase reporter assay using HEK293T cells was described18.
mRNA Analysis
Were extracted total RNA from whole liver or primary hepatocytes using the RNeasy kit (Qiagen), and measured mRNA levels as reported7.
Glucose production assay
We prepared primary hepatocytes as described18. Briefly, we seeded 5 × 105 cells per well in a 6-well dish with M199 medium (Invitrogen). Adenoviruses infected (M.O.I. of 5 for each CRYi and 10 for control USi) for 120 min prior to additional incubation for 42–45 h. We treated cells with glucagon (10 μg ml−1, Sigma) for 5 h, washed, and incubated with glucose production buffer (saline supplemented with 20 mM sodium lactate and 2 mM sodium pyruvate) for additional 2 h. We collected the supernatant for measuring glucose concentration (BioVision), and normalized the readings with whole-cell protein amount.
Glucose Tolerance Tests (GTT)
We fasted mice overnight and injected IP with glucose (1 g kg−1 body weight, Sigma). We measured blood glucose levels before the injection, and at 15, 30, 45, 60, and 90 min after injection.
Pyruvate Tolerance Tests (PTT)
We fasted mice for 24 h and injected IP with sodium pyruvate (2 g kg−1, Sigma). We measured blood glucose before the injection, and at 20, 40, 60, and 100 min after injection.
Cell fractionation
We collected and washed HEK293T cells, and resuspended them in lysis buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, and proteinase inhibitors), and centrifuged at 500 × g for 5 min. Supernatants were ultracentrifuged at 40,000 × g for 30 min. The resulting supernatant was allocated as cytosol fraction and the pellet was resuspended in extraction buffer (75 mM Tris-HCl, pH 7.4, 1 mM EDTA, 10% glycerol and 12.5 mM MgCl2, and proteinase inhibitors) as membrane fraction. We measured protein concentrations, adjusted to 1 μgμl−1, and stored them in −80° C freezer.
Adenyl cyclase activity assay
We mixed equal volume of membrane and cytosol fractions, incubated at 37° C for 5 min, and then mixed with one volume reaction buffer (5 mM Tris-HCl, pH 7.4, 17.5 mM MgCl2, 120 μM ATP, 3 μM GTP, 1.5 μM FAD, 60 mM phosphocreatine and 0.375 Unit Creatine phosphokinase) in the presence or absence of isoproterenol (100 μM) or FSK (1 μM) for reaction at 37° C for 30 min, stopped by mixing with HCl and neutralized by NaOH. We measured cAMP levels by ELISA kit (R&D). For the pre-clearance of HA-Cry1, cytosol fractions from Ad-HA-Cry1 infected cells were immuno-precipitated by HA-specific antibody or mouse IgG beads at 4° C for 1 h.
GST pull-down
We prepared GST-Cry1 and control GST-GFP proteins with Glutathione-Sepharose beads (GE Healthcare) as described19. We dissolved membrane pellets in pull-down buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 2mM Dithiothreitol, and proteinase inhibitors), and mixed with 10μg GST proteins. After 2 h incubation at 4° C and washes, we suspended pellets in SDS loading buffer, and applied 1% input samples for quantification.
Immunoprecipitation
We conducted immunoprecipitation assays using whole cell lysates.
Statistical analyses
We performed all studies at least three independent occasions. We reported results as mean and s.e.m., and considered differences statistically significant when P < 0.05.
Supplementary Material
Acknowledgments
We thank L. Vera, A. Luzader, E. Rodrigo, and X. Li for adenoviral injections, J. Altarejos and M. Lindstrom for mouse blood collection and glucose measurement, and N. Goebel for preparing primary hepatocytes. We also thank M. Yamout and P. E. Wright at The Scripps Research Institute for reagents, and N. Gekakis, D. Welsh and S. Wang for reading the manuscript. This is manuscript #080809 of Genomics Institute of the Novartis Research Foundation, and was supported in part by grants from US National Institutes of Health (R01 GM074868 and R01 MH051573 to SAK; R01 DK083834 and R01 DK049777 to MM). SAK is a co-founder of ReSet Therapeutics and is a member of its Scientific Advisory Board. YL is supported by “One Hundred Talents” Program of Chinese Academy of Sciences (No. 2010OHTP08) and Shanghai Pujiang Program (No. 10PJ1411200) in China.
Footnotes
Author Contributions
EEZ and SAK conceived the project. EEZ, YL, MM and SAK designed the research. EEZ, YL, RD, PYP, ACL, TH, DAN, XS, SL and YK performed the experiments. EEZ and YL analyzed the data. EEZ, YL, DAB, MM and SAK wrote the paper.
Author Information
All mouse works were conducted under regulations of Institutional Animal Care and Use Committee at Salk, GNF and CAS. The authors declare that there is no competing financial interest with this work. Reprints and permissions information is available at http://npg.nature.com/reprintsandpermissions.
References
- 1.Nordlie RC, Foster JD, Lange AJ. Regulation of glucose production by the liver. Annual Review of Nutrition. 1999;19:379–406. doi: 10.1146/annurev.nutr.19.1.379. [DOI] [PubMed] [Google Scholar]
- 2.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kida K, et al. The circadian change of gluconeogenesis in the liver. The Journal of Biochemistry. 1980;88:1009–1013. doi: 10.1093/oxfordjournals.jbchem.a133051. [DOI] [PubMed] [Google Scholar]
- 5.Gelling RW, et al. Lower blood glucose, hyperglucagonemia, and pancreatic α cell hyperplasia in glucagon receptor knockout mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1438–1443. doi: 10.1073/pnas.0237106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramsey KM, Marcheva B, Kohsaka A, Bass J. The clockwork of metabolism. Annual Review of Nutrition. 2007;27:219–240. doi: 10.1146/annurev.nutr.27.061406.093546. [DOI] [PubMed] [Google Scholar]
- 7.Dentin R, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 8.Vollmers C, et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proceedings of the National Academy of Sciences. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 10.Ueda HR. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulivarthy SR, et al. Reciprocity between phase shifts and amplitude changes in the mammalian circadian clock. Proceedings of the National Academy of Sciences. 2007;104:20356–20361. doi: 10.1073/pnas.0708877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Chen P, Vaughan J, Lee KF, Vale W. Urocortin 3 regulates glucose-stimulated insulin secretion and energy homeostasis. Proceedings of the National Academy of Sciences. 2007;104:4206–4211. doi: 10.1073/pnas.0611641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Horst GTJ, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 16.O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dentin R, Hedrick S, Xie J, Yates J, III, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 18.Sato TK, et al. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanada K, Harada Y, Sakai M, Todo T, Fukada Y. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes to Cells. 2004;9:697–708. doi: 10.1111/j.1356-9597.2004.00758.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.