Abstract
The inappropriate expression of the c-MET cell surface receptor in many human solid tumors necessitates the development of companion diagnostics to identify those patients who could benefit from c-MET targeted therapies. Tumor tissues are formalin-fixed and paraffin embedded (FFPE) for histopathological evaluation, making the development of an antibody against c-MET that accurately and reproducibly detects the protein in FFPE samples an urgent need. We have developed a monoclonal antibody, designated MET4, from a panel of MET-avid monoclonal antibodies, based on its specific staining pattern in FFPE preparations of normal human prostate tissues. The accuracy of MET4 immunohistochemistry (MET4-IHC) was assessed by comparing MET4-IHC in FFPE cell pellets with immunoblotting analysis. The technical reproducibility of MET4-IHC possessed a percentage coefficient of variability (%CV) of 6.25% in intra-assay and inter-assay testing. Comparison with other commercial c-MET antibody detection reagents demonstrated equal specificity and increased sensitivity for c-MET detection in prostate tissues. In two cohorts of ovarian cancers and gliomas, MET4 reacted with ovarian cancers of all histological subtypes (strong staining in 25%) and with 63% of gliomas. In addition, MET4 bound c-Met on the surfaces of cultured human cancer cells and tumor xenografts. In summary, the MET4 monoclonal antibody accurately and reproducibly measures c-MET expression by IHC in FFPE tissues and can be used for molecular imaging in-vivo. These properties encourage further development of MET4 as a multipurpose molecular diagnostics reagent to help to guide appropriate selection of patients being considered for treatment with c-MET-antagonistic drugs.
Keywords: c-MET, monoclonal antibody, ovarian cancer, glioma, molecular diagnostics, immunohistochemistry
Introduction
The c-MET receptor tyrosine kinase (RTK) regulates cellular proliferation, migration, and differentiation during development and tissue homeostasis in response to its ligand, hepatocyte growth factor/scatter factor (HGF/SF) (1, 2). c-MET is also one of the most frequently activated oncogenes in human cancer (http://www.vai.org/met/) and, thus, represents an attractive therapeutic target. In its activated state, the c-MET receptor controls growth, invasion and metastasis of cancer cells through multiple signal transduction pathways (3) and c-MET activation occurs in many ways. c-MET oncogenic activity increases through mutations such as in the kinase or juxtamembrane domains (4–7), through overexpression (8, 9), or through binding to HGF/SF (10, 11). Activating mutations of c-MET in the germline, which stimulate ligand-independent activation, are the cause of hereditary papillary renal carcinomas (7). In cancerous cells, selective amplification of the mutated c-MET allele further enhances overall c-MET kinase activity (12). Kinase-activating mutations in c-MET are also observed in sporadic renal, lung, head and neck, hepatocellular carcinoma, non-small cell lung cancer (NSCLC), gastric cancer and melanoma (9, 13–16).
The level of c-MET expression predicts the aggressiveness of a number of cancer types and has been associated with poor prognosis in many cancers, including those of the breast, stomach, cervix, liver, and of the head and neck (17–22). In breast cancer, c-MET expression is observed in cancers associated with increased cell proliferation (23, 24). Furthermore, amplification of the c-MET locus has been detected in gastric, metastatic colorectal and esophageal adenocarcinoma (9, 25). However, c-MET overexpression can also occur through transcriptional activation of the c-MET gene, without amplification (26) and these cancers could also be affected in their growth rate through c-MET inhibition. In ductal breast cancer, simultaneous expression of Syndecan-1, E-cadherin and c-MET correlates with enhanced angiogenesis and lymphangiogenesis (27).
A major advance occurred recently in the development of inhibitors of c-MET with remarkably favorable pharmacodynamic properties and low toxicity. These inhibitors prevent c-MET autophosphorylation, delay the growth of xenografts (15, 28–30) and are in phase I clinical trials. Thus, when used to treat cancers that may have an active c-MET axis, these drugs could indeed benefit many cancer patients, provided that accurate pre-treatment discrimination of inappropriate c-MET-expressing from nonexpressing cancers can be achieved. Unfortunately, ex vivo patient stratification through immunohistochemical evaluation of c-MET expression in formalin-fixed and paraffin embedded (FFPE) surgical samples is currently problematic. This is due to a limited number of validated monoclonal antibodies to c-MET that work in FFPE (31).
This study reports the development of an antibody, called MET4, which reacts with an epitope in the extracellular domain of c-MET in FFPE tissues using conventional antigen retrieval methods. We compare MET4 reactivity with the reactivity with commercial antiserum commonly used for detection of c-MET in tissue sections. In addition, we analyze ovarian cancers and gliomas, which are known to express c-MET and for which current therapies are limited. Moreover, we provide evidence that MET4 reacts strongly with c-MET as expressed on the surfaces of cultured human cancer cells as well as by human tumor xenografts raised in immunocompromised mice. The data from this study strongly indicate that the MET4 antibody could prove valuable as a multipurpose companion diagnostic reagent to the growing repertoire of c-MET inhibitory drugs.
Materials and Methods
Commercial antibodies
The following c-MET-avid antibodies were purchased: MET C-28 (Lot Numbers C082 and C1207, Santa Cruz, Hercules, CA). DL-21 (cat# 05–238 MILLIPORE), 3D4 (Zymed ®, Invitrogen, Carlsbad, CA), MAB3729 (Chemicon, MILLIPORE). Negative and control antibodies for IHC were obtained from Vector Laboratories (Burlingame, CA) (mouse) or from Jackson Laboratories (Bar Harbor, ME) (rabbit).
Monoclonal antibody generation and validation
Recombinant MET 25–567H and MET 928 proteins were prepared as previously described (32, 33) and recombinant fusion protein MET-IgG was purchased from R&D systems (Minneapolis, MN).
Mouse monoclonal antibodies were produced by injecting BALB/c mice intraperitoneally (IP) with native and SDS-denatured Met 25–567H in complete Freund’s adjuvant, followed by two additional injections with incomplete Freund’s adjuvant. The final injection was given IP and intravenously (IV) without adjuvant. Polyclonal antisera from immunized mice were tested by indirect immunofluorescence with formalin-fixed MKN45 (MET-positive) and NIH3T3 (MET-negative) cells. Spleen cells were fused with P3X63AF8/653 myeloma cells using standard techniques 4 days after final injection. Hybridoma cells were screened for reactivity against Met by ELISA and immunofluorescent staining. For screening, ten 96-well plates were coated with 2 μg/ml of MET25–567H in coating buffer (0.2M Na2CO3/NaHCO3, pH 9.6; 50 μl per well) overnight at 4° C. The plates were then blocked with PBS containing 1% BSA (200 μl/well) overnight at 4° C. Fifty microliters of hybridoma supernatant were added to wells for 1.5 h at room temperature (RT). Plates were washed twice in washing buffer (PBS with 0.05% Tween-20), and alkaline phosphatase-coupled goat anti-mouse IgG (Sigma) was added (50 μl/well) at 1:2000 dilution for 1.5 h at RT. After washing four times in washing buffer, phosphatase substrate CP-nitrophenyl phosphate (Kirkegaard & Perry Laboratories) was added for 30 min, and absorbance was measured at 405 nm. A total of 34 hybridomas were selected having strong reactivity with MET25–567H (A405nm > 2.0). The qualifying hybridomas were further tested against MET 928 by ELISA. Of the original panel of 34 clones, 14 hybridoma clones produced antibodies that reacted with both MET25–567H and MET928. By testing multiple forms of the receptor, we expected to select antibodies directed against the most stable epitopes. Therefore these 14 hybridomas were further evaluated by immunofluorescent staining against human c-MET positive MKN45 gastric carcinoma cells co-cultivated with c-MET negative NIH3T3 cells plated on a 96-well format. Cells were incubated overnight and then washed and fixed with 10% formalin. Fifty microliters of hybridoma supernatant were added to wells for 1.5 h at 37°C. Plates were washed twice in washing buffer (PBS with 0.05% Tween-20), and Rhodamine red-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Lab) was added (30 μl/well) at 1:100 dilution for 1.5 h at 37°C. After washing, cells were examined under a fluorescence microscope. Five positive clones were picked and expanded. Supernatants of these five clones were validated by immunohistochemistry on formalin-fixed normal human prostate tissue sections. Clone 8G6 (designated MET4) gave the strongest signal and was sub-cloned twice and validated by ELISA against three recombinant MET proteins. Monoclonal antibody was produced using a bioreactor and MET4 was purified using protein-G affinity column by FPLC.
Immunohistochemistry
Cell pellets
NIH 3T3, S114 (NIH 3T3 cells transfected with the human genes for HGF/SF and MET), SW-1783, U118, U87, U373 (human brain glioblastoma from ATCC). MCF-7 (breast cancer), OV-90, TOV-112D, IGROV1 and TOV-21G (ovarian adenocarcinoma) cells were obtained from ATCC and cultured according to their media specifications. 1847, A2780 and OVCAR10, cell lines (ovarian adenocarcinomas) were obtained from the Pacific Ovarian Cancer Research Consortium (Seattle, WA). After fixation in 10% neutral buffered formalin (Anatech Ltd., Battle Creek, MI) for 30 minutes, cell layers were scraped, centrifuged for 5 minutes at 2,000 rpm and all the liquid was removed. HistoGel (Richard-Allan Scientific) was melted according to manufacturer’s instructions and equilibrated to 42°C. Cell pellets were suspended in an equal volume of HistoGel and solidified on ice for 5 min. Formalin fixation of HistoGel embedded cell pellet was continued overnight at 4°C. Thereafter, cell pellets were placed in 70% ethanol at 4°C until processing. HistoGel pellets were processed for 8 hours in a tissue processor and embedded in paraffin.
Human tissues
Formalin-fixed and paraffin embedded sections of ovarian cancer were obtained from the Pacific Ovarian Cancer Research Consortium under an IRB approved protocol. Tissue microarrays from gliomas were purchased from Cybrdi (Frederick, MD). Tissues on slides were stained with MET4 and other antibodies within 48 hours of sectioning or stored in a nitrogen atmosphere. Antigen retrieval was performed with Target Retrieval Solution, pH9 (DAKO, Denmark) for 20 minutes in a steaming water bath. The slides were loaded on the Dako Autostainer. All of the following incubations were performed at room temperature on the Autostainer: Endogenous peroxidase activity was blocked for 8 minutes with 3% hydrogen peroxide; a protein blocking step using Dako Serum-Free Protein Block was then completed for 10 minutes. The monoclonal mouse anti-human MET4 antibody was diluted in Tris buffer/1%BSA 1: 300 for staining of prostate tissues, 1:150 for staining of TMA slides and 1:500 for staining of cell pellets. The MET C-28 polyclonal antibody was diluted 1:300, 3D4 1:500, AMB3729 1:500 and DL-21 1:10 for staining of prostate tissues. Other antibody dilutions are indicated in Table 3. For the isotype control, purified mouse IgG was diluted to match the MET4 concentration. Biotinylated anti-mouse or anti-rabbit IgG (Vector Labs) secondary antibodies were diluted at 1:200 and were applied for 30 minutes. Then, Peroxidase-conjugated Streptavidin (Jackson ImmunoResearch) was used at 1:2,000 for 30 minutes. Finally, Dako Liquid DAB+ Substrate Chromagen System was applied for 7 minutes. The slides were counterstained with Dako Automation Hematoxylin for 2 minutes, dehydrated and coverslipped.
Table 3.
Comparison of Immunoreactivity of c-MET Antibodies in FFPE Tissues
| MET4 (1:150) | DL21 (1:10) | MAB3729 (1:500) | 3D4 (1:500) | |
|---|---|---|---|---|
| Basal cells | 4+ | 2+ | 4+ | 3+ |
| Luminal cells (membrane) | 4+ | 0 | 1+ | 2+ |
| Luminal cells (cytoplasm) | 0 | 0 | 3+ | 0 |
| Epithelial nuclei | 0 | 0 | 2+ (30%) | 0 |
| Stromal cells | 1+ | 0–1+ | 3+ | 2+ |
| Endothelial cells | 4+ | 1+ | 4+ | 0–1+ |
| Inflammatory cells (B and T cells) | 3+ | 0 | 1+ | 0 |
| Inflammatory cells (other) | Granular cells in the prostate stroma 4+ | Granular cells in the prostate stroma 4+ | 0 | 2+ |
For each cell pellet, a negative control was included in all IHC experiments and stained with a mouse isotype control. As a control for the MET C-28 polyclonal antibody, normal rabbit serum was used as a negative control for which no signal was detected (data not shown).
The scoring of immunohistochemically stained sections was done as described (34). Briefly, a cumulative score was derived by multiplying the percentage of positive cells by the staining intensity. Cumulative scores were divided into categories from 0 to 3 and grouped by histologic subtype (ovarian cancer) or tumor grade (glioma).
Western blot analysis
Subconfluent cells were lyzed in RIPA supplemented buffer with protease and phosphatase inhibitors (Roche U.S. Pharmaceuticals, Nutley, NJ) as described (35). Protein lysates from cell lines were measured using a Bradford assay. Samples were measured in the same assay using BSA standards ranging from 0 to 12 μg/ml. Samples were diluted such that 1–2 μl of sample provided measurements within the linear range of the standard curve. Samples (50 μg) were resolved on a 4–15% gradient SDS–PAGE and transferred onto Immobilon™-P PVDF (Millipore, Billerica, MA) or nitrocellulose transfer membranes (Bio-Rad, Hercules, CA). Membranes for the formalin-fixed Western blot were fixed in 2% neutral buffered formalin Z-fix (Anatech Ltd., Battle Creek, MI) for 20 minutes and washed in PBS. Antigen retrieval was achieved with Target Retrieval Solution, pH9 (Dako, Carpinteria, CA) for 20 minutes in a steamer (Black and Decker). The membrane was blocked with 5% milk blocking buffer at room temperature and incubated with MET C-28 in 5% BSA at 1:250 dilution for 1.5 h at room temperature or at 1:1000 dilution over night; MET4 was used at 1:1000 and c-Met clone:3D4 at 1:500 for Western blotting. Donkey anti-rabbit IgG–HRP (Amersham) was used at 1:5,000 or AlexaFluor 680 goat anti-mouse IgG (Molecular Probes) was used at 1:10,000. Protein bands were detected by a chemiluminescence reagent (Pierce) or Odyssey® Infrared Imaging System (LI-COR Biosciences, Lincoln, Nebraska). Images scanned on the Odyssey® Infrared Imaging System were scanned using the 700nm channel at an intensity of 5.0. The signal intensity of the c-MET specific bands was measured using the 1D gel analysis module in the ImageQuant TL software. Background was subtracted using the “rolling ball” tool.
Digital image analysis of IHC-stained cell pellet sections
Slides with sections of cell pellets stained with MET4 were spectrally imaged using a CRI Nuance camera system (www.cri-inc.com). 6– 10 images were collected from each slide. Emission was measured between 420nm and 720nm in 20nm increments. The resulting image cubes were converted to optical density units, and were mathematically unmixed into their individual DAB and hematoxylin components using spectrums deduced from control specimens and saved in a spectral library. The DAB stain was pseudo-colored red to increase contrast. The hematoxylin was pseudo colored blue and the images were converted to pseudo-fluorescent format for quantification.
The spectrally unmixed images were then quantified with software developed by our laboratory that identified the mean DAB optical density as well as the number of pixels above background (as determined using the auto-thresholding algorithm of Ridler and Calvard (36). The cell count was determined by counting the nuclei (as identified by hematoxylin counter stain) using the connected component algorithm (37) on pixels whose hematoxylin optical density was above background. A count of total nuclei was determined after using an auto high pass threshold filter. The threshold for positive MET4 staining was set using an intensity threshold filter value of 171 intensity units. This threshold parameter was defined using c-MET negative control cell lines (A2780, OVCAR10 and TOV-112D) and restricting the number of positive pixels to 0.1%. This is similar to the PPA statistics (38). These counts were then used to determine the percent positive MET4 cells for each sample pellet.
Statistical analysis
For statistical analysis, the Pearson’s correlation coefficient was calculated using Microsoft© Excel software. Components of variance for biological variability within the TOV21G cell pellet and technical variability, which comprises variability due to antibody staining and image acquisition were estimated from a mixed-model analysis by employing the MIXED procedure in SAS.
In vitro and in vivo binding studies with MET4
MET-overexpressing human cancer cell lines
SK-LMS-1/HGF cells (human leiomyosarcoma cell line autocrine for human c-Met and human HGF/SF) (39–41) and the human osteosarcoma cell line, MNNG-HOS, was cultured in DMEM containing 10% FBS and maintained at 37 °C under an atmosphere of 5% CO2.
Fluorescence-activated cell sorting (FACS) analysis
Human Met-expressing cancer cell lines SK-LMS-1/HGF (leiomyosarcoma) and MNNG-HOS (osteosarcoma) were grown in culture, detached and suspended, and subjected to FACS analysis after incubation with MET4 followed by fluorescein-conjugated anti-mouse IgG1 (secondary antibody), performed using a FACS Calibur flow cytometer and the CellQuest analysis program (Becton Dickinson).
Tumor xenografts and nuclear imaging with MET4
SK-LMS-1/HGF and MNNG-HOS cells were used for xenograft induction. Female athymic nude (nu/nu) mice at about two months of age received subcutaneous injections of cell suspension (5 ×105 cells) in the posterior aspect of the right thigh. Tumors developed for 3–4 weeks, reaching 0.5 cm or more in greatest dimension by external caliper measurement before imaging. Mice were housed in small groups and given ad libitum access to mouse chow and drinking water under conditions approved by the institutional animal care committee. For nuclear imaging experiments, MET4 was equilibrated in 0.25M sodium phosphate buffer, pH 7, stored frozen in small aliquots, and thawed just prior to labeling. Protein radioiodination was performed as previously described (42, 43). 125I was purchased as NaI (480–630 MBq [13–17 mCi] per μg iodine) from Amersham Corp. (Arlington Heights, IL).
Host mice were imaged and scintigrams were analyzed by established methods (42, 43). In brief, each tumor xenograft-bearing mouse received I-125-MET4 (~160 μCi, ≤ 0.1 ml; injected within 24 hours of radiolabeling) intravenously by tail vein injection under light inhalation anesthesia. Just prior to each imaging session, each mouse was given up to 13 mg/kg xylazine and 87 mg/kg ketamine subcutaneously in the interscapular region. Serial posterior whole-body gamma camera images of each mouse were acquired beginning as early as one hour following I-125-MET4 injection and continuing for several days postinjection. Sedated mice were placed in groups on top of an inverted camera head with a protective layer over the collimator and taped to the layer to maintain optimum limb extension. Images of I-125 activity were acquired on a remanufactured ZLC-Scintron camera (MiE America) with a low-energy, high-sensitivity collimator. Acquisitions were obtained over a period of 15–30 min.
Relative activity was determined by computer-assisted region-of-interest (ROI) analysis for each tumor, for total body, and for appropriate background regions at each imaging time point. These data are expressed as background- and decay-corrected activity ratios. Graphical and statistical analysis of the converted data utilized the program Excel (Microsoft).
Results
Development of an anti-MET monoclonal antibody for detection of c-MET in formalin-fixed tissues
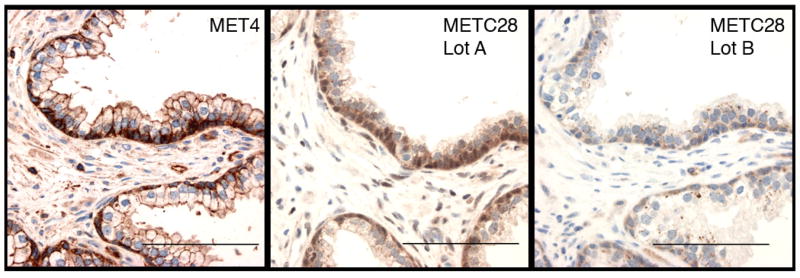
The limitations of available antibodies to measure c-MET expression in immunohistochemical studies led to our development of a monoclonal antibody (mAb) for measurement of c-MET protein expression in formalin-fixed tissues. Polyclonal antisera are commonly affected by lot-to-lot variability from differences in the animals in which they are raised. When we tested several lots of the polyclonal rabbit antisera to Met that is most commonly used for c-MET analysis (MET C-28), we observed lot-to-lot differences in prostate tissue as previously shown (31) (Figure 1). While lot A showed the expected staining of basal prostate epithelial cells, lot B revealed punctate cytoplasmic staining in secretory cells. To overcome the problems with polyclonal antibodies, we generated monoclonal antibodies (mAbs) that are easier to standardize for a clinical application. The immunogen was designed to obtain c-MET antibodies that bind to the extracellular domain (ecto-domain). To identify monoclonal antibodies directed against the most prevalent and least variable epitopes, our selection process included multiple forms of the ecto-domain of c-MET, including MET 25–567H, MET 928, MET-IgG, and c-MET on the surface of MKN45 cells (Table 1). As a final screen to identify mAbs that specifically bind to formalin-treated Met receptor, we used sections of archival normal prostate tissues (Table 1). Hybridoma supernatant “MET4” gave the strongest specific staining, consistent with the distinct subcellular pattern of c-MET receptor expression in prostate (44–46) (Figure 1). MET4 was further validated by staining co-cultures of c-MET-reactive MKN45 cells and non-reactive NIH3T3 cells, that were fixed in 10% formalin. The immunofluorescent staining was specifically observed along the membrane of MKN45 cells and coincides with immunoreactivity of MET C-28 (Figure 2A).
Figure 1. Specificity of MET4 immunohistochemistry.
Sections of FFPE normal prostate tissue were stained with MET4 or with two different lots of MET C-28 (400X). MET4 expression specifically occurs in basal epithelial cells, intercellular plasma membrane of luminal prostate cells and endothelial cells.
Table 1.
Selection Process for Isolation of the MET4 Hybridoma*
| Hybridoma selection | Negative selection criteria | Positive hybridoma clones for next screen |
|---|---|---|
| HAT medium survival after splenocyte fusion | 960 | |
| ELISA positive against Met25–567H | 34 | |
| ELISA positive against both Met25–567H and Met928 | 14 | |
| ELISA positive against 3 proteins: Met25–567H, Met928 and Met-IgG | 7 | |
| Positive in formalin-fixed MKN45 cells | Negative in formalin-fixed NIH3T3 cells | 5 |
| Positive by IHC in prostate basal epithelial cells, endothelial cells, plasma cells, faint in prostate stromal cells | Negative in prostate secretory cells (cytoplasm), extracellular matrix | 2/5 |
Mouse monoclonal antibodies were produced by injecting BALB/c mice i.p. with native and SDS-denatured Met 25–567H in complete Freund’s adjuvant, followed by two additional injections with incomplete Freund’s adjuvant. The final injection was given i.p. and i.v. without adjuvant. Polyclonal antisera from immunized mice were tested by indirect immunofluorescence with formalin-fixed MKN45 (c-MET positive) and NIH3T3 (c-MET negative) cells prior to splenocyte fusion.
Figure 2. Comparison of MET4 and MET C-28 reactivity.

A Immunofluorescent staining with MET4 and MET C-28. Gastric cancer MKN45 cells (c-MET positive) and NIH3T3 cells (c-MET negative) were co-cultured and fixed with formalin. Panels represent MET4 staining, C-28 staining, MET4 and C-28 overlay and a brightfield image (400X). B. Representative Western blot of ovarian cancer and glioma cell lines probed with MET C-28. Equal amounts of protein were loaded in each lane and the loading confirmed by staining with beta-tubulin. RNA expression results commensurate to the observed protein expression are shown in Supplementary Figure 5. C. Immunohistochemical staining with MET4 in FFPE cell pellets. A brown color indicates positive c-MET receptor expression. MET4 was diluted 1:500. (200X). The bar demarks 100 microns.
Avidity of MET4 for Met in formalin-fixed cells
To confirm the reactivity of MET4 with c-MET, we probed a Western blot with MET4. To approximate the conditions of formalin-fixation in tissues, Western blot membranes were treated with 10% formalin and underwent the same type of antigen retrieval as we applied to tissue sections. MET4 did not react with c-MET in IGROV1 ovarian cancer cells in Western blots, which express large amounts of the antigen. However, exposing the membrane to formalin and steam to mimic IHC conditions of antigen retrieval facilitated MET4 binding to c-MET (Supplementary Figure 4). MET4 did not react in a specific fashion with the A2780 cell line, which does not express MET RNA (Supplementary Figure 5). The immunofluorescence and Western blotting results demonstrate that MET4 binds to a specific epitope on c-MET that is sensitive to denaturation and that is re-established and enhanced by formalin-fixation and by antigen retrieval.
To further validate the specificity of MET4 in formalin-fixed cell preparations, we analyzed two panels of cell lines (Figure 2B). Cell lines were selected based on c-MET expression levels. The first panel consisted of 7 ovarian cancer cell lines. These cell lines express variable amounts of c-MET or none at all: A2780, OVCAR10 and TOV-112D do not express MET RNA or protein. The absence of MET RNA was confirmed by PCR (Supplementary Figure 5). OV-90 and TOV21G express some c-MET, while IGROV1 cells express high levels. We also examined a series of cell pellets comprised predominantly of glioblastoma tumor cell lines (U118, DBTRG, SW1783, U87, U373), NIH3T3 cells transfected with human c-MET (S114) and two human c-MET negative cell lines (NIH3T3 and MCF7). To evaluate the specificity of MET4 in formalin-fixed preparations, we processed HistoGel-embedded cell pellets of each cell line in the exact same way as human tissues. Cell pellets on slides were stained with MET4 and the reactivity was compared to MET C-28 reactivity in a Western blot. MET4 reacted with only the formalin-fixed cell pellets that were positive for c-MET by Western blot analysis (Figure 2B and C), but not with those negative for c-MET.
Reproducibility of MET4 immunohistochemical measurements
Since the results demonstrate that the novel MET4 antibody specifically binds c-MET in FFPE cell preparations, we tested its reproducibility in TOV21D cells, which express c-MET at intermediate levels. The assay reproducibility is a critical factor in the development of an antibody for ex vivo clinical tests. We analyzed the consistency of MET4 staining intensities in FFPE cell pellets amongst replicate stains on the same day (intra-assay reliability) and slides stained on three different days (inter-assay reliability) (Table 2). In addition, we compared the staining results from two separate lots of MET4 antibody. The two lots of purified antibody, showed good correlation. However, we noticed that in general, images taken at different areas of the cell pellet possess significant variability in staining intensity, which reduced the correlation coefficient in the lot-to-lot comparison (correlation coefficient of σ = 0.75). To obtain an accurate assessment of intra- and inter-assay reproducibility, we therefore calculated the intraclass correlation coefficient. This statistical method separates biological and technical variabilities and revealed that the biological variability amounted to 98% of the total variability and the technical variability only to 2%. Consequently, the %CV attributable to technical variability amounted to a %CV = 6.8% for the intra-assay and a %CV = 6.2% for the inter-assay variability. These data demonstrate that MET4-IHC possesses excellent reliability and lot-to-lot consistency for analysis of formalin-fixed cell preparations.
Table 2.
Reproducibility and Consistency of MET4 Staining
| MET4 lot-to-lot consistency (Correlation of lot1 and lot2) | Intra-assay %CV (average from 3 data sets) | Inter-assay %CV (average form 3 days) | ||
|---|---|---|---|---|
| technical | biological | technical | biological | |
| σ = 0.75 | 6.8 % | 33.2% | 6.5% | 14.5% |
Comparison of MET4 to other commercial c-MET reactive antibodies
Recently, several monoclonal antibodies against c-MET have been developed and demonstrated to bind c-MET in formalin-fixed tissues (Table 3). All specifically bind to c-MET in Western blots (31 and data not shown). While MET4 and DL21 bind to extracellular epitopes on the c-MET receptor, 3D4 and MAB3729 react with the intracellular portion of c-MET. After titrating each antibody to optimize its signal-to-noise ratio, we observed significant differences in cell type specific immunoreactivity between MET4 and the other antibodies. In FFPE prostate tissues, all antibodies react with c-Met in basal cells, however MET4 best displays c-MET expression in luminal epithelial cells at the basal-lateral plasma membrane, suggesting that MET4 possesses a higher avidity than the other antibodies. In addition to prostate epithelial cells, endothelial cells and subpopulations of inflammatory cells express c-Met. Stromal cells are weakly and diffusely positive for c-MET staining and c-Met is detected in cultures of primary prostate stromal cells by immunoblotting (data not shown).
The staining with DL21 is weak, even at a high antibody concentration. The strongest immunoreactivity was observed in granular inflammatory cells, which are dispersed in the stroma. Compared to prostate epithelial cells, endothelial cells react weaker with DL21 than with MET4. The MAB3729 antibody strongly stains basal, but not luminal prostate epithelial cells. In some glands, this antibody also stains the cytoplasm and nuclei, a staining pattern not observed with the other antibodies. The prostate stroma is highly positive. The 3D4 antibody reactivity in the prostate epithelium is most similar to MET4. However, the stroma is more reactive, and endothelial cells are rarely positive.
In summary, in correlation with independent measurements by immunoblotting, c-MET is expressed in basal epithelial, stromal and endothelial cells in the prostate. Using these cell types to evaluate the specificity of c-MET antibodies revealed higher specificity or affinity of MET4 than of the other c-MET reactive antibodies. Consequently, MET4 possessed a greater dynamic range and was less sensitive to susceptible to suboptimal tissue fixation and slide storage conditions (data not shown).
c-MET expression in ovarian cancers and gliomas
To evaluate the specificity of MET4-IHC in archival FFPE tissues, we analyzed sections of ovarian cancer tissues and a TMA of gliomas. MET4 possessed high staining specificity for ovarian cancer cells, endothelial cells, a population of cells in the ovarian stroma, and of plasma cells. Nuclear staining was not observed (Figure 3A). Histologic subtypes of ovarian cancers included serous, endometrioid and clear cell, but not mucinous cancers, with all cancers in FIGO stage III - IV. All 29 ovarian cancers showed MET4-IHC reactivity (Table 4). No significant statistical differences were observed in MET4 staining intensity across subtypes of ovarian cancer. Clear cell cancers, which are considered the most aggressive subtype, did not demonstrate greater reactivity than other histologic subtypes. Endothelial cells are MET4 positive and were used as an internal reference to confirm adequate tissue quality and antigen retrieval for MET4 and to standardize staining intensities of MET4 between slides.
Figure 3. MET4 reactivity in human ovarian cancers and gliomas.
Sections of ovarian cancers (A) or a glioma tissue microarray (B) were stained with MET4 at 1:150 dilution. Images were taken at 400X magnification. Negative and positive controls of prostate tissues were included in each staining. The bar demarks 100 microns.
Table 4.
MET4 Reactivity in Ovarian Cancers
| Score | Frequency (pts) | Percentage | Histologic Subtype | ||
|---|---|---|---|---|---|
| Serous (% S) | Endometriod (% EM) | Clear cell (% CC) | |||
| 0 | |||||
| 1 | 9 | 30 | 3 (23%) | 4 (36%) | 2 (40%) |
| 2 | 12 | 45 | 6 (46%) | 5 (45%) | 1 (25%) |
| 3 | 7 | 25 | 4 (31%) | 2 (18%) | 1 (25%) |
Sections of ovarian cancer were stained with MET4. The overall score represents the average staining intensity of the section on a categorical scale of 0 – 3. (S – papillary serous carcinoma, EM – endometriod carcinoma, CC – clear cell carcinoma)
To evaluate the specificity of MET4-IHC and expression of c-MET in brain tumors, we stained a tissue array of gliomas. The tissue microarray contained 18 gliomas in triplicate cores. In addition, there were three samples of normal brain tissues (Table 5, Figure 3B). Neurons and reactive astrocytes stained strongly with MET4, while oligodendrocytes did not. Surprisingly, endothelial cells varied in c-MET expression, including endothelial cells of tumor vasculature. Five of 18 gliomas were c-MET negative, while 3/18, all three were high-grade gliomas, stained very strongly. Altogether, 63% of gliomas expressed c-MET, and the expression level increased with tumor grade.
Table 5.
MET4 Reactivity of Gliomas (54 cores)
| Score | Frequency (Cores) | Percent (Cores) | Frequency (Patients) | Percent (Patients) |
|---|---|---|---|---|
| 0 | 21 | 34 | 5 | 37 |
| 1 | 22 | 35 | 7 | 36 |
| 2 | 13 | 21 | 3 | 16 |
| 3 | 6 | 10 | 3 | 11 |
| Total | 62 | 100 | 18 | 100 |
A tissue microarray of 18 gliomas on triplicate cores was stained with the MET4 antibody. MET4 reactivity was assessed by multiplying the relative staining intensity and the percentage of positive cells. The overall score is expressed on a categorical scale from 0 to 3.
In vitro and in vivo binding studies
To evaluate the suitability of MET4 for binding studies in vitro and in vivo, we performed FACS analysis on established c-MET-overexpressing human cancer cell lines grown in culture (Figure 4) and nuclear imaging studies of I-125-MET4 uptake and retention by c-MET-overexpressing human tumor xenografts raised in immunocompromised mice (Figure 5).
Figure 4. MET4 binding to c-MET-expressing human cancer cell lines as evaluated by fluorescence-activated cell sorting (FACS) analysis.
Human c-MET expressing cancer cell lines SK-LMS-1/HGF (leiomyosarcoma) and MNNG-HOS (osteosarcoma) were grown in culture, detached and suspended, and subjected to FACS analysis after incubation with MET4 followed by fluorescein-conjugated anti-mouse IgG1 (secondary antibody). The solid histogram represents the fluorescence intensity distribution of cells in the absence of primary or secondary antibody; green represents cells incubated with second antibody in the absence of MET4; and the red histogram shows the complete mixture (cells plus MET4 and secondary antibody). A significant shift in the fluorescent intensity signal (from solid or green to red) indicates avid binding of MET4 to the cell surface.
Figure 5.
Figure 5A: Images of I-125-MET4 uptake by c-MET-expressing human tumor xenografts in nude mice. Serial total body scintigrams (nuclear images) in posterior projection of single mice bearing subcutaneous xenografts in the right thigh of either SK-LMS-1/HGF (leiomyosarcoma) or MNNG-HOS (osteosarcoma), and injected intravenously with I-125-MET4 (~160 μCi nominal dose) are shown. Images are shown from 5–6 hours to 5–6 days postinjection. Activity in the lower right of each image represents tumor-associated radioactivity.
Figure 5B: Uptake and clearance of I-125-MET4 in human tumor xenograft-bearing and control immunocompromised mice. c-MET monoclonal antibody designated MET4 was labeled with radioactive iodide (I-125) by the chloramine-T method. Within 24 hours I-125-MET4 was injected at a nominal dose of 160 μCi intravenously via a lateral tail vein into female athymic nude mice (nu/nu) bearing subcutaneous xenografts of the c-MET- and HGF/SF-expressing human leiomyosarcoma SK-LMS-1/HGF in their right thighs, as well as into two immunocompromised mice without tumors, one nude and one with severe-combined immunodeficiency (SCID). Serial total body nuclear images of each mouse were acquired on a Siemens ZLC-Scintron unit between 1 hour and 8 days postinjection for tumor-bearing mice, by which time tumor size mandated euthanasia; and between 1 hour and 30 days postinjection for control mice. Images were evaluated by qualitative and quantitative region-of-interest (ROI) analysis. Data are shown as counts per minute for each ROI [total body, liver/lung (reflecting blood pool activity), thyroid, and tumor] as a function of time postinjection for n = 7 tumor-bearing mice (left panel) and for 1 control nude mouse and 1 control SCID mouse (right panel).
Just as we have observed by FACS analysis with other c-MET-avid immunological probes (42, 47), incubation of MET-overexpressing cell lines with MET4 and fluorescent second antibody resulted in significant upward shifts of fluorescence intensity (Figure 4), indicating high avidity of MET4 for an epitope exposed at the cell surface.
Nuclear images of c-MET-overexpressing human tumor xenografts in host nude mice injected with I-125-MET4 are qualitatively and quantitatively comparable to the best tumor images we have obtained with any other c-MET- or HGF/SF-avid antibody (42, 43, 48) (Figure 5A).
Moreover, by biodistribution analysis (Figure 5B), MET4 appears to be an exceptionally durable antibody for in vivo applications. MET4 has an unusually long biological half-life for total body activity compared to other full-length antibodies, about 150 hours (6.25 days) in tumor-bearing mice and in biphasic fashion for control mice with a “fast” component of about 200 hours (8.3 days) and a “slow” component of about 400 hours (16.6 days), whereas, most full-length antibodies in animals and in humans exhibit a biological half-life of 2–4 days.
Discussion
In this paper we describe the generation of a multipurpose c-MET monoclonal antibody and show examples of its potential usefulness measurement of c-MET expression by human cancers.
Molecular diagnostic tools that detect expression and determine the activation state of therapeutic targets of novel kinase inhibitors are urgently needed if we are to improve our ability to select appropriate treatments for individual cancer patients. Drugs that bind cell surface receptors or penetrate into cells and inhibit receptor and non-receptor kinases have immense promise in the oncology clinic, yet detection of the corresponding targets in human cancers by ex vivo or in vivo approaches provides a major challenge. Besides the Hercept test for quantitative measurement of Her-2/Neu expression, no validated diagnostic tests for receptor or non-receptor protein kinase protein expression are available. The difficulty in developing diagnostic assays, in particular for FFPE tissues as the most commonly available tissue preparation from patient cancers, results from low level expression and labile activation state of kinases. Overexpression is commonly used as a surrogate marker for kinase activation. A better way to identify active kinases and to increase the sensitivity of identifying drug responsive tumors is through phosphorylation measurements, since autophosphorylation is critical for kinase activity. Unfortunately, measurements of protein phosphorylation in FFPE tissues are not yet sufficiently robust for clinical assay. In the meantime, there is a compelling need to confirm the expression of drug targets in individual tumors in order to achieve better pretreatment patient stratification, a critically important factor for demonstrating therapeutic success in early-phase clinical trials.
The c-MET receptor tyrosine kinase (RTK) is particularly difficult to measure in FFPE tissues because of limited reagent availability and/or insufficient sensitivity and lack of validation of commercially obtainable antibodies. Moreover, the stability of c-MET epitopes during formalin-fixation or storage of FFPE tissue blocks appears to be poor. We spent several years developing strategies for generating and rigorously testing FFPE-suitable c-MET antibodies before obtaining MET4. The MET4 hybridoma was chosen based on a stringent screening assay in FFPE prostate tissues using, after four different antigen sources, a rigorous final selection of established specific expression of c-MET in FFPE prostate epithelial and endothelial cells (Figure 1, Table 1). We believe this screening approach greatly enhanced our chances of obtaining an antibody that recognizes a popular extracellular domain c-MET epitope that is preserved in FFPE tissues. MET4 also reacted stronger with c-MET in cell-cell junctions at the basolateral cell membrane, compared to other c-MET antibodies (Table 3). Thus, the MET4 epitope is either more available, or MET4 possesses high affinity than other antibodies. MET4 staining in different cell types appears to be faithful when compared to independent measurements of c-MET abundance by immunoblotting. Further, MET4 staining was reproducible in replicate stains on the same day (%CV = 6.8), on different days (%CV = 6.2) and when comparing two separate lots of MET4 (Table 2). Thus, the initial characterization of MET4 warrants further development of this monoclonal antibody as a diagnostic reagent for ex vivo measurement of c-MET in clinical samples.
Gonzatti-Haces et al. (49) first employed a rabbit polyclonal anti-MET C-28 antibody to identify c-MET. Most studies on c-MET expression in FFPE tissues employ various lots of anti-MET C-28 polyclonal antibody (Santa Cruz, Hercules, CA). A systematic comparison of commercial MET antibodies, including three monoclonal antibodies and two separate lots of MET C-28 with the AQUA™ technology clearly demonstrated a greater than expected amount of variability in a breast cancer tissue microarray study (31). One of the antibodies, DO-24, bound to the extracellular domain of MET, while the others recognized intracellular epitopes. Despite the fact that all antibodies bound to c-MET by immunoblotting, their reactivity in FFPE tissues was inconsistent between breast cancer samples from the same patient. The most consistent results were obtained when comparing MAB3729 from Chemicon and a single lot of MET C-28. MAB3729 possessed good reproducibility and a correlation coefficient of 0.94 for cores from the same cell line on adjacent TMA slides. When comparing two different lots of MET C-28, a considerable lot-to-lot variability was noted (31). However, side-by-side comparison of MAB3729 with MET4 demonstrates greater specificity of MET4.
MET4 reacts with an epitope within the extracellular 25 to 567 amino acid segment of the c-MET receptor protein (32). Interestingly, the MET4 binding site is sensitive to denaturation by boiling in SDS sample buffer, but is reestablished by either heat retrieval or formalin fixation (Supplementary Figure 4). The same heat retrieval is normally used with formalin-fixed tissues and vastly improves antibody reactivity (50). Boiling hydrolyzes bonds that are generated by formalin crosslinking and also allows refolding of proteins. Interestingly, both formalin fixation and heat retrieval were effective in re-establishing the MET4 binding site within the denatured c-MET receptor protein.
In breast cancer, antibodies that bind to the cytoplasmic domain of c-MET also react with nuclear epitopes, due to cleavage and translocation of the protein to the nucleus (51). Nuclear expression of c-MET measured with the MAB3729 antibody in sections of breast cancer TMAs was associated with a decrease in 5-year survival from 75% to 65% (31). Additional studies of nuclear c-MET expression are needed to evaluate its role as a biomarker in other cancer types.
While MET activation through somatic gene mutation or overexpression has been clearly observed in certain cancer types, they appear to be infrequent in primary ovarian cancers or gliomas. Previous analyses of c-MET expression in gliomas demonstrated increased Met expression in grade IV glioblastoma similar to the observation in this study and overall frequencies of high c-MET expression or c-MET gene amplifications of 10% and 30%, accordingly (52, 53). Overexpression of c-MET through duplication of the c-MET gene in double minute chromosomes was noted in 3/18 grade IV and 1/18 grade II gliomas (54). We observed a similar level of c-MET overexpression in high-grade gliomas as reported in these previous studies. In contrast to gliomas, all ovarian cancers expressed c-MET, and 7/28 had high levels of c-MET expression. A previous study observed lower c-MET expression in endometroid ovarian cancer and a lower fraction of “intense” c-MET staining across the other histologic subtypes (55). Potentially, the heightened reactivity of MET4 is due to increased sensitivity and binding affinity compared to other c-MET antibodies. The mechanisms responsible for high c-MET expression and the activation state of c-MET in ovarian cancer are unknown. In ovarian cancer cell lines it is unclear, whether high c-MET expression occurs because of amplification of the c-MET gene, increased transcription, or post-translational retention of c-MET protein.
We propose that the true relevance of c-MET overexpression in ovarian cancer, gliomas, and other cancer types—as well as its relationship to the therapeutic efficacy of c-MET inhibitory drugs— would best be evaluated in one or more clinical studies that correlate treatment responses with c-MET expression levels. Finally, we cannot confirm the in vivo usefulness of MET4 in human patients from our nuclear imaging and biodistribution results with mice bearing c-MET-overexpressing human tumor xenografts. MET4 could potentially be a robust agent for radioimmunoscintigraphy and radioimmunotherapy.
Supplementary Material
Acknowledgments
This work was supported in part by the Jay and Betty Van Andel Foundation of the Van Andel Research Institute and by funding (to GVW, MG, and RH) from the Michigan Life Sciences Corridor. Support to the B. S. K. was provided from the Prostate (P50 CA97186) and the Ovarian SPORE (P50 CA083636) grant.
At the Van Andel Research Institute we thank David Nadziejka for manuscript editing, Michelle Bassett for assistance preparing the manuscript, and Rich West, Sara Kunz, and Jennifer Vogel for assistance with FACS analysis. At the Fred Hutchinson Cancer Research Center we thank Kim Adolphsen in the histopathology shared resource for immunohistochemical staining and Dale McLerran for the statistical analysis.
References
- 1.Birchmeier C, Birchmeier W, Gherardi E, et al. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 2.Rosario M, Birchmeier W. How to make tubes: signaling by the Met receptor tyrosine kinase. Trends Cell Biol. 2003;13:328–335. doi: 10.1016/s0962-8924(03)00104-1. [DOI] [PubMed] [Google Scholar]
- 3.Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene. 2000;19:5582–5589. doi: 10.1038/sj.onc.1203859. [DOI] [PubMed] [Google Scholar]
- 4.Jeffers M, Fiscella M, Webb CP, et al. The mutationally activated Met receptor mediates motility and metastasis. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:14417–14422. doi: 10.1073/pnas.95.24.14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–6281. [PubMed] [Google Scholar]
- 6.Peschard P, Fournier TM, Lamorte L, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell. 2001;8:995–1004. doi: 10.1016/s1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 8.Di Renzo MF, Olivero M, Giacomini A, et al. Overexpression and amplification of the met/HGF receptor gene during the progression of colorectal cancer. Clin Cancer Res. 1995;1:147–154. [PubMed] [Google Scholar]
- 9.Kuniyasu H, Yasui W, Kitadai Y, et al. Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem Biophys Res Commun. 1992;189:227–232. doi: 10.1016/0006-291x(92)91548-5. [DOI] [PubMed] [Google Scholar]
- 10.Naldini L, Weidner KM, Vigna E, et al. Scatter factor and hepatocyte growth factor are indistinguishable ligands for the MET receptor. Embo J. 1991;10:2867–2878. doi: 10.1002/j.1460-2075.1991.tb07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosen EM, Knesel J, Goldberg ID. Scatter factor and its relationship to hepatocyte growth factor and met. Cell Growth Differ. 1991;2:603–607. [PubMed] [Google Scholar]
- 12.Zhuang Z, Park WS, Pack S, et al. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nat Genet. 1998;20:66–69. doi: 10.1038/1727. [DOI] [PubMed] [Google Scholar]
- 13.Di Renzo MF, Olivero M, Martone T, et al. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene. 2000;19:1547–1555. doi: 10.1038/sj.onc.1203455. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Han SU, Cho H, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene. 2000;19:4947–4953. doi: 10.1038/sj.onc.1203874. [DOI] [PubMed] [Google Scholar]
- 15.Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–1488. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- 16.Puri N, Ahmed S, Janamanchi V, et al. c-Met is a potentially new therapeutic target for treatment of human melanoma. Clin Cancer Res. 2007;13:2246–2253. doi: 10.1158/1078-0432.CCR-06-0776. [DOI] [PubMed] [Google Scholar]
- 17.Baykal C, Ayhan A, Al A, et al. Overexpression of the c-Met/HGF receptor and its prognostic significance in uterine cervix carcinomas. Gynecol Oncol. 2003;88:123–129. doi: 10.1016/s0090-8258(02)00073-2. [DOI] [PubMed] [Google Scholar]
- 18.Huang TJ, Wang JY, Lin SR, et al. Overexpression of the c-met protooncogene in human gastric carcinoma--correlation to clinical features. Acta Oncol. 2001;40:638–643. doi: 10.1080/028418601750444204. [DOI] [PubMed] [Google Scholar]
- 19.Kang JY, Dolled-Filhart M, Ocal IT, et al. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–1105. [PubMed] [Google Scholar]
- 20.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, et al. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Muzio L, Farina A, Rubini C, et al. Effect of c-Met expression on survival in head and neck squamous cell carcinoma. Tumour Biol. 2006;27:115–121. doi: 10.1159/000092716. [DOI] [PubMed] [Google Scholar]
- 22.Tsarfaty I, Alvord WG, Resau JH, et al. Alteration of Met protooncogene product expression and prognosis in breast carcinomas. Anal Quant Cytol Histol. 1999;21:397–408. [PubMed] [Google Scholar]
- 23.Lengyel E, Prechtel D, Resau JH, et al. C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer. 2005;113:678–682. doi: 10.1002/ijc.20598. [DOI] [PubMed] [Google Scholar]
- 24.Tolgay Ocal I, Dolled-Filhart M, D’Aquila TG, et al. Tissue microarray-based studies of patients with lymph node negative breast carcinoma show that met expression is associated with worse outcome but is not correlated with epidermal growth factor family receptors. Cancer. 2003;97:1841–1848. doi: 10.1002/cncr.11335. [DOI] [PubMed] [Google Scholar]
- 25.Di Renzo MF, Poulsom R, Olivero M, et al. Expression of the Met/hepatocyte growth factor receptor in human pancreatic cancer. Cancer Res. 1995;55:1129–1138. [PubMed] [Google Scholar]
- 26.Tsuda M, Davis IJ, Argani P, et al. TFE3 fusions activate MET signaling by transcriptional up-regulation, defining another class of tumors as candidates for therapeutic MET inhibition. Cancer Res. 2007;67:919–929. doi: 10.1158/0008-5472.CAN-06-2855. [DOI] [PubMed] [Google Scholar]
- 27.Gotte M, Kersting C, Radke I, et al. An expression signature of syndecan-1 (CD138), E-cadherin and c-met is associated with factors of angiogenesis and lymphangiogenesis in ductal breast carcinoma in situ. Breast Cancer Res. 2007;9:R8. doi: 10.1186/bcr1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens T, Schmidt NO, Eckerich C, et al. A novel one-armed anti-c-Met antibody inhibits glioblastoma growth in vivo. Clin Cancer Res. 2006;12:6144–6152. doi: 10.1158/1078-0432.CCR-05-1418. [DOI] [PubMed] [Google Scholar]
- 29.Puri N, Khramtsov A, Ahmed S, et al. A selective small molecule inhibitor of c-Met, PHA665752, inhibits tumorigenicity and angiogenesis in mouse lung cancer xenografts. Cancer Res. 2007;67:3529–3534. doi: 10.1158/0008-5472.CAN-06-4416. [DOI] [PubMed] [Google Scholar]
- 30.Zou HY, Li Q, Lee JH, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 31.Pozner-Moulis S, Cregger M, Camp RL, et al. Antibody validation by quantitative analysis of protein expression using expression of Met in breast cancer as a model. Lab Invest. 2007;87:251–260. doi: 10.1038/labinvest.3700515. [DOI] [PubMed] [Google Scholar]
- 32.Gherardi E, Coffer A. Purification and characterization of scatter factor. Exs. 1991;59:53–62. doi: 10.1007/978-3-0348-7494-6_4. [DOI] [PubMed] [Google Scholar]
- 33.Gherardi E, Youles ME, Miguel RN, et al. Functional map and domain structure of MET, the product of the c-met protooncogene and receptor for hepatocyte growth factor/scatter factor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12039–12044. doi: 10.1073/pnas.2034936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, Adenekan B, Chen L, et al. Syndecan-1 expression in locally invasive and metastatic prostate cancer. Urology. 2004;63:402–407. doi: 10.1016/j.urology.2003.08.036. [DOI] [PubMed] [Google Scholar]
- 35.Gmyrek GA, Walburg M, Webb CP, et al. Normal and malignant prostate epithelial cells differ in their response to hepatocyte growth factor/scatter factor. Am J Pathol. 2001;159:579–590. doi: 10.1016/S0002-9440(10)61729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridler, Calvard Picture threshorlding using an iterative selection method. SMC. 1978;8:630–632. [Google Scholar]
- 37.Russ J. The image processing handbook. 2002. p. 732. [Google Scholar]
- 38.Altstock RT, Stein GY, Resau JH, et al. Algorithms for quantitation of protein expression variation in normal versus tumor tissue as a prognostic factor in cancer: Met oncogene expression, and breast cancer as a model. Cytometry. 2000;41:155–165. [PubMed] [Google Scholar]
- 39.Jeffers M, Rong S, Vande Woude GF. Enhanced tumorigenicity and invasion-metastasis by hepatocyte growth factor/scatter factor-met signalling in human cells concomitant with induction of the urokinase proteolysis network. Molecular and cellular biology. 1996;16:1115–1125. doi: 10.1128/mcb.16.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 41.Park M, Dean M, Cooper CS, et al. Mechanism of met oncogene activation. Cell. 1986;45:895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- 42.Hay RV, Cao B, Skinner RS, et al. Radioimmunoscintigraphy of human met-expressing tumor xenografts using met3, a new monoclonal antibody. Clin Cancer Res. 2003;9:3839S–3844S. [PubMed] [Google Scholar]
- 43.Hay RV, Cao B, Skinner RS, et al. Radioimmunoscintigraphy of tumors autocrine for human met and hepatocyte growth factor/scatter factor. Mol Imaging. 2002;1:56–62. doi: 10.1162/15353500200200006. [DOI] [PubMed] [Google Scholar]
- 44.Humphrey PA, Zhu X, Zarnegar R, et al. Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am J Pathol. 1995;147:386–396. [PMC free article] [PubMed] [Google Scholar]
- 45.Knudsen BS, Edlund M. Prostate cancer and the met hepatocyte growth factor receptor. Adv Cancer Res. 2004;91:31–67. doi: 10.1016/S0065-230X(04)91002-0. [DOI] [PubMed] [Google Scholar]
- 46.van Leenders G, van Balken B, Aalders T, et al. Intermediate cells in normal and malignant prostate epithelium express c-MET: implications for prostate cancer invasion. Prostate. 2002;51:98–107. doi: 10.1002/pros.10073. [DOI] [PubMed] [Google Scholar]
- 47.Jiao Y, Zhao P, Zhu J, et al. Construction of human naive Fab library and characterization of anti-met Fab fragment generated from the library. Molecular biotechnology. 2005;31:41–54. doi: 10.1385/mb:31:1:041. [DOI] [PubMed] [Google Scholar]
- 48.Hay RV, Cao B, Skinner RS, et al. Nuclear imaging of Met-expressing human and canine cancer xenografts with radiolabeled monoclonal antibodies (MetSeek) Clin Cancer Res. 2005;11:7064s–7069s. doi: 10.1158/1078-0432.CCR-1004-0014. [DOI] [PubMed] [Google Scholar]
- 49.Gonzatti-Haces M, Seth A, Park M, et al. Characterization of the TPR-MET oncogene p65 and the MET protooncogene p140 protein-tyrosine kinases. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:21–25. doi: 10.1073/pnas.85.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi SR, Cote RJ, Taylor CR. Antigen retrieval techniques: current perspectives. J Histochem Cytochem. 2001;49:931–937. doi: 10.1177/002215540104900801. [DOI] [PubMed] [Google Scholar]
- 51.Pozner-Moulis S, Pappas DJ, Rimm DL. Met, the hepatocyte growth factor receptor, localizes to the nucleus in cells at low density. Cancer Res. 2006;66:7976–7982. doi: 10.1158/0008-5472.CAN-05-4335. [DOI] [PubMed] [Google Scholar]
- 52.Koochekpour S, Jeffers M, Rulong S, et al. Met and hepatocyte growth factor/scatter factor expression in human gliomas. Cancer Res. 1997;57:5391–5398. [PubMed] [Google Scholar]
- 53.Moon YW, Weil RJ, Pack SD, et al. Missense mutation of the MET gene detected in human glioma. Mod Pathol. 2000;13:973–977. doi: 10.1038/modpathol.3880177. [DOI] [PubMed] [Google Scholar]
- 54.Fischer U, Muller HW, Sattler HP, et al. Amplification of the MET gene in glioma. Genes Chromosomes Cancer. 1995;12:63–65. doi: 10.1002/gcc.2870120111. [DOI] [PubMed] [Google Scholar]
- 55.Sawada K, Radjabi AR, Shinomiya N, et al. c-Met overexpression is a prognostic factor in ovarian cancer and an effective target for inhibition of peritoneal dissemination and invasion. Cancer Res. 2007;67:1670–1679. doi: 10.1158/0008-5472.CAN-06-1147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.