Translation in Yeast
The process of translation initiation is highly conserved from yeast to mammalian cells. It is therefore not surprising that yeast has played an important role in deciphering diverse aspects of this fascinating step in gene expression. The importance of the yeast system relies particularly on the ease with which one can obtain knock-out/knock-in mutant strains and the possibility to perform genetic screens to isolate mutants or suppressors of mutations. Such screens have, in some instances, revealed genes encoding translation initiation factors. Often, the combination of yeast genetics and biochemistry has contributed to the understanding of the function of these factors. With its ease in transformation and gene replacement, yeast is a wonderful tool to create conditional mutants to test and unravel the function of translation initiation factors (Fig. 1, scheme of translation initiation). We have selected here a few examples, among many, in which the yeast system has proven useful in the analysis of translation initiation.
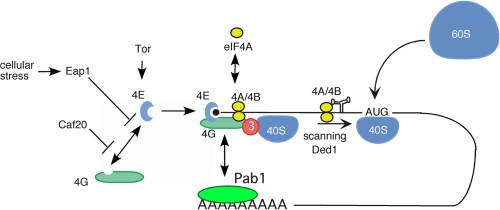
FIGURE 1.
Translation initiation. Shown are initiation factors (members from the eIF4 family, eIF3, Ded1, and Pab1 and their regulatory proteins) discussed in the text that are involved in mRNA 5′-UTR recognition and subsequent scanning to the initiator AUG codon of 40 S ribosomes.
Laying the Way for the Scanning Ribosome
The yeast system has played a pioneering role in establishing the scanning mechanism (for an excellent review, see Ref. 1). Long before transformation and molecular genetic methods were available, Fred Sherman showed, by using initiator codon mutations and revertant and pseudorevertant analysis in CYC1 encoding iso-1-cytochrome c, that the initiator codon specifies the initiation site and that the first AUG is used as the start codon to initiate translation (2).
The next step was to identify transacting factors required for the faithful recognition of the initiator codon. Using the yeast system, the laboratory of Thomas Donahue identified genes required for initiator codon selection. They mutated the anticodon of tRNAiMet from UAC to UCC, which allowed for initiation at an AGG codon, indicating that the initiator tRNA is responsible for initiator codon recognition (3). To identify transacting factors, the AUG initiator codons of both the HIS4 gene and a HIS4-lacZ reporter gene were modified to AUU, and histidine prototroph and blue suppressors (SUI for suppressor of initiator codon mutants) were selected (4). The analysis of the suppressor genes revealed the identity of eIF2α (5), eIF2β (4), eIF2γ and eIF5 (6), and eIF1 (7), all eukaryotic initiation factors known to play an important role in the scanning and AUG recognition process.
Related studies by Alan Hinnebusch and his laboratory on GCN4 expression provided further insights into the scanning process (8). The yeast GCN4 gene encodes a transcription activator required for the induction of >30 amino acid biosynthesis genes. The particularity of this gene is that the 5′-UTR contains four small upstream open reading frames (uORFs)2 that, according to the scanning model and consistent with the results discussed above, are inhibitory for Gcn4 translation (Fig. 2). In a landmark paper, Peter Mueller from the Hinnebusch laboratory showed that deletion of the uORFs permitted efficient translation of the GCN4 mRNA (9). Further genetic analysis of GCN4 expression allowed the identification of a multitude of gcn (general control noninducible, similar to the absence of the transcription factor itself) and gcd (general control derepressed, similar to the absence of the uORF) mutant genes that are involved in the scanning and initiator codon recognition process and thereby in regulation of GCN4 expression. The rationale behind this control system is that the migration distance required by the 40 S ribosomal subunit to acquire reinitiation competence after translation of the first uORF determines whether translation occurs at a proximal downstream uORF or at the GCN4 initiator codon located farther downstream. The outcome of these experiments showed that eIF2α phosphorylation greatly delayed active ternary complex (eIF2·GTP·tRNAiMet) formation. When eIF2α is phosphorylated by Gcn2 kinase, the GDP-GTP exchange by eIF2B is slowed down, and reinitiation is much less efficient. Therefore, the small ribosomal subunit continues scanning after translation of the first uORF until it reaches the GCN4 initiator codon. Under normal conditions, e.g. when Gcn2 is not activated by uncharged tRNAs, and therefore, eIF2α is not phosphorylated, reinitiation will occur at the fourth uORF, precluding initiation at the GCN4 initiator codon (Fig. 2).
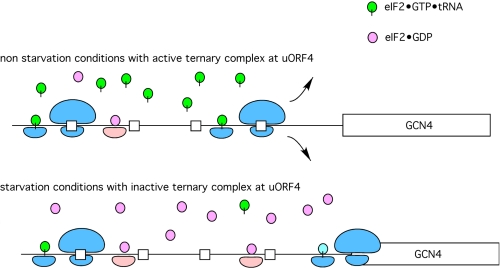
FIGURE 2.
Scanning at GCN4 mRNA. Under normal conditions, the product of the initiation pathway, eIF2·GDP, is recycled to eIF2·GTP by the aid of a guanine nucleotide exchange factor, eIF2B. With high levels of the ternary complex available (eIF2·GTP·tRNAiMet), ribosomes that translate uORF1 will rapidly reacquire a new ternary complex and initiate at uORF4, followed by complete release of the ribosomal subunits from the mRNA and no synthesis of Gcn4. Under conditions of starvation, uncharged tRNA activates Gcn2, leading to phosphorylation of the α-subunit of eIF2. This phosphorylated form of eIF2 complexed with GDP binds tightly to the nucleotide exchange factor and thereby blocks nucleotide exchange, resulting in low levels of ternary complexes. The absence of a large pool of ternary complexes delays the pairing of the ternary complex with an AUG start codon until the AUG for Gcn4 is reached (i.e. the upstream uORFs are bypassed). This results in an increase in Gcn4 expression relative to normal or non-starvation conditions.
Factor Discovery and Analysis
The power of genetic analysis using the yeast Saccharomyces cerevisiae has contributed to the identification and cloning of several genes involved in translation, as indicated by the following examples.
Prt1, a Subunit of eIF3
A landmark step in the identification of genes encoding translation initiation factors was the isolation of a series of 400 temperature-sensitive mutants by Leland H. Hartwell (10). Among these mutants, some displayed rapid cessation of protein synthesis under nonpermissive temperature conditions (37 °C) (11). The temperature-sensitive mutants were tested for incorporation of precursors for DNA, RNA, and protein synthesis (their reduced capability of polysome formation) (12). Mutant ts-187/prt1-1 was used to create cell extracts that were temperature-sensitive for the interaction of the ternary complex with the 40 S subunit. Finally, the temperature-sensitive growth phenotype permitted the cloning and analysis of PRT1, the gene encoding the second largest subunit of eIF3 (13).
Interestingly, the genetic mapping of cdc− (cell division cycle) mutants revealed the allelic nature of temperature-sensitive prt1-1 and cdc63-1 clones (14). Cell division cycle mutants were isolated as cells that can survive otherwise lethal conditions due to their arrest at G1 (15). Whereas prt1-1 mutants arrest randomly under nonpermissive temperature conditions during the cell cycle, cdc63-1 mutants preferentially arrest at START during the cell cycle (14). This indicates that subtle differences in the activity of one translation initiation factor can have different outcomes on cell cycle progression. Interestingly, in the original cdc63 mutant, protein synthesis seemed to be unaffected. Importantly, the interconnection of protein synthesis and cell cycle regulation has been later confirmed in many other instances (16, 17). This knowledge was then further developed in the mammalian system, where it was shown that eIF4E and its inhibitory partner, 4E-BP (eIF4E-binding protein), act as a proto-oncogene and as an oncogene inhibitor, respectively (18).
Yeast has also been widely used to search for in vivo protein-protein interactions with the yeast two-hybrid system (19). One well documented example showing the potential of the two-hybrid system has led to the discovery of the interactions of subunits of eIF3 (17).
eIF4E, the Cap-binding Protein
An additional example showing the advantages of studying translation initiation in yeast has been the analysis of the cap-binding protein eIF4E. The protein was originally purified by its virtue of binding to the m7GpppG cap structure of eukaryotic mRNAs (20). Antibodies were raised against the purified yeast protein and used to identify a cDNA clone in Escherichia coli. The resulting clone was then used to isolate the corresponding yeast gene (21).
A major breakthrough was the establishment of a cell-free eIF4E-dependent translation system (22). The development of such a system was based on the use of a temperature-sensitive eIF4E mutant (ts-4-2). A cell extract prepared from such a temperature-sensitive strain was sensitive to a short incubation at 37 °C, but importantly, translational activity could be restored by the addition of recombinant wild-type eIF4E protein purified from E. coli.
As for prt1-1/cdc63-1, the gene encoding eIF4E was independently isolated as the cell division cycle gene CDC33 (23). Moreover, it was later shown, in an analysis of eIF4E mutant cdc33-1, that the expression of a stable form of cyclin Cln3 can suppress its G1 arrest, consistent with the requirement of a high Cln3 expression, which is dependent on sufficient eIF4E activity at the transition from G1 to S phase (24).
Although the demonstration that eIF4E from other organisms such as mouse and Drosophila is able to complement a yeast eIF4E knock-out seems obvious today (25, 26), it was a crucial step to provide yeast with the “noblesse” in the translation initiation field (Fig. 3). Not all mammalian initiation factors complement the corresponding yeast factor despite high amino acid sequence homology, e.g. eIF4A (27).
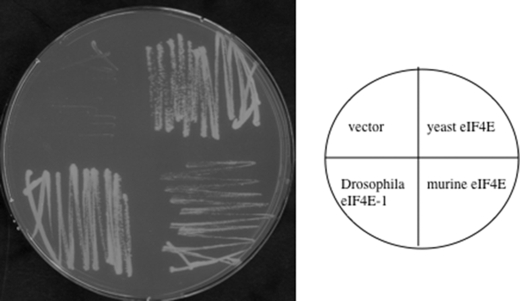
FIGURE 3.
Complementation of yeast by orthologous eIF4E proteins. Plasmid shuffling was carried out with haploid yeast strain RH2585, which carries the deletion Δcdc33::kanX, and the essential eIF4E activity is provided on plasmid pVTURA3. Cells were transformed with plasmids carrying different eIF4E versions (yeast, Drosophila, murine, and vector) and plated on yeast minimal medium containing 5-fluoroorotic acid to counterselect for the loss of the eIF4E-encoding plasmid. Note that Drosophila eIF4E and murine eIF4E complement the lack of yeast eIF4E, but murine eIF4E-transformed yeast cells show a slow growth phenotype.
eIF4A, Godfather of the DEAD Box Protein Family of RNA Helicases
Although eIF4A was first described in plant and mammalian systems (28–31), it was the discovery of yeast eIF4A that stimulated the work on the DEAD box protein family of RNA helicases (32). The first extensive mutational analysis of a DEAD box protein and the analysis of the effects on viability were carried out on yeast eIF4A (33). Some of the conditionally lethal eIF4A yeast mutants were later used for analysis in a yeast cell-free translation system (34) to confirm the ATPase dependence of the eIF4A function in translation initiation. Although eIF4A was known as a translation initiation factor, it was one of the first factors for which a deficiency in the in vitro enzymatic activity could be directly correlated with an in vivo phenotype. Based on its enzymatic activity, its high abundance, and the inhibitory effects of secondary structures in the 5′-UTR, it was suggested that eIF4A is required in the scanning process. Interestingly, in a factor-dependent extract, it was shown that an mRNA with a leader as short as 8 nucleotides was still dependent on the presence of eIF4A, indicating that it is probably required for additional steps in translation initiation other than scanning (35).
The RNA helicase activity of eIF4A is strongly stimulated when this DEAD box protein is present as a component of eIF4F, the cap-binding complex (consisting also of eIF4E and eIF4G). This is one of the first examples in which it has been shown that a DEAD box protein is stimulated by partner proteins, in this case, probably by increasing its local concentration at the cap structure of mRNAs. Similarly, in mammals, the activity of eIF4A is stimulated by eIF4B and eIF4H (36, 37). Yeast eIF4B was isolated by two independent and complementary approaches. Altmann et al. (38) isolated the yeast eIF4B gene by screening cDNA libraries with anti-cap-binding complex antibodies. At the same time, a genetic screen for a multicopy suppressor of a temperature-sensitive eIF4A mutation allowed the cloning of the yeast eIF4B gene (39). Later, it was shown that eIF4B also displays RNA-annealing activity (40). Intriguingly, TIF3, the gene encoding eIF4B, is not essential in yeast. The role(s) of eIF4B in translation initiation is not well defined, and it is not clear why yeast does not possess an eIF4H-like protein.
Despite many efforts in the analysis of eIF4A in yeast, plant, and mammalian cells, we still do not know the exact function of the free or eIF4F-bound form of eIF4A in translation initiation. An in-depth biochemical analysis combined with a genetic and mutational analysis would be welcomed.
Ded1/DDX3, a Splicing and Export Factor Required for Translation Initiation
The multipurpose protein Ded1 (DDX3 in humans) was originally identified as a suppressor of a prp8 splicing mutant (41, 42). Although this analysis classified Ded1 as a splicing factor, a splicing defect due to a ded1 mutation has not been reported to date. Nevertheless, an involvement in splicing was supported by a proteomic analysis of yeast that clearly identified Ded1 as a component of the spliceosome (43). Intriguingly, an analysis of the human homolog DDX3 gene revealed a function of this protein in the export of partially spliced HIV mRNA.
A genetic analysis in yeast established Ded1 as a translation initiation factor (44, 45). The analysis of two cold-sensitive mutations did not reveal a splicing defect but rather a strong reduction of [35S]methionine incorporation into de novo protein synthesis at the nonpermissive temperature. This defect was corroborated by polysome profile analysis. In accordance with its assumed function in translation initiation, indirect immunofluorescence microscopy revealed the majority of the protein to be localized in the cytoplasm (44). At the same time, a multicopy suppressor screen of temperature-sensitive eIF4E mutants allowed the isolation of the DED1 gene. Interestingly and in accordance with the suppressor phenotype of a deficient cap-binding complex, an analysis of synthetic lethal interactions between a ded1 mutant and diverse mutant translation initiation factors revealed a genetic interaction with other proteins of the eIF4 family, but not with Prt1 (a component of eIF3) or the splicing factor Prp28 (45).
The involvement of Ded1 in translation initiation was also confirmed by the establishment of a Ded1-dependent translation system in yeast (44). Later, it was shown that the Ded1 protein is required for optimal scanning of the small subunit for the AUG initiator codon (46). In this latter study, the long 5′-UTR of GCN4 without upstream initiator codons was fused to a luciferase reporter system. A mutation in DED1 considerably reduced the expression of the reporter construct compared with a construct with a short 5′-UTR. Interestingly, in this study, translation initiation factor eIF4A was not important for the scanning process, consistent with the previous finding that a short 5′-UTR still requires eIF4A in translation initiation (see above).
The Ded1 protein has been shown to be an efficient DEAD box RNA helicase (47) and has served since as a model in the analysis of DEAD box proteins. Such analyses using the yeast system have allowed the dissection of the role of many motifs conserved in the DEAD box RNA helicase family and the function of DEAD box proteins in the displacement of proteins from RNA. Importantly, the in vitro analysis of RNA displacement using the Ded1 protein has shed new light on the mode of action of DEAD box proteins. Experiments performed by the laboratory of Eckhard Jankowsky have shown that Ded1 requires binding to a single-stranded extension of an RNA duplex (48). However, contrary to classical helicases that require a single strand for loading before translocation, Ded1 does not translocate, and a covalent connection of the single-stranded extension with the duplex is not required. These results indicate that the single-stranded extension is required to increase the local concentration of the enzyme. Additionally, these studies provide evidence that the mechanism of duplex unwinding by DEAD box RNA helicases is distinct from that of the traditional DNA helicases. Clearly, further studies to integrate the biochemical data of Ded1 into our understanding of translation initiation are needed.
When the 5′-End Meets the 3′-End: Role of the Poly(A)-binding Protein in Translation Initiation
Both the 5′- and 3′-ends of an mRNA need to be intact to allow efficient gene expression. The 5′-end is necessary to assemble the cap-binding complex, and the poly(A) tail at the 3′-end protects the mRNA against degradation. The 3′-end of eukaryotic mRNAs carries a poly(A) tail that is a binding site for the poly(A)-binding protein Pab1. Deletion of the PAB1 gene is lethal, indicating an important role in the maintenance of gene expression. A genetic screen allowed the isolation of suppressors of a pab1 knock-out (49). Interestingly, some of these suppressors (spb, suppressor of pab1) affected proteins of the 60 S ribosomal subunit (Spb2/RPL46) or its biogenesis (Spb4, a DEAD box protein required for 60 S biogenesis) (50). Thus, at least on a genetic level, a communication of the 3′-end of the mRNA with the large ribosomal subunit was shown to be involved during translation (see also review by Sachs and Wahle (51) on poly(A) as a stimulating element).
Further translation experiments performed by the laboratory of Alan Sachs showed in vitro that Pab1 binds to the poly(A) tail and thereby stimulates the binding of 40 S subunits to the mRNA (52). This clearly reinforced the idea of a communication of the 3′-end of the mRNA with the 5′-end. Because affecting 60 S subunit biogenesis leads to an excess of 40 S subunits (e.g. Ref. 50), it has been suggested that suppression of PAB1 is effective through a mass effect of an elevated 40 S subunit concentration.
In accordance with these data, Tarun and Sachs (52) further showed that the Pab1 protein interacts with yeast eIF4G (Tif4632). Finally, this interaction and the circularization of the mRNA by atomic force microscopy were beautifully shown by the laboratory of Alan Sachs (53). Clearly, this important concept, which is nowadays found in basic textbooks, was promoted by genetic screens in yeast.
Analysis of Translation-mediated Control of Gene Expression
Translation initiation is an energy-consuming process that is subject to multiple regulatory circuits that act mostly at the initiation level. The best studied example is the repression of general translation by amino acid starvation and the simultaneous induction of GCN4 (9, 54) required for the coordinated expression of amino acid biosynthesis genes. Other examples are the regulation of translation initiation by the TOR (target of rapamycin) pathway (see below), the down-regulation of translation upon glucose withdrawal, and the addition of exogenous effectors such as caffeine (55).
Translation Regulation by Cellular Stress
It seems logical that a eukaryotic cell diminishes its main biosynthetic activities if reduced secretion occurs. However, the first secretion mutants (sec) were isolated on the basis of continued protein synthesis and a concomitant increase in buoyant density (56). Nevertheless, Deloche et al. (57) have shown that, in certain vesicular transport mutants, translation initiation is attenuated. When using a sec4 or end3 mutant, they showed that, after a shift to nonpermissive temperature, polysomes relocate to the monosomal 80 S peak, and incorporation of radiolabeled methionine is attenuated. This effect has been shown not to be due to an arrest of ribosome biogenesis or tRNAiMet synthesis. Importantly, the shift to nonpermissive temperature or cell treatment with chlorpromazine (a drug that alters the structure of lipid membranes) does not completely inhibit but causes an attenuation of translation initiation for a prolonged period of time. This allows the cells to cope with a defect in vesicular transport. Further analysis showed that translational attenuation is mediated by Gcn2, a kinase that phosphorylates and inactivates eIF2α, and Eap1, an eIF4E-binding protein. A mutant eIF2α protein that cannot be phosphorylated (Sui2-S51A) or the deletion of EAP1 abolishes the attenuation effect induced by chlorpromazine or by a sec4 eap1 double mutant.
A genetic analysis of an essential membrane-spanning protein involved in inserting the spindle pole body into the nuclear membrane has revealed an interesting translational control mechanism (58). Whereas the secretion defect described above results in translational attenuation of bulk mRNA, a defective spindle pole body results in a very specific inhibition of the nuclear pore membrane protein POM34 mRNA. A battery of very elegant experiments, including multicopy suppressor analysis, identification of synthetic lethal interactions, and co-immunoprecipitations, identified a complex composed of the mRNA-binding protein Scp160, the 4E-BP Eap1, the endoplasmic reticulum-associated Smy2 protein, and Asc1 (G-protein β-subunit) that is involved in this regulation. Thus, a protein complex is stimulated by a defect in spindle pole body insertion into the nuclear membrane to direct the translation regulator Eap1 to act on a specific mRNA. Importantly, mutations that affect the interaction of Eap1 with its target protein, eIF4E, abolish this regulation. Moreover, genetic analysis of the second yeast eIF4E-binding protein Caf20 (see below) did not result in the same regulatory defects.
Translation Initiation Inhibited by Metabolic Changes or by Exogenous Compounds
Glucose withdrawal results in an immediate strong translational arrest that is not accompanied by transcriptional inhibition (59). This inhibitory effect is not mediated by phosphorylation of eIF2α or the activation of Tap42, a phosphatase activator that acts downstream of the TOR kinase pathway (see below). Interestingly, defects in glucose repression, the induction of hexose transporters, or in the cAMP-dependent protein kinase pathways abolish the inhibitory effects of glucose withdrawal. Therefore, the translational inhibition by glucose withdrawal is most likely not simply due to an energy limitation effect but is dependent on glucose signaling pathways.
Other studies have used the yeast system to follow the effect of fusel alcohols or volatile anesthetics on translation. In the case of fusel alcohols (by-products of cellular metabolism), it has been shown that certain strains are inhibited for translation upon the addition of butanol (60). Genetic crosses allowed the identification of the GCD1 gene as the one involved in the translational repression of BUTs strains. GCD1 encodes a subunit of eIF2B, the guanine exchange factor required for eIF2-GDP to eIF2-GTP recycling. Accordingly, the addition of butanol in BUTs strains slightly induces the GCN4 response in a Gcn2 (eIF2α kinase)-independent way. Interestingly, fusel alcohols induce pseudohyphal growth in BUTs strains but not in BUTr strains. Similarly, volatile anesthetics such as isoflurane also inhibit translation initiation (61). These examples of translational fine-tuning nicely illustrate the application of yeast genetics to the study of translational control.
TOR Signaling Affects Cap-dependent Translation Initiation
In the early 1990s, the laboratory of Michael Hall identified the yeast genes encoding Tor1 and Tor2 as targets of the immunosuppressant rapamycin (62). Ever since, a vast number of publications have described the importance of these evolutionary conserved kinases in signal transduction pathways that allow the cells to monitor proper nutritional conditions in their environment before undergoing cell division.
In yeast, the presence of rapamycin or the absence of the kinases Tor1 and Tor2 arrests growth in the early G1 phase of the cell cycle due to an inhibition of translation initiation, inducing starvation (63). An important cause for the observed decrease in cap-dependent translation is the rapamycin-induced degradation of initiation factor eIF4G (71).
In mammals, rapamycin blocks the phosphorylation of 4E-BPs, a family of small acidic eIF4E-binding proteins that block cap-dependent translation. Their interaction with eIF4E is regulated by their phosphorylation status: hyperphosphorylation of 4E-BPs, which is accomplished by mTOR (mammalian TOR), leads to their dissociation from eIF4E, allowing cap-dependent translation (64–66).
In the meantime, other proteins involved in protein synthesis such as S6 kinase and eIF4B have been identified as phosphorylation targets of mTOR. In yeast, although there also exists a small 4E-BP named Caf20, which inhibits cap-dependent protein synthesis (67), the regulation of its interaction with eIF4E has not yet been established.
Conclusion
During the last 20 years, yeast has served as an invaluable system to study the mechanism and regulation of translation initiation. Now that most genes involved in translation initiation have been identified and their gene products characterized, one could ask if yeast still represents a useful system to study this fundamental process.
Previous studies indicate that this will be indeed the case, e.g. to study differentiation when cells are submitted to nutritional deprivation leading to morphological changes such as invasive growth (68). This ability was overlooked for a long time, as laboratory yeast strains had lost this property, but this is not the case for many yeast strains found “free” in nature or also aggressive yeast strains that cause many problems in hospitals after surgical intervention (69).
A recent study shows that several gene products involved in yeast invasive growth become translated in a cap-independent manner (70). Pivotal in this process is the presence of a poly(A) stretch in the 5′-UTR of involved mRNAs that serves as a binding site for the poly(A)-binding protein Pab1 and allows internal initiation under conditions in which overall cap-dependent translation is low due to nutritional restrictions. The cap-independent translation of mRNAs such that for Flo8, a transcription factor required for invasive growth, is fundamental for this physiological adaptation.
We anticipate that studies carried out in yeast will serve as an indispensable system allowing also for conclusions on the differentiation process in higher cells. Indeed, yeast will very likely be an early focus for systems biology analyses that examine gene expression and enzyme regulation as a function of physiological state because of the existing genetic information available, the ease of performing knock-out/knock-in gene replacements, and the ability to use the well established genetic tools to screen for different genetic interactions.
Acknowledgments
We are grateful to Thomas Dever and Claudio De Virgilio for comments on the manuscript, and we thank Hans Trachsel, a pioneer in the translation initiation field.
This work was supported by the Swiss National Science Foundation and the Universities of Bern and Geneva. This is the second article in the Thematic Minireview Series on Protein Synthesis. This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- uORF
- upstream open reading frame.
REFERENCES
- 1.Hinnebusch A. G. (2006) in Landmark Papers in Yeast Biology (Linder P., Shore D., Hall M. N. eds) pp. 85–107, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2.Sherman F., Stewart J. W., Schweingruber A. M. (1980) Cell 20, 215–222 [DOI] [PubMed] [Google Scholar]
- 3.Cigan A. M., Feng L., Donahue T. F. (1988) Science 242, 93–97 [DOI] [PubMed] [Google Scholar]
- 4.Donahue T. F., Cigan A. M., Pabich E. K., Valavicius B. C. (1988) Cell 54, 621–632 [DOI] [PubMed] [Google Scholar]
- 5.Cigan A. M., Pabich E. K., Feng L., Donahue T. F. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 2784–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H. K., Yoon H., Hannig E. M., Donahue T. F. (1997) Genes Dev. 11, 2396–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon H. J., Donahue T. F. (1992) Mol. Cell. Biol. 12, 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnebusch A. G. (2005) Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 9.Mueller P. P., Hinnebusch A. G. (1986) Cell 45, 201–207 [DOI] [PubMed] [Google Scholar]
- 10.Hartwell L. H. (1967) J. Bacteriol. 93, 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartwell L. H., McLaughlin C. S. (1968) J. Bacteriol. 96, 1664–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartwell L. H., McLaughlin C. S. (1969) Proc. Natl. Acad. Sci. U.S.A. 62, 468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanic-Joyce P. J., Singer R. A., Johnston G. C. (1987) J. Biol. Chem. 262, 2845–2851 [PubMed] [Google Scholar]
- 14.Hanic-Joyce P. J. (1985) Genetics 110, 591–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bedard D., Johnston G., RA S. (1981) Curr. Genet. 4, 205–214 [DOI] [PubMed] [Google Scholar]
- 16.Lazaris-Karatzas A., Montine K. S., Sonenberg N. (1990) Nature 345, 544–547 [DOI] [PubMed] [Google Scholar]
- 17.Verlhac M. H., Chen R. H., Hanachi P., Hershey J. W., Derynck R. (1997) EMBO J. 16, 6812–6822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rousseau D., Gingras A. C., Pause A., Sonenberg N. (1996) Oncogene 13, 2415–2420 [PubMed] [Google Scholar]
- 19.Fields S., Song O. (1989) Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 20.Altmann M., Edery I., Sonenberg N., Trachsel H. (1985) Biochemistry 24, 6085–6089 [DOI] [PubMed] [Google Scholar]
- 21.Altmann M., Handschin C., Trachsel H. (1987) Mol. Cell. Biol. 7, 998–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altmann M., Sonenberg N., Trachsel H. (1989) Mol. Cell. Biol. 9, 4467–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner C., Nakayama N., Goebl M., Tanaka K., Toh-e A., Matsumoto K. (1988) Mol. Cell. Biol. 8, 3556–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danaie P., Altmann M., Hall M. N., Trachsel H., Helliwell S. B. (1999) Biochem. J. 340, 135–141 [PMC free article] [PubMed] [Google Scholar]
- 25.Altmann M., Müller P. P., Pelletier J., Sonenberg N., Trachsel H. (1989) J. Biol. Chem. 264, 12145–12147 [PubMed] [Google Scholar]
- 26.Hernández G., Altmann M., Sierra J. M., Urlaub H., Diez del Corral R., Schwartz P., Rivera-Pomar R. (2005) Mech. Dev. 122, 529–543 [DOI] [PubMed] [Google Scholar]
- 27.Prat A., Schmid S. R., Buser P., Blum S., Trachsel H., Nielsen P. J., Linder P. (1990) Biochim. Biophys. Acta 1050, 140–145 [DOI] [PubMed] [Google Scholar]
- 28.Grifo J. A., Tahara S. M., Leis J. P., Morgan M. A., Shatkin A. J., Merrick W. C. (1982) J. Biol. Chem. 257, 5246–5252 [PubMed] [Google Scholar]
- 29.Edery I., Humbelin M., Darveau A., Lee K. A., Milburn S., Hershey J. W., Trachsel H., Sonenberg N. (1983) J. Biol. Chem. 258, 11398–11403 [PubMed] [Google Scholar]
- 30.Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. (1985) J. Biol. Chem. 260, 7651–7658 [PubMed] [Google Scholar]
- 31.Seal S. N., Schmidt A., Marcus A. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 6562–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., Nishi K., Schnier J., Slonimski P. P. (1989) Nature 337, 121–122 [DOI] [PubMed] [Google Scholar]
- 33.Schmid S. R., Linder P. (1991) Mol. Cell. Biol. 11, 3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blum S., Mueller M., Schmid S. R., Linder P., Trachsel H. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 6043–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blum S., Schmid S. R., Pause A., Buser P., Linder P., Sonenberg N., Trachsel H. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7664–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richter-Cook N. J., Dever T. E., Hensold J. O., Merrick W. C. (1998) J. Biol. Chem. 273, 7579–7587 [DOI] [PubMed] [Google Scholar]
- 37.Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. (1990) Mol. Cell. Biol. 10, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altmann M., Müller P. P., Wittmer B., Ruchti F., Lanker S., Trachsel H. (1993) EMBO J. 12, 3997–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coppolecchia R., Buser P., Stotz A., Linder P. (1993) EMBO J. 12, 4005–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altmann M., Wittmer B., Méthot N., Sonenberg N., Trachsel H. (1995) EMBO J. 14, 3820–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jamieson D. J., Beggs J. D. (1991) Mol. Microbiol. 5, 805–812 [DOI] [PubMed] [Google Scholar]
- 42.Jamieson D. J., Rahe B., Pringle J., Beggs J. D. (1991) Nature 349, 715–717 [DOI] [PubMed] [Google Scholar]
- 43.Stevens S. W., Ryan D. E., Ge H. Y., Moore R. E., Young M. K., Lee T. D., Abelson J. (2002) Mol. Cell 9, 31–44 [DOI] [PubMed] [Google Scholar]
- 44.Chuang R. Y., Weaver P. L., Liu Z., Chang T. H. (1997) Science 275, 1468–1471 [DOI] [PubMed] [Google Scholar]
- 45.de la Cruz J., Iost I., Kressler D., Linder P. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. (2004) Mol. Microbiol. 51, 987–1001 [DOI] [PubMed] [Google Scholar]
- 47.Iost I., Dreyfus M., Linder P. (1999) J. Biol. Chem. 274, 17677–17683 [DOI] [PubMed] [Google Scholar]
- 48.Yang Q., Jankowsky E. (2006) Nat. Struct. Mol. Biol. 13, 981–986 [DOI] [PubMed] [Google Scholar]
- 49.Sachs A. B., Davis R. W. (1990) Science 247, 1077–1079 [DOI] [PubMed] [Google Scholar]
- 50.de la Cruz J., Kressler D., Rojo M., Tollervey D., Linder P. (1998) RNA 4, 1268–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sachs A., Wahle E. (1993) J. Biol. Chem. 268, 22955–22958 [PubMed] [Google Scholar]
- 52.Tarun S. Z., Jr., Sachs A. B. (1995) Genes Dev. 9, 2997–3007 [DOI] [PubMed] [Google Scholar]
- 53.Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. (1998) Mol. Cell 2, 135–140 [DOI] [PubMed] [Google Scholar]
- 54.Hinnebusch A. G. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 6442–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wanke V., Cameroni E., Uotila A., Piccolis M., Urban J., Loewith R., De Virgilio C. (2008) Mol. Microbiol. 69, 277–285 [DOI] [PubMed] [Google Scholar]
- 56.Novick P., Schekman R. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deloche O., de la Cruz J., Kressler D., Doère M., Linder P. (2004) Mol. Cell 13, 357–366 [DOI] [PubMed] [Google Scholar]
- 58.Sezen B., Seedorf M., Schiebel E. (2009) Genes Dev. 23, 1559–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashe M. P., De Long S. K., Sachs A. B. (2000) Mol. Biol. Cell 11, 833–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashe M. P., Slaven J. W., De Long S. K., Ibrahimo S., Sachs A. B. (2001) EMBO J. 20, 6464–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palmer L. K., Shoemaker J. L., Baptiste B. A., Wolfe D., Keil R. L. (2005) Mol. Biol. Cell 16, 3727–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heitman J., Movva N. R., Hall M. N. (1991) Science 253, 905–909 [DOI] [PubMed] [Google Scholar]
- 63.Barbet N. C., Schneider U., Helliwell S. B., Stansfield I., Tuite M. F., Hall M. N. (1996) Mol. Biol. Cell 7, 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pause A., Belsham G. J., Gingras A. C., Donzé O., Lin T. A., Lawrence J. C., Jr., Sonenberg N. (1994) Nature 371, 762–767 [DOI] [PubMed] [Google Scholar]
- 65.Lin T. A., Kong X., Haystead T. A., Pause A., Belsham G., Sonenberg N., Lawrence J. C., Jr. (1994) Science 266, 653–656 [DOI] [PubMed] [Google Scholar]
- 66.Beretta L., Gingras A. C., Svitkin Y. V., Hall M. N., Sonenberg N. (1996) EMBO J. 15, 658–664 [PMC free article] [PubMed] [Google Scholar]
- 67.Altmann M., Schmitz N., Berset C., Trachsel H. (1997) EMBO J. 16, 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. (1992) Cell 68, 1077–1090 [DOI] [PubMed] [Google Scholar]
- 69.Saville S. P., Lazzell A. L., Bryant A. P., Fretzen A., Monreal A., Solberg E. O., Monteagudo C., Lopez-Ribot J. L., Milne G. T. (2006) Antimicrob. Agents Chemother. 50, 3312–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gilbert W. V., Zhou K., Butler T. K., Doudna J. A. (2007) Science 317, 1224–1227 [DOI] [PubMed] [Google Scholar]
- 71.Berset C., Trachsel H., Altmann M. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]