Abstract
Endocytosis is a conserved cellular process in which nutrients, lipids, and receptors are internalized and transported to early endosomes, where they are sorted and either channeled to degradative pathways or recycled to the plasma membrane. MICAL-L1 and EHD1 are important regulatory proteins that control key endocytic transport steps. However, the precise mechanisms by which they mediate transport, and particularly the mode by which they connect to motor proteins, have remained enigmatic. Here we have identified the collapsin response mediator protein-2 (Crmp2) as an interaction partner of MICAL-L1 in non-neuronal cells. Crmp2 interacts with tubulin dimers and kinesin and negatively regulates dynein-based transport in neuronal cells, but its expression and function in non-neuronal cells have remained poorly characterized. Upon Crmp2 depletion, we observed dramatic relocalization of internalized transferrin (Tf) from peripheral vesicles to the endocytic recycling compartment (ERC), similar to the effect of depleting either MICAL-L1 or EHD1. Moreover, Tf relocalization to the ERC could be inhibited by interfering with microtubule polymerization, consistent with a role for uncoupled motor protein-based transport upon depletion of Crmp2, MICAL-L1, or EHD1. Finally, transfection of dynamitin, a component of the dynactin complex whose overexpression inhibits dynein activity, prevented the relocalization of internalized Tf to the ERC upon depletion of Crmp2, MICAL-L1, or EHD1. These data provide the first trafficking regulatory role for Crmp2 in non-neuronal cells and support a model in which Crmp2 is an important endocytic regulatory protein that links MICAL-L1·EHD1-based vesicular transport to dynein motors.
Keywords: Dynein, Endosomes, Intracellular Trafficking, Membrane Recycling, Membrane Trafficking, Microtubules, Molecular Motors, Protein-Protein Interactions, Receptor Recycling, RNA Silencing
Introduction
The complexity of the endocytic pathways has broadened in recent years upon discovery of the involvement of the four C-terminal Eps 15 homology domain (EHD)3 proteins (reviewed in Ref. 1). Since the single Caenorhabditis elegans ortholog (known as RME-1) was originally identified as a regulator of yolk receptor recycling (2), its closest mammalian homolog, EHD1, was found to regulate the recycling of receptors that traverse both the clathrin-dependent (3, 4) and the clathrin-independent (5, 6) internalization pathways. Despite similarities to the Ras family of GTP-binding proteins (5, 7), EHD proteins bind and hydrolyze ATP (7–9), a function necessary for their localization to tubular and vesicular membranes (5, 7, 10).
Proteins that contain EH domains interact with the tripeptide asparagine-proline-phenylalanine (NPF) (11). Recent studies have determined that the EH domains of the C-terminal EHD proteins have a highly positively charged surface (12) and selectively interact with NPF motifs followed by clusters of acidic residues (13, 14). Moreover, EHD proteins coordinate endocytic transport with Rab proteins through their interactions with common effectors that contain such NPF motifs (9, 15). More recently, it was demonstrated that EHD1 interacts with the NPF-containing MICAL family protein, MICAL-L1, a Rab8 effector that localizes to EHD1-containing tubules and regulates endocytic transport and recycling (16).
Movement of cytoplasmic vesicles from the cell periphery to the ERC depends upon motor protein-mediated transport along microtubules (17). A growing number of recent studies provide evidence of direct and indirect connections between Rab proteins and cytoplasmic dynein (18–22), but to date, the mode by which EHD proteins are connected to dynein remains unknown. Here we identify the collapsin response mediator protein-2 (Crmp2) as a novel interaction partner of MICAL-L1 and demonstrate its function in the regulation of endocytic transport in non-neuronal cells, where it serves as a critical link between MICAL-L1·EHD1 vesicles and dynein motors.
EXPERIMENTAL PROCEDURES
Recombinant DNA Constructs
Crmp2 cDNA was obtained from a human brain cDNA library and cloned into a pHA-CMV vector (Clontech). siRNA-resistant HA-Crmp2 was generated using the QuikChange site-directed mutagenesis kit (Stratagene), to produce a series of “silent mutations” within the regions recognized by the four oligonucleotides (Dharmacon). GFP-MICAL-L1 has been described previously (Sharma et al. (16)). MICAL-L1 CH and LIM domains were subcloned into pGEX-6p-2 vector (GE Healthcare). p50 dynamitin-GFP was kindly provided by Dr. Julie Donaldson.
Gene Knockdown by Silencing RNA (siRNA)
Custom design oligonucleotide duplexes targeting human EHD1 (Naslavsky et al. (15)) and ON-TARGETplus SMARTpool siRNA targeting MICAL-L1 and Crmp2 (Dharmacon) were transfected for 72 h using Dharmafect (Dharmacon) as described previously (16).
Antibodies and Reagents
The following antibodies and reagents were used in this study: mouse anti-Crmp2 antibody clone C4G (American Research Products), mouse polyclonal anti-MICAL-L1 antibody (Novus Biologicals), mouse anti-α-tubulin antibody (Molecular Probes, Eugene, OR), mouse anti-human HLA-A, -B, and -C antibody (Leinco Technologies, Inc.), mouse anti-CD59 antibody (a kind gift from Dr. Vaclav Horejsi), mouse anti-HA antibody (Covance), mouse anti-GFP antibody (Roche Applied Science), rabbit anti-HA (Bethyl Laboratories, Inc.), Alexa Fluor 568-conjugated goat anti-mouse antibody (Molecular Probes), Alexa Fluor 488-conjugated goat anti-mouse antibody (Molecular Probes), Alexa Fluor 568-conjugated goat anti-rabbit antibody (Molecular Probes), horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc.), Alexa Fluor 568-conjugated human transferrin (Invitrogen), and nocodazole (LKT Laboratories, Inc.).
Immunofluorescence and Uptake Assays
HeLa cells were grown on coverslips, transfected with FuGENE 6 (Roche Applied Science), and fixed with 4% (v/v) paraformaldehyde as described previously (16). In some cases, cytosolic proteins were extracted by treatment for 1 min with 0.05% w/v saponin before fixation. Cells were immunostained as described (16), and images were obtained using an LSM5 Pascal confocal microscope (Carl Zeiss) with a 63× 1.4 numerical aperture objective with appropriate filters. Transferrin uptake experiments were done essentially as described (16). CD59 and major histocompatibility complex class I (MHC I) uptake was performed in complete medium (DMEM plus serum) at 37 °C for either 15 min or 30 min, respectively, prior to acid rinse (0.5% acetic acid and 0.5 m NaCl, pH 3.0) for 1 min. Cells were fixed and immunostained with the appropriate fluorochrome-conjugated secondary antibody.
Protein Expression, Purification, Pulldowns, and Immunoprecipitations
GST-MICAL-L1 CH and LIM domains and GST constructs were purified and utilized for pulldown experiments as described previously (16).
RESULTS
Crmp2 Is an Interaction Partner of MICAL-L1
To identify potential links between the MICAL-L1·EHD1-based endocytic regulatory proteins and motor proteins, we generated a purified GST-MICAL-L1 protein (supplemental Fig. S1A) containing both the CH and the LIM domains to perform pulldown assays using homogenized bovine brain cytosol. Upon elution, specific bands from Coomassie Blue-stained gels were excised and subjected to the University of Nebraska Medical Center Mass Spectrometry and Proteomics Core Facility for identification using the LTQ Orbitrap. MASCOT search analysis yielded Crmp2 as a potential interaction partner. In neuronal cells, Crmp2 expression is required for axon outgrowth and axon-dendrite specification (23). Crmp2 has multiple binding domains, including a dihydropyrimidinase (DHPase) homology domain, a tubulin binding domain, a kinesin light chain-1 (KLC-1) binding domain, and two dynein heavy chain binding regions (24) (supplemental Fig. S1B). We have demonstrated that Crmp2 is not only expressed in neuronal cell lines such as SK-N-MC but also in a variety of non-neuronal cell lines (supplemental Fig. S1C).
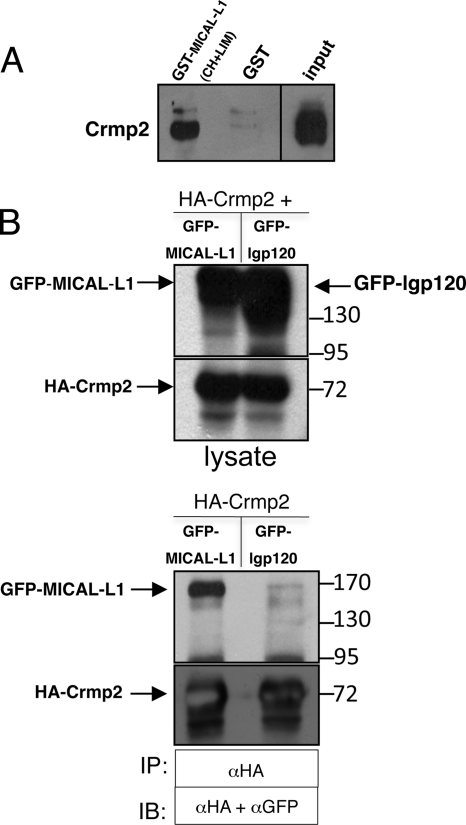
To confirm MICAL-L1 binding to Crmp2, pulldown assays were done using GST-MICAL-L1 CH and LIM domains, and Crmp2 antibodies were utilized for immunoblotting. As shown, GST-MICAL-L1 CH and LIM domains pulled-down Crmp2, whereas the negative control (GST protein only) did not (Fig. 1A). To determine whether this interaction could be detected in lysates from non-neuronal cells, we transfected HeLa cells with HA-Crmp2 along with either GFP-MICAL-L1 or GFP-lgp120 (a lysosomal protein and negative control) and performed co-immunoprecipitation assays. As demonstrated, HA-Crmp2 interacted with GFP-MICAL-L1 but not with GFP-lgp120 (Fig. 1B). We next hypothesized that if the Crmp2-MICAL-L1 interaction is functionally significant, we would anticipate a level of co-localization between the two proteins in cells. Unfortunately, available Crmp2 antibodies were not useful for immunofluorescence experiments, necessitating the overexpression of a tagged version of Crmp2. Initial co-staining experiments determined that overexpressed Crmp2 was largely cytosolic, complicating attempts to gauge its co-localization with the endogenous MICAL-L1. We have previously shown that upon overexpression of certain proteins, their high levels in the cytoplasm can often mask an association with underlying membrane structures, and non-membrane-associated proteins can be removed by a brief cytosolic extraction (25). Upon cytosol extraction, HA-Crmp2 was visualized in a partial co-localization with microtubules (supplemental Fig. S2, A–C), consistent with its known interaction with α-tubulin (26). To further validate these findings, we employed nocodazole to induce microtubule depolymerization prior to immunostaining (supplemental Fig. S2, D–F). As observed (arrows), a large number of punctae were positive for both Crmp2 and α-tubulin, providing additional evidence of their co-localization. In addition, HA-Crmp2 also displayed a small degree of overlap with MICAL-L1 on tubular membranes (supplemental Fig. S2, G–I) and punctae (supplemental Fig. S2, J–L, arrows), suggesting that a transient interaction between these two proteins may have a significant role in non-neuronal cells.
FIGURE 1.
Crmp2 is an interaction partner of MICAL-L1. A, a GST fusion protein containing only the MICAL-L1 CH and LIM domains and GST alone were each incubated with bovine brain cytosol. Precipitated proteins were immunoblotted with anti-Crmp2 antibody. B, HeLa cells were transfected with HA-Crmp2 and either GFP-MICAL-L1 or GFP-lgp120 (seen as a broad band), harvested, lysed, and incubated with immobilized anti-HA beads for 2 h. Lysates (top panels) and immunoprecipitates (bottom panels) were immunoblotted (IB) with either anti-HA or anti-GFP antibodies. IP, immunoprecipitation.
Depletion of Crmp2 Causes Accumulation of Transferrin at the ERC
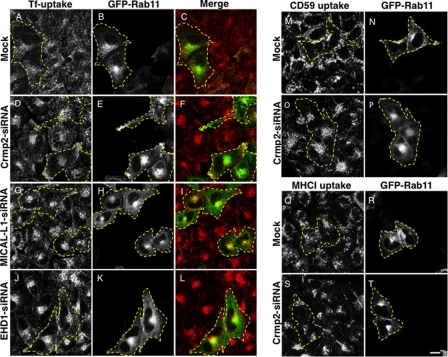
To investigate the functional role of Crmp2 and its interaction with MICAL-L1 in non-neuronal cells, we depleted Crmp2 with siRNA oligonucleotides. We observed that less than 10% of endogenous Crmp2 remained upon Crmp2-siRNA treatment, and the knockdown was specific as scrambled-siRNA oligonucleotides had no impact on Crmp2 expression levels (supplemental Fig. S3A). We then performed transferrin (Tf) uptake assays on mock- and Crmp2-siRNA-treated cells expressing GFP-Rab11 to mark recycling endosomes and the ERC, a tubulo-vesicular network of recycling endosomes localized to the perinuclear region of the cell (27). As observed after 5 min of uptake, in mock-treated cells, internalized fluorochrome-labeled transferrin (Tf-568) was still localized primarily to peripheral vesicles and endosomes, with very little Tf-568 having arrived at the perinuclear ERC (Fig. 2, A–C). However, in cells depleted of endogenous Crmp2, MICAL-L1, or EHD1, internalized Tf-568 rapidly reached and accumulated at the ERC region (Fig. 2, D–L). To ensure the specificity of Crmp2 deletion and that the changes in Tf transport were not due to off-target effects, we designed an siRNA-resistant Crmp2 construct. As demonstrated, this Crmp2 construct was highly expressed even in the presence of Crmp2-siRNA (supplemental Fig. S3B). Importantly, when Crmp2 was reintroduced into cells depleted of endogenous Crmp2, internalized Tf-568 no longer rapidly translocated to the ERC (supplemental Fig. S3, C–H; compare Crmp2-transfected cells (yellow dashed border) with untransfected cells in F). Moreover, the rapid transport of internalized receptors to the ERC upon Crmp2 depletion was a general phenomenon that also included proteins internalized via clathrin-independent pathways, such as CD59 (Fig. 2, M–P, see co-localization with Rab11 at the ERC in cells outlined by dashed borders) and MHC I (Fig. 2, Q–T, see co-localization with Rab11 at the ERC in cells outlined by dashed borders). These data suggest a novel role for Crmp2 in partnership with MICAL-L1 and EHD1 in regulating the transport of internalized cargo from early endosomes to the ERC.
FIGURE 2.
Crmp2 regulates vesicular transport in non-neuronal cells. HeLa cells were either mock-treated (A–C, M and N, and Q and R) or treated with Crmp2-siRNA (D–F, O and P, and S and T), MICAL-L1-siRNA (G–I), or EHD1-siRNA (J–L) oligonucleotides for 72 h. Cells were also transfected with GFP-Rab11 for the last 48 h of the siRNA treatment (to denote recycling endosomes and the ERC) and subjected to 5 min of Tf-568 uptake (A–L) or internalization of anti-CD59 antibodies (M–P) and anti-MHC I (Q–T) antibodies for either 15 or 30 min, respectively. Cells were then acid-washed to remove non-internalized surface antibody, fixed, and analyzed by confocal microscopy. Dashed borders depict GFP-Rab11-transfected cells. Bar, 10 μm.
Dynein Function Is Required for Crmp2·MICAL-L1·EHD1-based Vesicular Transport
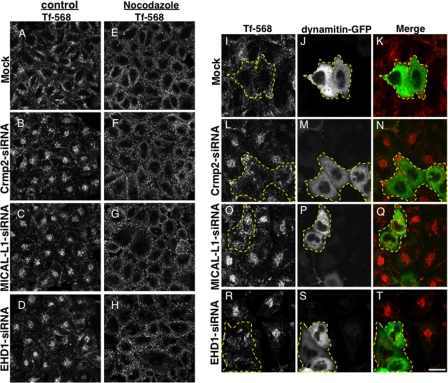
Intriguing new studies have demonstrated that Crmp2 is a modulator of microtubule dynamics (26, 28, 29) and a regulator of motor proteins (24, 30, 31). Based on our observations that the depletion of Crmp2, its interaction partner MICAL-L1, or EHD1 (which binds to MICAL-L1) affects Tf-568 intracellular transport, we hypothesized that Crmp2 might serve as a link between MICAL-L1·EHD1-based endocytic transport and dynein-based movement to the ERC along microtubules. Because dynein transports vesicles along microtubules, initially we determined whether intact microtubules are required for this rapid translocation of Tf to the ERC. The microtubule depolymerizing agent, nocodazole, was added to HeLa cells that were mock-treated or treated with Crmp2-siRNA, MICAL-L1-siRNA, or EHD1-siRNA (Fig. 3). Efficacy of microtubule depolymerization was demonstrated by anti-tubulin staining (data not shown). After 5 min of Tf-568 uptake, as expected in mock-treated cells, most of the Tf-568 was localized to the periphery of the cells, either in the absence or in the presence of nocodazole (Fig. 3, A and E). Upon siRNA treatment in the absence of nocodazole, Tf-568 accumulated at the ERC (Fig. 3, B–D). However, in the presence of nocodazole, the siRNA-treated cells exhibited an entirely peripheral distribution of internalized TF-568, with no accumulation at the ERC (Fig. 3, F–H). To test the idea that Crmp2 serves as a link between MICAL-L1·EHD1-based endocytic transport and dynein-based movement to the ERC, we utilized dynamitin-GFP, a protein that comprises part of the dynactin complex connecting organelle membranes to motor proteins (32) and whose overexpression inhibits dynein. Dynamitin-GFP was transfected into HeLa cells that were mock-treated or treated with Crmp2-siRNA, MICAL-L1-siRNA, or EHD1-siRNA, and the cells were then subjected to a 5-min uptake of Tf-568. As expected, in mock-treated cells, internalized Tf-568 was mostly localized to the periphery, and this did not change upon dynamitin-GFP expression (Fig. 3, I–K, dashed borders). Consistent with our earlier results, cells depleted of Crmp2, MICAL-L1, or EHD1 showed a rapid accumulation of Tf-568 at the perinuclear ERC (Fig. 3, L–T). However, in depleted cells that were also expressing dynamitin-GFP, rapid accumulation of Tf-568 at the ERC was prevented (Fig. 3, L–T, cells outlined by dashed borders). These data suggest that Crmp2 may link MICAL-L1·EHD1-based vesicular transport to cytoplasmic dynein.
FIGURE 3.
Dynein function is required for Crmp2·MICAL-L1·EHD1-based vesicular transport. HeLa cells were either mock-treated (A, E, and I–K) or treated with Crmp2-siRNA (B, F, and L–N), MICAL-L1-siRNA (C, G, and O–Q), or EHD1-siRNA (D, H, and R–T) oligonucleotides for 72 h. In E–H, cells were treated for 30 min with 66 μm nocodazole prior to 5 min of Tf-568 uptake. In I–T, cells were transfected with dynamitin-GFP for the last 48 h of the siRNA treatment, subjected to 5 min Tf-568 uptake prior to fixation, and analyzed by confocal microscopy. Dashed borders depict dynamitin-GFP-transfected cells. Bar, 10 μm.
DISCUSSION
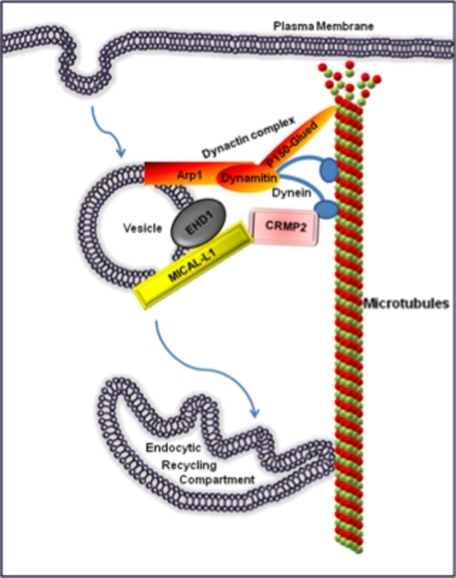
To date, little is known about the mechanisms linking EHD1 and MICAL-L1 to cytoplasmic motor proteins that transport vesicles along microtubules. Here we have identified Crmp2 as a novel interaction partner of MICAL-L1. Although Crmp2 has been attributed a role in proliferation events in non-neuronal cells (33), it has been characterized almost exclusively in neurons, where it regulates axonal growth and neuronal polarity (23, 34). Although Crmp2 has been described as a regulator of microtubule polymerization (29) and a regulator of both kinesin and cytoplasmic dynein (24, 30, 31), our study utilizing siRNA-based depletion of Crmp2 promotes a novel role for Crmp2 in coupling the MICAL-L1·EHD1 endocytic regulatory machinery with motor proteins, such as cytoplasmic dynein (Fig. 4). Remarkably, depletion of Crmp2 or either MICAL-L1 or EHD1 induces the same phenotype; internalized Tf is rapidly transported to the ERC, where it accumulates. Because Crmp2 is a negative regulator of dynein motors (24), a likely explanation is that the interaction between MICAL-L1 and Crmp2 links MICAL-L1·EHD1-containing peripheral vesicles with dynein and microtubules, allowing slow, regulated transport to the ERC. Thus, in the absence of any of these three proteins, an uncoupling occurs that would allow dysregulation of dynein and rapid transport of peripheral vesicles along microtubules to the ERC. Moreover, under these conditions, when dynein is inhibited by dynamitin overexpression, rapid transport of Tf to the ERC is blocked, providing further support for the model in which Crmp2 links MICAL-L1·EHD1 endocytic transport to dynein motors.
FIGURE 4.
Model depicting Crmp2 as a link between MICAL-L1·EHD1-associated endosomes and dynein motors. Depletion of Crmp2, MICAL-L1, or EHD1 causes a rapid movement of vesicles containing internalized receptors to the ERC along intact microtubules. Crmp2, which was described as a negative regulator of dynein (24), may modulate dynein-derived motility, thus decelerating movement of MICAL-L1·EHD1-decorated endosomes on route to the ERC. Depletion of Crmp2 would remove its constraining effect, thus resulting in faster delivery of cargo to the ERC. Overexpression of the dynactin subunit dynamitin retains the receptors in the cell periphery, pointing to dynein as the candidate motor protein functionally linked to MICAL-L1·EHD1-based vesicular transport.
Although we have not observed altered Tf trafficking upon Crmp2 overexpression,4 Arimura et al. (24) have demonstrated a degree of microtubule fragmentation. However, acute siRNA depletion studies are generally easier to interpret, and our data are consistent with the recent study of Arimura et al. (35), who demonstrated that in neuronal cells, siRNA depletion of Crmp2 led to altered TrkB receptor anterograde transport.
A number of recent studies have forged important connections between various Rab and motor proteins (19, 20, 36–38). In particular, the Rab11 effectors Rab11-FIP5 and Rab11-FIP3 link Rab11 to kinesin II and cytoplasmic dynein, (19, 38). However, to date, aside from a connection between EHD1 and Rab8a with myosin Vb (39) and the interaction between EHD1 and the lemur tyrosine kinase 2 and myosin VI (40), little attempt has been made to relate EHD proteins and motor proteins, and a relationship between EHDs and cytoplasmic dynein has not been described. Our study provides evidence for the first physical and functional link between MICAL-L1·EHD1-decorated endosomes and cytoplasmic dynein, via Crmp2, a novel MICAL-L1 interaction partner in non-neuronal cells. Although at present we might only speculate on the “advantage” of such regulation, one possibility is that kinetics of transport affect sorting at the endosome. This idea has been proposed by Mayor et al. (41), who demonstrated that delayed transport of glycosylphosphatidylinositol-anchored proteins (compared with Tf receptor) results in their targeting to distinct organelles. Our studies also raise the possibility of a more widespread role for “neuronal proteins” in the regulation of endocytosis in non-neuronal cells.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM074876 (to S. C. and N. N.) and R01GM087455 (S. C.) and National Center for Research Resources Grant P20 RR018759 (to N. N.). This work was also supported by a grant from the Nebraska Department of Health (to N. N.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
J. Rahajeng, S. S. P. Giridharan, N. Naslavsky, and S. Caplan, unpublished observations.
- EHD
- Eps 15 homology domain
- ERC
- endocytic recycling compartment
- Tf
- transferrin
- MICAL
- molecules interacting with CasL
- CH
- calponin homology
- LIM
- Lin11-Isl-1 and Mec-3.
REFERENCES
- 1.Grant B. D., Caplan S. (2008) Traffic 9, 2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grant B., Zhang Y., Paupard M. C., Lin S. X., Hall D. H., Hirsh D. (2001) Nat. Cell Biol. 3, 573–579 [DOI] [PubMed] [Google Scholar]
- 3.Lin S. X., Grant B., Hirsh D., Maxfield F. R. (2001) Nat. Cell Biol. 3, 567–572 [DOI] [PubMed] [Google Scholar]
- 4.Picciano J. A., Ameen N., Grant B. D., Bradbury N. A. (2003) Am. J. Physiol. Cell Physiol. 285, C1009–C1018 [DOI] [PubMed] [Google Scholar]
- 5.Caplan S., Naslavsky N., Hartnell L. M., Lodge R., Polishchuk R. S., Donaldson J. G., Bonifacino J. S. (2002) EMBO J. 21, 2557–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jović M., Naslavsky N., Rapaport D., Horowitz M., Caplan S. (2007) J. Cell Sci. 120, 802–814 [DOI] [PubMed] [Google Scholar]
- 7.Daumke O., Lundmark R., Vallis Y., Martens S., Butler P. J., McMahon H. T. (2007) Nature 449, 923–927 [DOI] [PubMed] [Google Scholar]
- 8.Lee D. W., Zhao X., Scarselletta S., Schweinsberg P. J., Eisenberg E., Grant B. D., Greene L. E. (2005) J. Biol. Chem. 280, 17213–17220 [DOI] [PubMed] [Google Scholar]
- 9.Naslavsky N., Rahajeng J., Sharma M., Jovic M., Caplan S. (2006) Mol. Biol. Cell 17, 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jović M., Kieken F., Naslavsky N., Sorgen P. L., Caplan S. (2009) Mol. Biol. Cell 20, 2731–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salcini A. E., Confalonieri S., Doria M., Santolini E., Tassi E., Minenkova O., Cesareni G., Pelicci P. G., Di Fiore P. P. (1997) Genes Dev. 11, 2239–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieken F., Jović M., Naslavsky N., Caplan S., Sorgen P. L. (2007) J. Biomol. NMR 39, 323–329 [DOI] [PubMed] [Google Scholar]
- 13.Kieken F., Sharma M., Jovic M., Giridharan S. S., Naslavsky N., Caplan S., Sorgen P. L. (2010) J. Biol. Chem. 285, 8687–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henry G. D., Corrigan D. J., Dineen J. V., Baleja J. D. (2010) Biochemistry 49, 3381–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naslavsky N., Boehm M., Backlund P. S., Jr., Caplan S. (2004) Mol. Biol. Cell 15, 2410–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma M., Giridharan S. S., Rahajeng J., Naslavsky N., Caplan S. (2009) Mol. Biol. Cell 20, 5181–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kardon J. R., Vale R. D. (2009) Nat. Rev. Mol. Cell Biol. 10, 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horgan C. P., Hanscom S. R., Jolly R. S., Futter C. E., McCaffrey M. W. (2010) Biochem. Biophys. Res. Commun. 394, 387–392 [DOI] [PubMed] [Google Scholar]
- 19.Horgan C. P., Hanscom S. R., Jolly R. S., Futter C. E., McCaffrey M. W. (2010) J. Cell Sci. 123, 181–191 [DOI] [PubMed] [Google Scholar]
- 20.Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., Neefjes J. (2009) J. Cell Biol. 185, 1209–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wanschers B., van de Vorstenbosch R., Wijers M., Wieringa B., King S. M., Fransen J. (2008) Cell Motil. Cytoskeleton 65, 183–196 [DOI] [PubMed] [Google Scholar]
- 22.Sun Y., Shestakova A., Hunt L., Sehgal S., Lupashin V., Storrie B. (2007) Mol. Biol. Cell 18, 4129–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimura N., Menager C., Fukata Y., Kaibuchi K. (2004) J. Neurobiol. 58, 34–47 [DOI] [PubMed] [Google Scholar]
- 24.Arimura N., Hattori A., Kimura T., Nakamuta S., Funahashi Y., Hirotsune S., Furuta K., Urano T., Toyoshima Y. Y., Kaibuchi K. (2009) J. Neurochem. 111, 380–390 [DOI] [PubMed] [Google Scholar]
- 25.Caplan S., Hartnell L. M., Aguilar R. C., Naslavsky N., Bonifacino J. S. (2001) J. Cell Biol. 154, 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y., Ihara Y. (2000) J. Biol. Chem. 275, 17917–17920 [DOI] [PubMed] [Google Scholar]
- 27.Maxfield F. R., McGraw T. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 28.Chae Y. C., Lee S., Heo K., Ha S. H., Jung Y., Kim J. H., Ihara Y., Suh P. G., Ryu S. H. (2009) Cell. Signal. 21, 1818–1826 [DOI] [PubMed] [Google Scholar]
- 29.Fukata Y., Itoh T. J., Kimura T., Ménager C., Nishimura T., Shiromizu T., Watanabe H., Inagaki N., Iwamatsu A., Hotani H., Kaibuchi K. (2002) Nat. Cell Biol. 4, 583–591 [DOI] [PubMed] [Google Scholar]
- 30.Kawano Y., Yoshimura T., Tsuboi D., Kawabata S., Kaneko-Kawano T., Shirataki H., Takenawa T., Kaibuchi K. (2005) Mol. Cell. Biol. 25, 9920–9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura T., Watanabe H., Iwamatsu A., Kaibuchi K. (2005) J. Neurochem. 93, 1371–1382 [DOI] [PubMed] [Google Scholar]
- 32.Valetti C., Wetzel D. M., Schrader M., Hasbani M. J., Gill S. R., Kreis T. E., Schroer T. A. (1999) Mol. Biol. Cell 10, 4107–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tahimic C. G., Tomimatsu N., Nishigaki R., Fukuhara A., Toda T., Kaibuchi K., Shiota G., Oshimura M., Kurimasa A. (2006) Biochem. Biophys. Res. Commun. 340, 1244–1250 [DOI] [PubMed] [Google Scholar]
- 34.Inagaki N., Chihara K., Arimura N., Ménager C., Kawano Y., Matsuo N., Nishimura T., Amano M., Kaibuchi K. (2001) Nat. Neurosci. 4, 781–782 [DOI] [PubMed] [Google Scholar]
- 35.Arimura N., Kimura T., Nakamuta S., Taya S., Funahashi Y., Hattori A., Shimada A., Ménager C., Kawabata S., Fujii K., Iwamatsu A., Segal R. A., Fukuda M., Kaibuchi K. (2009) Dev. Cell 16, 675–686 [DOI] [PubMed] [Google Scholar]
- 36.Fukuda M., Kuroda T. S., Mikoshiba K. (2002) J. Biol. Chem. 277, 12432–12436 [DOI] [PubMed] [Google Scholar]
- 37.Valente C., Polishchuk R., De Matteis M. A. (2010) Nat. Cell Biol. 12, 635–638 [DOI] [PubMed] [Google Scholar]
- 38.Schonteich E., Wilson G. M., Burden J., Hopkins C. R., Anderson K., Goldenring J. R., Prekeris R. (2008) J. Cell Sci. 121, 3824–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roland J. T., Kenworthy A. K., Peranen J., Caplan S., Goldenring J. R. (2007) Mol. Biol. Cell 18, 2828–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chibalina M. V., Seaman M. N., Miller C. C., Kendrick-Jones J., Buss F. (2007) J. Cell Sci. 120, 4278–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayor S., Sabharanjak S., Maxfield F. R. (1998) EMBO J. 17, 4626–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.