Abstract
Three multiprotein systems are known for iron-sulfur (Fe-S) cluster biogenesis in prokaryotes and eukaryotes as follows: the NIF (nitrogen fixation), the ISC (iron-sulfur cluster), and the SUF (mobilization of sulfur) systems. In all three, cysteine is the physiological sulfur source, and the sulfur is transferred from cysteine desulfurase through a persulfidic intermediate to a scaffold protein. However, the biochemical nature of the sulfur source for Fe-S cluster assembly in archaea is unknown, and many archaea lack homologs of cysteine desulfurases. Methanococcus maripaludis is a methanogenic archaeon that contains a high amount of protein-bound Fe-S clusters (45 nmol/mg protein). Cysteine in this archaeon is synthesized primarily via the tRNA-dependent SepRS/SepCysS pathway. When a ΔsepS mutant (a cysteine auxotroph) was grown with 34S-labeled sulfide and unlabeled cysteine, <8% of the cysteine, >92% of the methionine, and >87% of the sulfur in the Fe-S clusters in proteins were labeled, suggesting that the sulfur in methionine and Fe-S clusters was derived predominantly from exogenous sulfide instead of cysteine. Therefore, this investigation challenges the concept that cysteine is always the sulfur source for Fe-S cluster biosynthesis in vivo and suggests that Fe-S clusters are derived from sulfide in those organisms, which live in sulfide-rich habitats.
Keywords: Archaebacteria, Homocysteine, Iron-Sulfur Protein, Methionine, Sulfur, Methanococcus, Archaea, Cysteine
Introduction
Iron-sulfur (Fe-S) clusters play critical roles in a broad range of biological processes, such as electron transfer reactions, substrate binding, nonredox catalysis, sulfur donation, and sensing of redox status for regulatory processes (1, 2). Fe-S clusters can be synthesized in vitro on an apoprotein with iron salts and sulfide under anaerobic conditions (3, 4). However, because of their toxicity, the cellular concentrations of free iron and sulfide are usually much lower than the levels required for abiotic cluster formation. In vivo Fe-S cluster assembly uses several complex protein systems, which construct nascent clusters on scaffold proteins and then transfer them into recipient apoproteins. Three Fe-S cluster assembly systems, NIF (nitrogen fixation), the ISC (iron-sulfur cluster), and the SUF (mobilization of sulfur), are conserved among bacteria, and many organisms possess more than one system (5, 6).
Cysteine is the sulfur source for Fe-S cluster biogenesis in prokaryotes and eukaryotic mitochondria and chloroplasts. Cysteine desulfurase, a pyridoxal 5′-phosphate (PLP)2-dependent enzyme, initiates Fe-S cluster formation by mobilizing the sulfur atom from cysteine and transferring it to Fe-S clusters. A persulfide intermediate is formed on a conserved cysteinyl residue at the active site and functions as the proximal sulfur donor for Fe-S cluster biosynthesis. Four cysteine desulfurase homologs are known as follows: NifS, IscS, SufS, and CsdA (5, 7).
The sulfur source for Fe-S cluster biosynthesis in archaea awaits further investigation. The methanogenic archaeon Methanococcus maripaludis produces cysteine primarily on tRNACys via the SepRS/SepCysS pathway (8). This organism possesses two pathways for charging tRNACys. When exogenous cysteine is available, tRNACys is aminoacylated with cysteine by a canonical cysteinyl-tRNA synthetase (CysRS) (9, 10). In a second pathway, cysteine is synthesized de novo in two steps (8). In the first step, tRNACys is aminoacylated with O-phosphoserine (Sep) by O-phosphoseryl-tRNA synthetase (SepRS). In the second step, Sep-tRNACys is converted to Cys-tRNACys by Sep-tRNA:Cys-tRNA synthase (SepCysS). However, the sulfur source for this reaction is not known. A ΔsepS mutant is a cysteine auxotroph (8). This raises two questions. First, do cells contain an alternative pathway to generate free cysteine? Second, if not, what is the sulfur source for the biosynthesis of Fe-S clusters and other sulfur-containing compounds in cells? Moreover, homologs of cysteine desulfurase are absent in many Methanococcus species, other Methanococcales, and many nonmethanogenic archaea (supplemental Fig. S1). Therefore, no characterized enzymes are currently known that specifically catalyze the release of sulfur from cysteine in these organisms.
Here, unlike in all other previously studied organisms, we demonstrate that the sulfur in Fe-S clusters in the methanogenic archaeon Methanococcus maripaludis does not originate from cysteine. Instead, it is derived from sulfide, which is abundant in the anaerobic environment where the organism was isolated (11). This discovery also sheds light on the Fe-S cluster biogenesis in other organisms living in sulfide-rich environments and early life forms on the anoxic earth when sulfide was abundant.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
Two M. maripaludis strains, the wild-type strain S2 and the ΔsepS mutant (a cysteine auxotroph lacking the O-phosphoseryl-tRNA synthetase (8)), were used in this study. Cultures were grown in McNA medium (a minimal medium with 10 mm sodium acetate) reduced with 3 mm cysteine or dithiothreitol (DTT) as indicated (12). The 5-ml cultures were grown in 28-ml aluminum seal tubes pressurized to 276 kPa with H2/CO2 (4:1, v/v). The 100-ml cultures were grown in 1-liter bottles pressurized to 138 kPa with H2/CO2 (4:1, v/v). Before inoculation, 3 mm sodium sulfide was added as the sulfur source. The Escherichia coli strain K12 MG1655 was grown aerobically in Luria-Bertani (LB) medium.
Quantification of Protein-bound Inorganic Sulfide, Iron, and Cysteinyl and Methionyl Residues
The inorganic acid-labile sulfide content in cell extracts was determined by an adaptation of the methylene blue method (13, 14). The assays were carried out in the anaerobic chamber with an atmosphere of 95% of N2 and 5% of H2. Cell-free extracts in 0.1 m potassium phosphate (pH 8.0) were passed twice through a 2-ml Sephadex G-25 column (Aldrich) to remove non-protein-bound sulfide. Precipitated FeS complexes from the medium were also trapped at the top of the column. To measure the sulfide content, 0.3 ml of 1% (w/v) freshly prepared zinc acetate and 10 μl of 12% sodium hydroxide were added and mixed with 200 μl of cell extract (0.2–0.4 mg of protein of M. maripaludis or 1–2 mg of protein of E. coli) in microcentrifuge tubes. The tubes were closed and left at room temperature for 20 min. Then 0.1 ml of 1% of N,N-dimethylphenylenediamine in 5 m of HCl and 40 μl of 11.5 mm FeCl3 in 1.2 m of HCl were added, and the solution was mixed rapidly. The samples were centrifuged at 16,000 × g for 10 min to remove denatured proteins. The absorbance of the supernatant was determined at 670 nm. Solutions of sodium sulfide were used as standards.
The iron content was determined by an adaptation of the o-phenanthroline method (15, 16). The cell extracts (0.1–0.2 mg of protein of M. maripaludis or 1–2 mg of protein of E. coli) were passed twice through a 2-ml Sephadex G-25 column and then deproteinized with 5% (w/v, final concentration) trichloroacetic acid (TCA). The supernatant (100 μl) was transferred to a microcentrifuge tube, and then 0.1 ml of 1 m sodium acetate/acetic acid buffer (pH 4.0), 0.1 ml of 0.3% (w/v) o-phenanthroline, and 0.1 ml of 10% (w/v) hydroxylamine hydrochloride were added. The mixture was incubated at 37 °C for 30 min. The absorbance was measured at 512 nm. Solutions of FeCl3·6H2O were used as standards.
The contents of cysteinyl and methionyl residues in M. maripaludis proteins were determined at the University of California, Davis, Amino Acid Analysis Facility after acid hydrolysis with 6 m HCl for 24 h at 110 °C. The protein concentrations were determined by BCA protein assay (Pierce) (17).
Quantification of Intracellular Free Cysteine, Homocysteine, Coenzyme M, and Cystathionine
The M. maripaludis wild-type (strain S2) cells were grown in 100 ml of McNA medium (reduced with 3 mm DTT) to an absorbance of ∼0.35 at 600 nm. Cells were harvested anaerobically by centrifugation at 3,000 × g for 15 min at room temperature. The following steps were carried out in an anaerobic chamber with an atmosphere of 95% N2 and 5% H2. The pellet was suspended in 1.5 ml of 10 mm acetic acid and incubated on ice for 15 min to lyse the cells, and the suspension was centrifuged in microcentrifuge tubes at 16,000 × g for 10 min. The supernatant (∼1.3 ml) was collected and divided into two 0.5-ml samples, to which 40 μl of buffer (1 m HEPES, 50 mm EDTA (pH 8.2)) was added. One sample that served as a negative control was supplemented with 4 mm N-ethylmaleimide. Thiols were quantified by a modification of the monobromobimane (mBBr) derivatization method (18). To each sample (∼500 μl), 20 μl of 50 mm mBBr (in acetonitrile) was added to a final concentration of 2 mm, and the solution was incubated for 5 min in the dark at room temperature. Samples were then deproteinized by addition of 0.6 ml of acetonitrile and incubated for 15 min at 60 °C. To stop the mBBr reaction, glacial acetic acid was added to the final concentration of 1% (v/v), and the samples were cooled on ice. The excess of unreacted mBBr was removed by extraction with 0.2 ml of chloroform, and the aqueous phase was collected. In this step, the sample was concentrated by extraction of acetonitrile into the chloroform fraction. Samples (20 μl) were analyzed on a Waters 2695 Separation HPLC System. The samples were separated on an Altima C-18 reversed phase column at 30 °C using a linear gradient of 5–95% of methanol with 0.25% acetic acid. Fluorescence detection was performed with a Shimadzu-RF-10A XL detector, using an excitation at 390 nm and emission at 480 nm. The standard curves were prepared with cysteine, homocysteine, and coenzyme M. The quantification of cystathionine in the free amino acid pool was performed as described previously (19) at the University of California, Davis, Amino Acid Analysis Facility.
Measurement of the Incorporation of [3,3-2H2]Cysteine into Cellular Proteins
M. maripaludis wild-type strain S2 and the ΔsepS mutant were grown in 5 ml of McNA medium with 3 mm DTT, 3 mm sodium sulfide, and 1.5 mm dl-[3,3,3′,3′-2H4]cystine to an absorbance of ∼0.5 at 600 nm. The cells were collected by centrifugation at 2,460 × g for 20 min at room temperature. The cell pellet was suspended, incubated with 0.4 ml of 5% TCA for 10 min at room temperature, and then centrifuged to collect denatured proteins. This process was repeated two more times to ensure the complete removal of free cysteine from the precipitated proteins. The resulting sample was dissolved in 200 μl of 0.5 m Tris-HCl buffer (pH 8.6) containing 6 m urea and 16 mm DTT, and then the protein bound cysteinyl residues were alkylated with 10 μl of methyl iodide. The samples were then dialyzed for 15 h against 3× 1 liter of water and evaporated. This process removed urea and also any remaining free cysteine and methionine not removed in the TCA washing. The protein samples were then acid-hydrolyzed (6 m HCl, 110 °C for 12 h under nitrogen), and the S-methylcysteine was assayed by gas chromatography-mass spectrometry (GC-MS) as described previously (20).
Measurement of the Incorporation of [34S]Sulfide into Thiol-containing Amino Acids and Fe-S Clusters
Elemental sulfur with the following isotopic abundances, 32S, 3.48 atom %; 33S, 2.23 atom %; 34S, 92.59 atom %; 36S, 1.70 atom % (Monsanto Research Corp.), was used in the synthesis of [34S]sulfide by reduction with DTT. 34S (4 mg) was heated to 120 °C for 10 min in an aluminum seal tube containing 2 ml of McNA medium with 0.2 m DTT. To volatilize the sulfide, 2 ml of 1 m HCl was added, and the solution was incubated for 30 min. After the removal of the liquid, the H2S gas was trapped in 0.5 ml of 0.5 m NaOH. Final concentration of sulfide was determined upon reaction with 1 mm 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). An extinction coefficient of ϵ412 = 14.15 mm−1 cm−1 was used for calculations (21).
The wild-type or ΔsepS mutant cells were grown in McNA medium containing 1.5 mm [34S]sulfide and 3 mm l-cysteine with the natural abundance of 34S (4.2 atom % 34S). All the following steps were carried out anaerobically. Cells from 40 ml of overnight grown cultures with an absorbance of ∼1 at 600 nm were harvested by centrifugation at 2,460 × g for 20 min at room temperature and washed twice with McNA medium reduced with 3 mm coenzyme M. The cell pellets were suspended in 0.5 ml of 50 mm PIPES buffer (pH 6.8) containing 1 mm DTT. Cells (∼10 mg dry weight) were lysed by repeated (2×) freezing (−20 °C) and thawing. About 10 units of DNase I (Promega) was added to digest genomic DNA. The cell extract was passed through a 2-ml Sephadex G-25 column to remove free sulfide, insoluble iron sulfides, and other low molecular weight compounds. The cell extract (∼1 ml) was then concentrated using a Centricon 10-kDa filtration unit (Millipore). The concentrated samples (70 μl) were transferred to 1.5-ml microcentrifuge tubes, which were placed in 10-ml glass serum bottles, closed with butyl stoppers, sealed, flushed with N2 for 5 min, and frozen at −20 °C. As controls, 1 ml of media was collected immediately after inoculation of culture and again after harvesting the cells. To test the possibility that homocysteine or cystathionine supplied the sulfur for the Fe-S clusters, ΔsepS cells were grown with 3 mm homocysteine plus 3 mm cysteine plus 1.5 mm [34S]sulfide or 5 mm cystathionine plus 1.5 mm [34S]sulfide.
The 34S isotopic abundance in Fe-S clusters was analyzed by GC-MS. The acid-labile sulfide was released by treatment of the sample with 1 m HCl and trapped in 1 m NaOH as described previously (22). The resulting sulfide was converted into dibenzyl sulfide and assayed by GC-MS to determine the isotopic abundance of the 32S/34S in the M+ = 214 m/z ion as described previously (23). The apparatus for trapping the sulfide used a septate 1.0-ml glass vial containing a 2 × 4-mm glass test tube and a stirring bar. The septate vial was flushed with argon, and a 50–200-μl sample was injected into the bottom of the vial. Then 15 μl of 1 m NaOH was added to the absorbent tube, followed by addition of 0.1 volume of 1 m HCl. After stirring at room temperature for 1 h, 40 μl of 50% methanol in water was added to the absorbent tube, and the sample was transferred to the 1-ml septate glass vial flushed with argon. Then 2 μl of distilled benzyl chloride was added, and the vial was heated for 1 h at 60 °C. The samples were then evaporated to dryness with a stream of nitrogen gas and extracted with 0.5 ml of methylene chloride. The methylene chloride was separated from water by passing the solution through a small plug of absorbent cotton and concentrated by evaporation to 3–6 μl for GC-MS analysis.
The proteins remaining in the acidified samples were dissolved in 400 μl of Tris-HCl buffer (pH 8.6) containing 6 m urea and 16 mm DTT, and the cysteinyl residues were alkylated with methyl iodide as described above. The methylcysteine and methionine in proteins were assayed for 34S abundance as described previously (24).
Enzyme Assays
All enzyme assays were carried out in an anaerobic chamber with an atmosphere of 95% N2 and 5% H2. The protein concentrations were determined by BCA protein assay (Pierce) (17).
Cystathionine γ-cleavage activity in M. maripaludis cell extracts were determined by following the production of cysteine by reaction with the acid-ninhydrin reagent (25). Assay mixture in a total volume of 0.3 ml contained 0.1 m Tris-HCl (pH 8.0), 10 μm PLP, and cell-free extract (0.3 mg of total protein). The reactions were initiated by addition of 10 mm l-cystathionine and incubated for 30 min at 37 °C. The reactions were stopped by addition of 0.3 ml of glacial acetic acid and 0.3 ml of 2.5% (w/v) ninhydrin in 60% (v/v) acetic acid and 40% (v/v) concentrated HCl. The samples were heated in boiling water for 10 min for color development, and the absorbance was determined at 560 nm after cooling. Solutions of cysteine were used as standards. Kinetic constants in cell extracts were determined with 0.5, 1, 2, 4, 8, and 10 mm l-cystathionine.
Cystathionine β-cleavage activity was determined by following the production of Hcy by reaction with DTNB (21). Assay mixture in a total volume of 0.2 ml contained 0.1 m Tris-HCl (pH 8.0), 10 μm PLP, and cell-free extract (0.2 mg of total protein). The reactions were initiated by addition of 10 mm l-cystathionine and incubated for 30 min at 37 °C. The reactions were stopped by heating in boiling water for 10 min, followed by reaction with 50 μl of 0.1 m DTNB. The samples (20 μl) were analyzed for Hcy and Cys on an Altima C-18 reversed phase column with a Waters 2695 Separation HPLC system at 25 °C using a linear gradient of 5–95% of methanol with 0.25% acetic acid, and the peaks of the 2-nitro-5-thiobenzoate derivatives of Hcy and Cys were monitored at 330 nm. Kinetic constants in cell extracts were determined with 1.25, 2.5, 5, 7.5, and 10 mm l-cystathionine.
Cysteine desulfurase activity was determined by the methylene blue method (14). Assay mixtures in a total volume of 0.8 ml contained 0.1 m Tris-HCl (pH 8.0), 0.2 m NaCl, 1 mm DTT, 50 μm PLP, and cell-free extract (0.2 mg of total protein). The reactions were initiated by addition of 1 mm l-cysteine and incubated for 30 min at 37 °C. The reactions were stopped by addition of 100 μl of 1% N,N-dimethylphenylenediamine in 5 m of HCl and 100 μl of 11.5 mm FeCl3 in 1.2 m HCl. The samples were centrifuged at 16,000 × g for 10 min, and the absorbance of the supernatant was determined at 670 nm. Kinetic constants in cell extracts were determined with 0.05, 0.1, 0.2, 0.5, and 1 mm cysteine and DTT.
Homoserine sulfhydrylase activity was determined by following the production of Hcy after the reaction with DTNB (21). Assay mixture in a total volume of 0.2 ml contained 0.25 m Tris-HCl (pH 8.0), 10 mm MgSO4, 0.2 m KCl, 10 mm homoserine, 50 mm ATP, 50 mm acetyl-CoA or succinyl-CoA, 10 μm PLP, and cell-free extract (0.2 mg of total protein). The reactions were initiated by the addition of 10 mm sodium sulfide and incubated for 1 h at 37 °C. The reactions were stopped by heating in boiling water for 10 min, followed by reaction with DTNB and separation with HPLC as described above for the cystathionine β-cleavage activity assay. The kinetic data were analyzed using SigmaPlot 10.0 with the enzyme kinetics module (Systat Software Inc.) fitted with the Michaelis-Menten equation for cystathionine cleavage activities and a Ping Pong Bi Bi mechanism for cysteine desulfurase activity.
RESULTS
Fe-S Clusters Are Abundant in Methanococci
A prior bioinformatics analysis suggested that Fe-S cluster-containing proteins are abundant in methanogens (26). To test this hypothesis, the amount of Fe-S clusters in M. maripaludis was determined by measurements of protein-bound inorganic sulfide and iron and compared with that found in aerobically grown E. coli (Table 1). The inorganic sulfide content agreed with the measurement in Methanocaldococcus jannaschii (31 nmol/mg protein) (20), and the iron content agreed with that in whole cells of Methanococcus voltae (30–94 nmol/mg protein) (27). Therefore, methanococci possessed ∼15-fold higher amounts of Fe-S clusters than aerobically grown E. coli. If the Fe-S clusters were mostly low potential [4Fe-4S] or [2Fe-2S] clusters (28) and coordinated by cysteinyl residues at a 1:1 stoichiometry, ∼ 40% of the total cysteinyl residues in M. maripaludis were associated with Fe-S clusters.
TABLE 1.
The levels of protein-bound inorganic sulfide and iron and cysteinyl and methionyl residues in M. maripaludis
| Chemicals | Amount in the cella |
|
|---|---|---|
| M. maripaludisb | E. colic | |
| nmol/mg protein | ||
| Inorganic sulfide | 45 ± 9 | 2.5 ± 0.4 |
| Iron | 50 ± 3 | 6.7 ± 0.7 |
| Cysteinyl residue | 102 ± 7 | 158d |
| Methionyl residue | 202 ± 11 | 265d |
a Values are means ± 1 S.D. obtained from measurements of three independent cultures.
b M. maripaludis strain S2 was grown anaerobically at 37 °C in McNA medium.
c E. coli strain K12 MG1655 was grown aerobically at 37 °C in LB medium.
d Values are from Ref. 51.
SepRS/SepCysS Pathway Is the Primary Pathway for Cysteine Biosynthesis
The ΔsepS mutant of M. maripaludis was a cysteine auxotroph, suggesting that this pathway is necessary for cysteine biosynthesis in this organism (8). However, it is possible that the mutant was a pseudoauxotroph and possessed an alternative cysteine biosynthetic pathway that was insufficient to support growth (29). Three lines of evidence demonstrated that the SepRS/SepCysS pathway provides the predominant, if not sole, source of cysteine.
First, exogenous 2H-labeled cysteine was incorporated into cellular proteins with little dilution in the ΔsepS mutant. When the ΔsepS mutant cells were grown with 3 mm sulfide, 3 mm DTT, and 1.5 mm [3,3,3′,3′-2H4]cystine, >99% of the cysteine in cellular proteins was 2H-labeled, suggesting that the mutant synthesized <1% of its cysteine. On the other hand, in the wild-type cells, ∼45% of the cysteine in proteins was 2H-labeled under the same growth condition. Thus, about half of the cysteine was biosynthesized by the cells, and half was assimilated directly from the medium.
Second, in the ΔsepS mutant, 34S-labeled sulfide was poorly incorporated into cysteinyl residues in cellular proteins. Presumably, if the cells could synthesize cysteine de novo from sulfide, the cysteinyl residues would contain high levels of 34S. The ΔsepS mutant was grown in medium containing 34S-enriched sulfide (92.6 atom % of 34S) and cysteine with the natural abundance of 34S (4.2 atom % of 34S). Cysteine recovered from the medium after growth retained its isotopic signatures and did not interchange its sulfur atoms with sulfide during cultivation (data not shown). Similarly, at the beginning of growth, the measured enrichment of 34S-labeled sulfide in the medium was 95 ± 2%, and it decreased to 82 ± 8% after growth. This decrease could be explained by the release of a small amount of unlabeled sulfide from cysteine, possibly via cysteine desulfurase (see below) or cysteine desulfidase activity, which converts cysteine into pyruvate, ammonia, and sulfide (22). In cellular proteins, only a small amount of 34S was incorporated into cysteine (Table 2). Considering the abundance of 34S in cysteine and sulfide used as substrates for growth, less than 1.7–3.0% of the cysteinyl residues of the ΔsepS mutant were derived from sulfide. On the other hand, in the wild-type cells grown under the same conditions of 1.5 mm sulfide and 3 mm cysteine, the atom % of 34S in cysteinyl residues was 81.6%, suggesting that 87.6% of the cysteine was derived from sulfide, and the rest was assimilated from the medium. This agreed with the 2H-labeling experiment above suggesting that, in wild-type cells, cellular cysteine was partially synthesized from sulfide and partially assimilated from the medium. However, the percentage of synthesized cysteine was higher in the 34S-labeling experiment. This variation was probably caused by different growth conditions, such as higher concentration of l-cysteine and lower concentration of sulfide in the medium and higher absorbance of the culture at the end of the growth in the 34S-labeling experiment.
TABLE 2.
Incorporation of [34S]sulfide into Fe-S clusters and cysteinyl and methionyl residues in the M. maripaludis ΔsepS mutant
| Sulfur sources in the growth mediuma | Atom % of 34S |
||
|---|---|---|---|
| Fe-S | Cys | Met | |
| [32S]Cys + [34S]Na2Sb | 87 ± 4 | 8 ± 2 | 92 ± 3 |
| [32S]Cystathionine + [34S]Na2Sc | 84 | 5 | 5 |
| [32S]Cys + [32S]Hcy + [34S]Na2Sd | 90 | 9 | 7 |
a Cells were grown in media containing 1.5 mm [34S]sulfide, 3 mm l-cysteine, 5 mm l-cystathionine, and/or 3 mm l-homocysteine as indicated. The numbers represented the measured atom % of 34S in the derivatives of the indicated molecules. The 34S enrichment of Na2S used as the substrate was 92.6%. The natural abundance of 34S in the sulfur-containing amino acids was 4.2%.
b Values are the means of three independent experiments ± 1 S.D.
c Medium contained 1.5 mm coenzyme M as a reductant.
d Values are the means of two independent experiments.
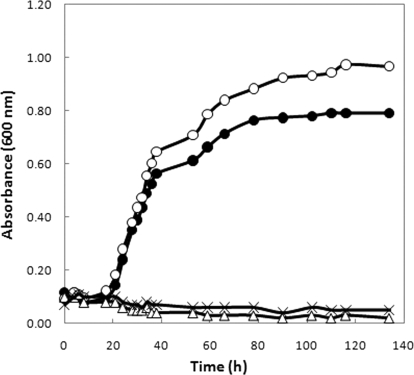
Third, the activity of cysteine production from l-cystathionine cleavage was examined. l-Cystathionine is an important intermediate in trans-sulfuration, and cysteine production from cystathionine γ-cleavage is employed by mammals, yeasts, fungi, and many bacteria such as actinomycetes (30). M. jannaschii cell extracts had activities of both cystathionine β-cleavage (produces homocysteine that is a precursor of methionine) and γ-cleavage (produces cysteine) (20), suggesting that trans-sulfuration with cystathionine could be an intermediate in the interconversion and biosynthesis of cysteine and methionine. Supporting this assumption, high levels of l-cystathionine (≥2 mm) can substitute for cysteine in the growth of the ΔsepS mutant (Fig. 1). When the ΔsepS mutant was grown with 34S-enriched sulfide and unenriched cystathionine, neither the cysteinyl nor the methionyl residues in proteins were 34S-enriched (Table 2), suggesting that cystathionine provided the sulfur for the biosynthesis of both of these amino acids under this growth condition. Therefore, cystathionine can be assimilated by the cells and function as an intermediate of sulfur metabolism. To examine the physiological significance of these reactions, the intracellular amount of cystathionine in wild-type cells grown without exogenous cystathionine was determined (Table 3). Assuming that proteins account for 60% of the cell dry weight and cells contain 2.5 μl of water per mg dry weight (31), the intracellular cystathionine concentration would be ∼0.1 mm. In M. maripaludis cell extracts, cystathionine γ-cleavage activity had an apparent Km(cystathionine) of 8.7 ± 0.5 mm and Vmax of 7.8 ± 0.3 nmol/min/mg protein (Table 4). Accordingly, at the physiological concentration of cystathionine, the cysteine biosynthetic activity would be ∼0.1 nmol/min/mg protein, which was 10-fold lower than the activity required for cysteine biosynthesis (1 nmol/min/mg protein) during growth. Therefore, cysteine production from cystathionine at physiological conditions was insignificant and would only generate a small amount of free cysteine, possibly as part of an organic sulfur salvage pathway in this organism. On the other hand, the homocysteine biosynthetic activity from cystathionine β-cleavage had an apparent Km(cystathionine) of 5 ± 1 mm and Vmax of 40 ± 5 nmol/min/mg protein (Table 4). With the physiological concentration of 0.1 mm cystathionine, the activity would be ∼1 nmol/min/mg protein, which was about half of the activity required for methionine biosynthesis (∼2 nmol/min/mg protein). Therefore, cystathionine β-cleavage may play a more important role in methionine biosynthesis.
FIGURE 1.
Growth of ΔsepS in McNA medium reduced with 3 mm DTT (×) and supplemented with 0.05 mml-cysteine (●), 5 mml-cystathionine (○), or 5 mml-homocysteine (Δ). The inoculums size was ∼107 cells per 5 ml of culture. All values were the averages of five cultures.
TABLE 3.
Contents of free sulfur-containing amino acids and coenzyme M in cells of M. maripaludis
| Compounds | Amount in the cella |
|---|---|
| nmol/mg dry wt | |
| Cysteine | 0.05 ± 0.01 |
| Homocysteine | 0.023 ± 0.004 |
| Cystathionine | 0.31 ± 0.01 |
| Coenzyme M | 0.43 ± 0.04 |
a Values are the means ± 1 S.D. obtained from two to five independent measurements. The free thiol contents (cysteine, homocysteine, and coenzyme M) were determined by derivatization with mBBr and fluorescence detection after HPLC separation. The free cystathionine content was determined using a lithium citrate-based amino acid analyzer.
TABLE 4.
Kinetic constants of l-cystathionine cleavage in M. maripaludis wild-type strain S2 cell extracts
| Reaction | Product determined | Km (l-cystathionine) | Vmax | R2a |
|---|---|---|---|---|
| mm | nmol/min/mg protein | |||
| γ-Cleavage | Cysteine | 8.7 ± 0.5 | 7.8 ± 0.3 | 0.99 |
| β-Cleavage | Homocysteine | 5 ± 1 | 40 ± 5 | 0.98 |
a R2 of Michaelis-Menten equation is given.
Free Cysteine Pool and Cysteine Desulfurase Activity in M. maripaludis
If cysteine was synthesized primarily on tRNA via the SepRS/SepCysS pathway, the intracellular levels of free cysteine would be expected to be very low. The free cysteine content was determined to be 0.05 nmol/mg dry weight (Table 3). Therefore, the intracellular concentration of free l-cysteine would be ∼20 μm. As an internal control for the loss in thiols during these measurements, the levels of coenzyme M were determined at the same time. The levels of coenzyme M were similar to those that had been reported previously (32). The expected intracellular concentration of free cysteine in bacteria ranges between 100 and 200 μm (33). Therefore, although free cysteine was present in M. maripaludis, the levels were much lower than those common in bacteria. Presumably, the free cysteine may be formed from l-cystathionine cleavage or hydrolysis of Cys-tRNACys and proteins.
Although cysteine desulfurase homologs have not been identified in the M. maripaludis genome, cysteine desulfurase activity was detected in cell-free extracts. The specific activity was 0.20 ± 0.02 nmol/min/mg protein, which was about half of the activity measured in aerobically grown E. coli cell extracts (0.45 ± 0.04 nmol/min/mg protein). This activity may come from an uncharacterized enzyme. This reaction in cell extracts had an apparent Km (cysteine) of 25 ± 3 μm and Vmax of 0.28 ± 0.05 nmol/min/mg protein. Therefore, the activity at the physiological concentration of cysteine (20 μm) would be about 0.1 nmol/min/mg protein and insufficient to support Fe-S cluster biosynthesis required for growth (∼0.5 nmol/min/mg protein). However, cysteine desulfurase activity may still be sufficient to serve as a major sulfur source for tRNA modification and vitamin biosynthesis.
Origin of Sulfur in Fe-S Clusters and Methionine
Even though a small pool of free cysteine exists in M. maripaludis, two lines of evidence indicated that cysteine is not the origin of sulfur in Fe-S clusters and methionine. First, when the ΔsepS mutant was grown with limiting amounts of cysteine (0.01 mm) in McNA medium, 80 μmol of cysteine was required for growth of 1 g cell dry weight. This value agreed well with the cysteine content of the whole cells of 102 μmol/g protein (or 61 μmol/g dry weight) (Table 1). In contrast, if cysteine was a general intermediate in sulfur assimilation, greater than 200 μmol of cysteine/g cell dry weight would be required for cysteine, Fe-S cluster, and methionine biosyntheses. Second, the incorporation of 34S-labeled sulfide into Fe-S clusters and methionine demonstrated that cysteine was a minor sulfur source for these compounds. Cells were grown with 34S-enriched sulfide and unlabeled cysteine as described above. The acid-labile sulfide, which was released from the Fe-S clusters present in cellular proteins, was highly 34S-enriched in both wild-type and the ΔsepS mutant cells (Table 2). In wild-type cells, the atom % of 34S in Fe-S clusters and methionyl residues was 90.7 and 90.5%, respectively. Given that the atom % of 34S sulfide in the medium declined from 92.6 to 82% during growth (see above), these results suggested that at least 98% of the sulfur in both Fe-S clusters and methionyl residues originated from 34S-enriched sulfide instead of cysteine assimilated from the medium. In contrast, 87.6% of the cellular cysteinyl residues originated from sulfide in the same experiment (see above). On the other hand, in the ΔsepS mutant cells, based upon the atom % of 34S (Table 2), at least 94 and 99% of the sulfur in Fe-S clusters and methionyl residues, respectively, originated from 34S-enriched sulfide instead of cysteine assimilated from the medium. Therefore, cysteine provided no more than 6 and 1% of the sulfur in Fe-S clusters and methionyl residues, respectively.
Whether sulfide could be a sulfur source for methionine biosynthesis through the direct sulfhydrylation of homoserine derivatives was examined. Yeast, fungi, and some bacteria can directly synthesize homocysteine from sulfhydrylation of acylated homoserine. Thus, they bypass cystathionine of the trans-sulfuration pathway for methionine biosynthesis (34). The homocysteine biosynthesis activities in M. maripaludis cell extracts with sulfide and homoserine plus succinyl-CoA, acetyl-CoA, or ATP as substrates were below the detection limit of 0.1 nmol/min/mg protein, suggesting that the direct sulfhydrylation reaction for homocysteine biosynthesis is probably absent. This observation agreed with the previous report that even though homocysteine can be synthesized from O-phosphohomoserine in M. jannaschii cell extracts, this reaction does not use sulfide as a sulfur source (20).
The possibility of homocysteine and cystathionine as sulfur sources for Fe-S cluster biosynthesis was also examined. When the ΔsepS mutant was grown with 34S-labeled sulfide and unlabeled cystathionine instead of cysteine, at least 90% of the sulfur in Fe-S clusters was derived from sulfide (Table 2). Homocysteine could not replace cysteine for the growth of ΔsepS (Fig. 1). Addition of unlabeled homocysteine to cells grown with 34S-enriched sulfide and unlabeled cysteine did not significantly dilute the enrichment of 34S in Fe-S clusters (Table 2). Therefore, l-cystathionine and l-homocysteine plus cysteine provided no more than 10 and 3%, respectively, of the sulfur for Fe-S cluster formation. However, under both growth conditions, the sulfur in methionyl residues was not enriched with 34S, suggesting that l-cystathionine and l-homocysteine, when present in the medium, provided 99 and 97%, respectively, of the sulfur for methionine biosynthesis (Table 2).
DISCUSSION
In summary, sulfur in Fe-S clusters and methionine in M. maripaludis originated from sulfide instead of cysteine. Moreover, sulfur assimilation in methanococci is different from previously studied prokaryotes in four aspects.
First, prokaryotes normally maintain cellular sulfide concentrations in the range of 20–160 μm (33). In contrast, methanococci inhabit anaerobic environments with high levels of sulfide (35) and are normally cultivated with 3–5 mm Na2S (27). At neutral pH, one-third of the sulfide remains in nonionized form (H2S), which may freely diffuse across the cell membrane (36). Therefore, the intracellular sulfide concentrations of methanococci are likely in the millimolar range. Methanococci are adapted to a high sulfide-tolerant lifestyle (37, 38), and they lack many targets for sulfide toxicity, such as the α,β-unsaturated fatty acid biosynthetic intermediates and cytochromes (36, 39). Considering that sulfide at millimolar concentrations is used frequently in vitro for the enzymatic and nonenzymatic reactions, it is reasonable that methanococci may use sulfide as a proximal sulfur donor in some reactions.
Second, enteric bacteria and plants synthesize cysteine via direct sulfhydrylation of O-acetylserine by O-acetylserine sulfhydrylase (40), and sulfide is the physiological sulfur donor for this reaction with Km values in the micromolar range (41, 42). In contrast, even though sulfide is presumably present at high levels in methanococci, it is probably not the direct sulfur donor for cysteine biosynthesis. Cysteine in methanococci is synthesized primarily on tRNACys via the SepRS/SepCysS pathway, and a persulfide group is proposed to be a proximal sulfur donor based upon the structural similarity of SepCysS with cysteine desulfurases (43–45). If persulfide groups were derived from cysteine in vivo, it would be a futile cycle to synthesize cysteine from persulfide groups. Therefore, methanococci presumably use a new biochemical process to generate persulfide groups for sulfur incorporation into cysteine.
Third, the known Fe-S cluster assembly system is incomplete in methanococci and many other archaea. It is especially interesting that methanococci and some other archaea living in solfataric hydrothermal systems lack homologs of cysteine desulfurases (supplemental Fig. S1), indicating that they may use a different mechanism for sulfur incorporation into Fe-S clusters. Indeed, only the sufBC and apbC/nbp35 genes involved in Fe-S biogenesis are conserved across archaea with completely sequenced genomes. Like the bacterial and eukaryotic ApbC/Nbp35 homologs, the M. maripaludis protein forms Fe-S clusters in vitro and may function as a Fe-S cluster carrier protein in cells (46). The suf system in bacteria is important under iron starvation and oxidative stress conditions, and it is the only Fe-S cluster assembly system in certain bacteria, such as Thermatoga maritima, Mycobacterium tuberculosis, and cyanobacteria (5, 47), suggesting that suf can function as the sole Fe-S cluster assembly system in some organisms. Recent biochemical studies showed that SufC is an ABC-type ATPase (48) and SufB is a persulfide acceptor, but it may also function as a scaffold protein (49). In most archaea sufB and sufC are arranged as neighboring genes. Mutagenesis of the genomic copy of sufB (MMP1169) and sufC (MMP1168) in M. maripaludis was only successful with sufBC expressed in trans on a shuttle vector (data not shown), suggesting that these proteins may be essential in methanococci. However, identification of other assembly components is required to understand the nature of the sulfur donor and how sufBC could function without other known Fe-S cluster biogenesis proteins.
Fourth, bacteria synthesize homocysteine, the precursor of methionine, by either trans-sulfuration with cystathionine as an intermediate or direct sulfhydrylation of O-succinylhomoserine or O-acetylhomoserine (34, 50). None of the homologs of homocysteine biosynthetic genes are present in the M. maripaludis genome. The trans-sulfuration route uses cysteine as a sulfur source for cystathionine biosynthesis. Because cysteine is not the sulfur source for methionine biosynthesis in M. maripaludis, this route either plays a minor part in methionine biosynthesis or there is a new reaction independent of cysteine for cystathionine biosynthesis. The sulfhydrylation route uses sulfide as a sulfur source for homocysteine biosynthesis. However, the direct sulfhydrylation activity with sulfide and homoserine derivatives as substrates was below the detection limit in M. maripaludis cell extracts. Therefore, a new biochemical process should be responsible for homocysteine and methionine biosynthesis in methanococci.
Supplementary Material
Acknowledgments
We thank Kim Harich for assistance with MS analysis and Walter Niehaus for assistance with editing this manuscript.
This work was supported by National Science Foundation Grant MCB 0231319 (to R. H. W.), a Department of Energy grant (to W. B. W.), and a Doctoral Completion Award from the University of Georgia Graduate School (to Y. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PLP
- pyridoxal-5′-phosphate
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
- mBBr
- monobromobimane.
REFERENCES
- 1.Beinert H. (2000) J. Biol. Inorg. Chem. 5, 2–15 [DOI] [PubMed] [Google Scholar]
- 2.Fontecave M. (2006) Nat. Chem. Biol. 2, 171–174 [DOI] [PubMed] [Google Scholar]
- 3.Herskovitz T., Averill B. A., Holm R. H., Ibers J. A., Phillips W. D., Weiher J. F. (1972) Proc. Natl. Acad. Sci. U.S.A. 69, 2437–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagen K. S., Reynolds J. G., Holm R. H. (1981) J. Am. Chem. Soc. 103, 4054–4063 [Google Scholar]
- 5.Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 6.Frazzon J., Fick J. R., Dean D. R. (2002) Biochem. Soc. Trans. 30, 680–685 [DOI] [PubMed] [Google Scholar]
- 7.Fontecave M., Ollagnier-de-Choudens S. (2008) Arch. Biochem. Biophys. 474, 226–237 [DOI] [PubMed] [Google Scholar]
- 8.Sauerwald A., Zhu W., Major T. A., Roy H., Palioura S., Jahn D., Whitman W. B., Yates J. R., 3rd, Ibba M., Söll D. (2005) Science 307, 1969–1972 [DOI] [PubMed] [Google Scholar]
- 9.Stathopoulos C., Kim W., Li T., Anderson I., Deutsch B., Palioura S., Whitman W., Söll D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14292–14297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T., Graham D. E., Stathopoulos C., Haney P. J., Kim H. S., Vothknecht U., Kitabatake M., Hong K. W., Eggertsson G., Curnow A. W., Lin W., Celic I., Whitman W., Söll D. (1999) FEBS Lett. 462, 302–306 [DOI] [PubMed] [Google Scholar]
- 11.Jones W. J., Paynter M. J. B., Gupta R. (1983) Arch. Microbiol. 135, 91–97 [Google Scholar]
- 12.Whitman W. B., Shieh J., Sohn S., Caras D. S., Premachandran U. (1986) Syst. Appl. Microbiol. 7, 235–240 [Google Scholar]
- 13.Beinert H. (1983) Anal. Biochem. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 14.Fogo J. K., Popowsky M. (1949) Anal. Chem. 21, 732–734 [Google Scholar]
- 15.Harvey A. E., Smart J. A., Amis E. S. (1955) Anal. Chem. 27, 26–29 [Google Scholar]
- 16.Mahler H. R., Elowe D. G. (1954) J. Biol. Chem. 210, 165–179 [PubMed] [Google Scholar]
- 17.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 18.Fahey R. C., Newton G. L., Dorian R., Kosower E. M. (1981) Anal. Biochem. 111, 357–365 [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson E. L., Liu Y., Rosas-Sandoval G., Porat I., Söll D., Whitman W. B., Leigh J. A. (2008) J. Bacteriol. 190, 2198–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White R. H. (2003) Biochim. Biophys. Acta 1624, 46–53 [DOI] [PubMed] [Google Scholar]
- 21.Ellman G. L. (1959) Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 22.Tchong S. I., Xu H., White R. H. (2005) Biochemistry 44, 1659–1670 [DOI] [PubMed] [Google Scholar]
- 23.White R. H. (1983) Biochem. Biophys. Res. Commun. 112, 66–72 [DOI] [PubMed] [Google Scholar]
- 24.White R. H. (1981) Anal. Biochem. 114, 349–354 [DOI] [PubMed] [Google Scholar]
- 25.Gaitonde M. K. (1967) Biochem. J. 104, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Major T. A., Burd H., Whitman W. B. (2004) FEMS Microbiol. Lett. 239, 117–123 [DOI] [PubMed] [Google Scholar]
- 27.Whitman W. B., Ankwanda E., Wolfe R. S. (1982) J. Bacteriol. 149, 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer J. (2008) J. Biol. Inorg. Chem. 13, 157–170 [DOI] [PubMed] [Google Scholar]
- 29.Shieh J., Mesbah M., Whitman W. B. (1988) J. Bacteriol. 170, 4091–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagasawa T., Kanzaki H., Yamada H. (1984) J. Biol. Chem. 259, 10393–10403 [PubMed] [Google Scholar]
- 31.Dybas M., Konisky J. (1992) J. Bacteriol. 174, 5575–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balch W. E., Wolfe R. S. (1979) J. Bacteriol. 137, 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessler D. (2006) FEMS Microbiol. Rev. 30, 825–840 [DOI] [PubMed] [Google Scholar]
- 34.Lee H. S., Hwang B. J. (2003) Appl. Microbiol. Biotechnol. 62, 459–467 [DOI] [PubMed] [Google Scholar]
- 35.Edmond J. M., Von Damm K. L., McDuff R. E., Measures C. I. (1982) Nature 297, 187–191 [Google Scholar]
- 36.Truong D. H., Eghbal M. A., Hindmarsh W., Roth S. H., O'Brien P. J. (2006) Drug Metab. Rev. 38, 733–744 [DOI] [PubMed] [Google Scholar]
- 37.Lloyd K. G., Edgcomb V. P., Molyneaux S. J., Böer S., Wirsen C. O., Atkins M. S., Teske A. (2005) Appl. Environ. Microbiol. 71, 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visser A., Hulshoff Pol L. W., Lettinga G. (1996) Wat. Sci. Tech. 33, 99–110 [Google Scholar]
- 39.Edgcomb V. P., Molyneaux S. J., Saito M. A., Lloyd K., Böer S., Wirsen C. O., Atkins M. S., Teske A. (2004) Appl. Environ. Microbiol. 70, 2551–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabeh W. M., Cook P. F. (2004) J. Biol. Chem. 279, 26803–26806 [DOI] [PubMed] [Google Scholar]
- 41.Hwang C. C., Woehl E. U., Minter D. E., Dunn M. F., Cook P. F. (1996) Biochemistry 35, 6358–6365 [DOI] [PubMed] [Google Scholar]
- 42.Tai C. H., Nalabolu S. R., Jacobson T. M., Minter D. E., Cook P. F. (1993) Biochemistry 32, 6433–6442 [DOI] [PubMed] [Google Scholar]
- 43.Fukunaga R., Yokoyama S. (2007) J. Mol. Biol. 370, 128–141 [DOI] [PubMed] [Google Scholar]
- 44.Hauenstein S. I., Perona J. J. (2008) J. Biol. Chem. 283, 22007–22017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheppard K., Yuan J., Hohn M. J., Jester B., Devine K. M., Söll D. (2008) Nucleic Acids Res. 36, 1813–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyd J. M., Drevland R. M., Downs D. M., Graham D. E. (2009) J. Bacteriol. 191, 1490–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huet G., Daffé M., Saves I. (2005) J. Bacteriol. 187, 6137–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eccleston J. F., Petrovic A., Davis C. T., Rangachari K., Wilson R. J. (2006) J. Biol. Chem. 281, 8371–8378 [DOI] [PubMed] [Google Scholar]
- 49.Layer G., Gaddam S. A., Ayala-Castro C. N., Ollagnier-de Choudens S., Lascoux D., Fontecave M., Outten F. W. (2007) J. Biol. Chem. 282, 13342–13350 [DOI] [PubMed] [Google Scholar]
- 50.Yamagata S. (1989) Biochimie 71, 1125–1143 [DOI] [PubMed] [Google Scholar]
- 51.Neidhardt F. C., Umbarger H. E. (1996) in Escherichia coli and Salmonella (Neidhardt F. C., Curtiss R., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E. eds) pp. 13–16, American Society for Microbiology, Washington, DC [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.