Abstract
Condensation of chromatin into higher order structures is mediated by intra- and interfiber nucleosome-nucleosome interactions. Our goals in this study were to determine the impact specific activator-dependent histone acetylation had on chromatin condensation and to ascertain whether acetylation-induced changes in chromatin condensation were related to changes in RNA polymerase II (RNAPII) activity. To accomplish this, an in vitro model system was constructed in which the purified transcriptional activators, Tax and phosphorylated CREB (cAMP-response element-binding protein), recruited the p300 histone acetyltransferase to nucleosomal templates containing the human T-cell leukemia virus type-1 promoter sequences. We find that activator-dependent p300 histone acetylation disrupted both inter- and intrafiber nucleosome-nucleosome interactions and simultaneously led to enhanced RNAPII transcription from the decondensed model chromatin. p300 histone acetyltransferase activity had two distinct components: non-targeted, ubiquitous activity in the absence of activators and activator-dependent activity targeted primarily to promoter-proximal nucleosomes. Mass spectrometry identified several unique p300 acetylation sites on nucleosomal histone H3 (H3K9, H3K27, H3K36, and H3K37). Collectively, our data have important implications for understanding both the mechanism of RNAPII transcriptional regulation by chromatin and the molecular determinants of higher order chromatin structure.
Keywords: Chromatin Structure, Gene Regulation, Histone Acetylase, Mass Spectrometry (MS), Nuclear Organization, Post-translational Modification, RNA Polymerase II, Transcription Regulation, Folding, Oligomerization
Introduction
The genomes of eukaryotes are assembled into a complex nucleoprotein architecture called chromatin. In the first level of chromatin structure, 147 bp of DNA are wrapped 1.65 times around an octamer of core histones to form the nucleosome (1). Nucleosomal arrays make up the next level of chromatin architecture and consist of histone octamers spaced repetitively at ∼20–60-bp intervals along a DNA molecule. When proteins other than core histones bind to nucleosomal arrays, chromatin is formed. Chromatin undergoes a series of hierarchical conformational changes, leading to formation of higher order levels of condensed structure (see below). The transcription of genes by RNA polymerase II (RNAPII)2 must occur within this dense chromatin environment. Hence, a long-standing question in the field is the extent to which higher order chromatin fiber architecture influences gene expression (Refs. 2–4; for review, see Refs. 5–10).
Nucleosomal arrays and chromatin in solution in vitro are in equilibrium between multiple architectural states; that is, a primary structure having an extended “beads-on-a-string” conformation, secondary folded structures resulting from intrafiber nucleosome-nucleosome interactions, and tertiary oligomeric structures resulting from reversible interfiber nucleosome-nucleosome interactions (5, 9). Folding and oligomerization both are induced by increasing concentrations of cations, e.g. 1–10 mm MgCl2. In addition, transitions between the different condensed states require the amino-terminal “tail” domains (NTDs) of the core histones (5, 11). Removal of the NTDs by proteolysis or recombinant methods disrupts both folding (12–16) and oligomerization (14–17). Thus, the ionic composition of the solution and the core histone NTDs are key points of control of higher order chromatin architecture in vitro.
Post-translational modification of the core histone NTDs occurs commonly in vivo (6, 18). A widely characterized modification is lysine acetylation catalyzed by histone acetyltransferases (HATs). Lysine acetylation has long been correlated with active transcription (19), although the molecular mechanisms through which acetylation functions are still being deciphered. Previous work has shown that core histone NTD acetylation causes decondensation of nucleosomal arrays. These earlier in vitro studies utilized nucleosomal arrays assembled with either bulk modified histones purified from cells treated with histone deacetylase inhibitors such as sodium butyrate (2, 20), chemically ligated histones (21), histone mutants that mimic lysine acetylation (e.g. Lys to Gln) (22–24), or genetically installed acetyllysine residues (25). These experiments have yielded important insight into how lysine acetylation affects nucleosomal array conformational dynamics. However, the biochemical structure/function relationships that link specific activator-dependent HAT acetylation, chromatin fiber architecture, and RNAPII-dependent transcription have not been established in vitro.
To define these relationships, a model chromatin system has been constructed and characterized in which purified transcriptional activator proteins (Tax, pCREB) recruit the p300 HAT to the human T-cell leukemia virus type-1 (HTLV-1) promoter embedded in an array of positioned nucleosomes. We find that activator-dependent p300 acetylation led to significantly enhanced RNAPII transcription from the model chromatin. Physicochemical analyses indicated that native unacetylated chromatin was extensively oligomeric under transcription conditions, whereas activator-dependent p300-acetylated chromatin was mostly monomeric and unfolded under the same conditions. Activator-dependent p300 HAT activity resulted in heterogeneous acetylation along the chromatin template and was highest on promoter-proximal nucleosomes. Mass spectrometry identified numerous p300 acetylation sites, including novel sites on nucleosomal H3 at positions Lys-9, -27, -36, and -37. Taken together, our data demonstrate that disruption of repressive nucleosome-nucleosome interactions by p300 acetylation is strongly correlated with enhanced transcription by RNAPII in vitro. Our studies also characterize changes in the higher order architecture of model chromatin fibers resulting from activator-dependent p300 histone acetylation.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human His6-tagged p300 was expressed in Sf9 cells from a baculovirus-based plasmid and purified as described (26). Recombinant Ser133-phosphorylated CREB327 (pCREB) (27, 28) and Tax-His6 (29) were purified as described (30). Nuclear extract from CEM cells was prepared as described (31). Recombinant Xenopus laevis core histones and Lys to Gln mutant core histones (23) were expressed and purified as described (32, 33). Histone octamers were prepared as described (32, 33).
Construction of HTLV-1 DNA Templates and Assembly into Nucleosomal Arrays
To construct the HTLV-1 DNA template used for structure/function studies, an 832-bp fragment containing the HTLV-1 promoter sequences, the G-less cassette, and pUC13 polylinker DNA was amplified by PCR. The PCR fragments were ligated at each end to DNA consisting of four 208-bp 5 S rDNA repeats (34). This 2496-bp HTLV-1 template was ligated into pUC19 to form pHTLV208-8. After digestion of this plasmid with HhaI, the excised HTLV-1 DNA template was purified by size exclusion chromatography as described (17, 35). Nucleosomal arrays were assembled by salt dialysis as described (17, 36) and analyzed by sedimentation velocity in TEN (10 mm Tris·HCl (pH 7.9), 0.25 mm EDTA, and 2.5 mm NaCl) and EcoRI digestion to determine the extent of nucleosome loading (17, 36).
For the acetylation site usage experiments, a biotinylated 588-bp fragment containing the HTLV-1 promoter linked to a G-less cassette was generated by PCR as described previously (37). DNA was assembled into nucleosomal arrays by mixing histone octamers and promoter DNA together at a molar ratio of ∼2.5 in a reaction containing 2 m NaCl in TE (10 mm Tris·HCl (pH 7.9) and 0.25 mm EDTA) followed by stepwise dilution of NaCl as described (28), except the chromatin was bound to streptavidin-coated magnetic beads in 0.3 m NaCl in TE after assembly.
Histone Acetylation
HTLV-1 nucleosomal arrays (77 nm) were incubated with purified Tax and pCREB (0.9 μm each) before the addition of p300 (160 nm) with and without acetyl-CoA (100 μm) in HAT reaction buffer (20 mm Tris-Cl (pH 7.5), 50 mm NaCl, 5% glycerol, 4 mm β-mercaptoethanol) and incubated at 30 °C for 60–120 min as indicated. HAT buffer was then exchanged for 10 mm Tris-Cl (pH 8.0) by passage through a BioSpin 6 (Bio-Rad) column, and samples were diluted into indicated buffers for subsequent physicochemical studies. For the acetylation density analysis, acetylated chromatin was prepared as above except [14C]acetyl-CoA (100 μm) (or 60 mCi/mmol) was used for labeling, and the reaction time at 30 °C was for 1 h. HAT buffer (including [14C]acetyl-CoA) was exchanged through a Biospin 6 column, chromatin samples were digested with 20 units of EcoRI (New England Biolabs) for 2 h at 30 °C, and digests were resolved on a native 1% agarose, 2% polyacrylamide mixed gel. Reaction products were analyzed by Coomassie staining followed by phosphorimaging. For acetylation site usage experiments, bead-bound nucleosomal arrays (0.25 μm) were incubated with Tax and pCREB (1.5 μm each) for 15 min at 4 °C. Binding reactions (200 μl final volume) were supplemented with p300 (0.25 μm) and acetyl-CoA (100 μm) or 100 μm [1-14C]acetyl (60 mCi/mmol) for 14C-labeling reactions and incubated in either HAT buffer or transcription buffer at 30 °C for 0–120 min, as indicated (60 min for mass spectrometry). Supernatants were removed, and template-bound proteins were resolved by SDS-PAGE and either analyzed by Coomassie staining and phosphorimaging or by matrix-assisted laser desorption/ionization-tandem time of flight (MALDI-TOF/TOF) mass spectrometry.
In Vitro Transcription
In vitro transcription reactions were performed as described (38), except HTLV-1 chromatin or free DNA templates (4 nm) were used in place of the bead-bound chromatin. Briefly, 30-μl reaction mixtures containing Tax (130 nm), pCREB (130 nm), p300 (10 nm), and/or acetyl-CoA (100 μm) as indicated in transcription buffer (1 mm dithiothreitol, 25 mm Tris-HCl (pH 7.9), 50 mm KCl, 6.25 mm MgCl2, 0.5 mm EDTA, 10% glycerol) were supplemented with ∼40 μg (total protein) of nuclear extract for 30 min at 30 °C. RNA synthesis was initiated by the addition of ATP (250 μm), CTP (250 μm), and 0.8 μm [α-32P]UTP (3000 Ci/mmol), and the reactions were incubated for 60 min at 30 °C. A 32P-labeled 622-bp DNA fragment was added to each reaction as a recovery standard before RNA extraction. Labeled HpaII-digested/pBR322 was used a molecular weight marker.
Analytical Ultracentrifugation
Salt-dependent folding was assayed by sedimentation velocity experiments using either a Beckman XL-A or XL-I ultracentrifuge as described (39). Boundaries were analyzed by the method of Demeler and van Holde (40) and van Holde and Weischet (41) using Ultrascan software. Data were plotted as boundary fraction versus sedimentation coefficient (s, corrected to 20 °C and water) to yield the diffusion corrected integral distribution of s20,w, G(s).
Differential Centrifugation
Salt-dependent oligomerization was assayed by differential centrifugation as described (39, 42). Data were plotted as the percentage of the initial absorbance (260 nm) that remained in the supernatant after centrifugation at 16,000 × g for 5 min at room temperature in a microcentrifuge.
Mass Spectrometry
Gel slices containing ∼2 μg of isolated histones were washed in 100 mm ammonium bicarbonate (Ambic), 50% acetonitrile, then in 100% acetonitrile followed by overnight digestion (37 °C) with Arg-C endoproteinase (Calbiochem or Roche) (0.5 μg) in 100 mm Tris-Cl (pH 7.6), 10 mm CaCl2, 5 mm DTT, 0.5 mm EDTA). Peptides were then twice-extracted with 50% acetonitrile, 0.1% trifluoroacetic acid (TFA), dried, and resuspended in 10 μl of 0.1%TFA and purified with a Zip-Tip (C18 Millipore) as described by the manufacturer. One μl of sample was then mixed with 1 μl of α-cyano-4-hydroxycinnamic acid (10 mg/ml in 50% acetonitrile, 0.1% TFA). The mixture was spotted on the MALDI target plate and dried. Samples were analyzed by an Ultraflex MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA) in positive ion, reflector mode using a 25-kV accelerating voltage. External calibration was performed using a peptide calibration mixture on a spot adjacent to the sample, and raw data were processed by FlexAnalysis software (Version 2.4, Bruker Daltonics). Protein identification was confirmed via data base search (SwissProt (Version 57.0_121609, X. laevis taxonomy filter, 3170 sequence entries) using the Mascot search engine (Version 2.3). Acetylation sites were manually validated for each MS/MS spectrum using the Sequence Editor and Biotools software (Version 3.0, Bruker Daltonics).
RESULTS
The HTLV-1 Promoter
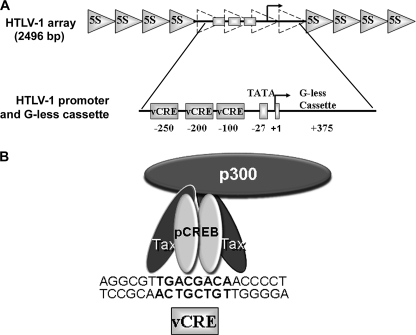
Our studies exploit the advantages of the HTLV-1 promoter to study activator-dependent p300 histone acetylation and its effects on transcription and higher order chromatin structure. The HTLV-1 promoter contains three viral cAMP-response DNA elements (vCREs), the TATA element, and the transcription start site (+1) linked to a G-less cassette (Fig. 1A). Purified Tax and Ser-133-phosphorylated CREB (pCREB) avidly bind a vCRE and recruit p300 to form a ternary complex (Fig. 1B) (30, 37, 43–45) (for review, see Ref. 46), which activates RNAPII-dependent transcription (47). p300 recruitment to the promoter and RNAPII transcriptional activation is associated with acetylation of core histones in vivo (48, 49) and in vitro (38, 44, 48, 50). Thus, the HTLV-1 promoter provides an excellent system to study the mechanistic basis for how activator-dependent p300 acetylation affects gene expression. In this regard it has recently been shown that under in vitro conditions, acetylated nucleosomes are evicted from the HTLV-1 promoter and require the coordinated action of Tax, pCREB, p300, the histone chaperone, Nap1, and acceptor DNA (28, 46). To minimize histone acetylation-dependent nucleosome eviction and focus on higher order chromatin structure, exogenous Nap1 and acceptor DNA have been purposefully omitted from all our experiments.
FIGURE 1.
Chromatin model system used to study activator-dependent p300 acetylation. A, shown is a schematic depicting the HTLV-1 chromatin model system. The HTLV-1 model system is based on a 2496-bp DNA fragment consisting of an HTLV-1 promoter, G-less cassette, and ∼150 bp of pUC13 polylinker DNA (totaling 832 bp) flanked on both sides by 4 repeats of the 208-bp 5 S rDNA sequence. The positions of the HTLV-1 promoter (three vCREs), and TATA box are indicated relative to the transcription start site (+1) linked to the G-less cassette. After reconstitution with core histones, shaded triangles indicate the approximate positions of the 5 S rDNA nucleosomes, whereas approximate locations of promoter nucleosomes (in a saturated array) are indicated by dashed triangles. Upon activator binding and p300 recruitment, the resulting HTLV-1 chromatin was used for both in vitro transcription and structural studies. B, shown is a schematic depicting p300 recruitment to the HTLV-1 promoter by activators (pCREB/Tax). Ser-133-phosphorylated CREB, Tax, and p300 form a ternary complex with a single vCRE consisting of a core CRE element (bold) flanked by 5′ G-rich and 3′ C-rich sequences within the HTLV-1 promoter (43, 46).
Activator-dependent p300 Acetylation Enhances Transcription and Disrupts Higher Order Chromatin Structures
Characterization of the in vitro relationships between activator-dependent HAT activity, higher order chromatin structure, and RNAPII transcription was made possible by constructing a new HTLV-1 promoter-based DNA template suitable for simultaneous studies of RNAPII transcription and chromatin condensation (Fig. 1A). An 832-bp fragment containing the natural HTLV-1 promoter and G-less cassette was fused at each end to four 208-bp tandem-repeated 5 S rDNA nucleosome positioning sequences. This created a 2496-bp DNA template with the potential to bind 12 nucleosomes; 8 positioned nucleosomes flanking the promoter region and 4 nucleosomes within the promoter region (Fig. 1A). In this regard the HTLV-1 template closely resembled the parent 208-12 DNA template, which consists of 12 tandem-repeated 208-bp 5 S rDNA nucleosome-positioning sequences (34). However, the HTLV-1 promoter DNA does not position nucleosomes (51). Therefore, as a control the HTLV-1 and the 208-12 DNAs were reconstituted in parallel with purified recombinant Xenopus histone octamers, and the salt-dependent condensation properties of the resulting nucleosomal arrays were determined (supplemental Fig. S1). We found that Mg2+-dependent folding (as judged by sedimentation velocity; supplemental Fig. S1A) and oligomerization (as judged by differential centrifugation; supplemental Fig. S1B) were essentially the same for the matched 208-12 and HTLV-1 arrays. Thus, insertion of the viral promoter sequences and linked G-less cassette did not alter the fundamental solution state condensation behavior of HTLV-1 nucleosomal arrays compared with the parental 208-12 arrays.
The presence of the embedded HTLV-1 promoter and G-less cassette allowed the transcriptional status of the model chromatin to be readily determined. We, therefore, measured RNAPII-dependent transcriptional output from unacetylated and activator-dependent p300-acetylated chromatin. Fig. 2A outlines our experimental design. Chromatin assembled with an average of 10–11 nucleosomes per DNA was used in the transcription reactions. The optimal ionic conditions were 50 mm KCl and 6.25 mm MgCl2, which have been standard for RNAPII in vitro transcription experiments for over 30 years (31). HTLV-1 DNA and nucleosomal arrays were exposed to combinations of purified activators, p300, and acetyl-CoA and nuclear extract as indicated (Fig. 2A). In the absence of acetyl-CoA (which is a required for the HAT activity of p300), both naked DNA and chromatin templates showed an ∼5-fold enhancement of transcription in response to activators and p300 (Fig. 2B). This likely reflects the acetylation-independent coactivator function of p300. In contrast, the addition of acetyl-CoA in the presence of activators and p300 caused a 14-fold total activation from the HTLV-1 chromatin, whereas the activation from the naked DNA remained at ∼5-fold (Fig. 2B). Thus, activator-dependent p300 acetylation was responsible for roughly two-thirds of the maximal transcriptional activation observed in our experiments. It is important to note that the in vitro transcription data were obtained under salt conditions that should induce substantial chromatin condensation (Refs. 5, 10, and 52; see supplemental Fig. S1). Hence, to properly interpret the biochemical mechanism(s) underlying the acetylation-dependent enhanced transcription observed in Fig. 2B, the higher order structure of unacetylated and p300-acetylated HTLV-1 chromatin must be determined under transcription conditions.
FIGURE 2.
Activator-dependent p300 acetylation enhances transcription and disrupts higher order secondary and tertiary chromatin structures. A, shown is a flowchart describing the experimental design. B, in vitro transcription of HTLV-1 chromatin and free DNA is shown. Transcription was assayed in the presence (+) and absence (−) of purified activators (pCREB/Tax), p300, and acetyl-CoA. Transcript levels were normalized to basal transcription for free DNA or chromatin in the absence of activators and acetyl-CoA (set at 1.0). The positions of RNA transcript (right), recovery standard (32P-labeled DNA) (right), and DNA size markers (left) are indicated. C, shown is oligomerization of HTLV-1 chromatin under transcription conditions. HTLV-1 nucleosomal arrays were incubated with activators (pCREB/Tax) and p300 with (+) and without (−) acetyl-CoA in HAT buffer before buffer exchange and differential centrifugation in transcription buffer. Naked HTLV-1 DNA template (free DNA) in transcription buffer was used as a control. Total A260 was corrected for the contribution from residual acetyl-CoA. Data are expressed as the percentage of initial absorbance (260 nm) that remained in the supernatant after centrifugation (% in supernatant). Error bars indicate the standard deviation of at least three independent oligomerization assays. D, shown is sedimentation velocity analysis of p300-acetylated HTLV-1 arrays in low salt TEN (□) and high salt transcription (■) buffers. Shown are the sedimentation coefficient distributions, G(s). E, HTLV-1 nucleosomal arrays in TEN (▴) were subjected to sedimentation velocity in 1.4 mm MgCl2 and compared with unacetylated HTLV-1 chromatin (□) and chromatin that was acetylated by p300 and activators (■). Shown are the sedimentation coefficient distributions, G(s).
For the physicochemical experiments, nuclear extract was omitted from the samples due to its interference with the measurement of the A260 of the chromatin. HTLV-1 chromatin was bound to activators and p300 in the absence or presence of acetyl-CoA. The unacetylated and activator-dependent p300-acetylated chromatin was then characterized by sedimentation techniques to determine the extent of condensation (Fig. 2A). The conformational state(s) of p300-acetylated chromatin in these experiments, thus, correlate with chromatin architecture before RNAPII preinitiation complex recruitment. The structure of the chromatin in the presence of preinitiation complex or after transcription elongation was not investigated here. Extrapolation of previous studies of salt effects on chromatin condensation (5, 14) suggests that native unacetylated HTLV-1 chromatin may be all or partially oligomeric under transcription conditions. We, therefore, first assayed for tertiary fiber interactions. HTLV-1 DNA, unacetylated chromatin, and activator-dependent p300-acetylated chromatin in transcription buffer were centrifuged for 5 min in a microcentrifuge. Results indicated that only ∼10% of the unacetylated chromatin remained in the supernatant after a brief microcentrifugation compared with ∼75% of the p300-acetylated HTLV-1 chromatin under the same conditions (Fig. 2C). Thus, the data in Fig. 2C indicate that under the ionic conditions of the transcription experiments, unacetylated HTLV-1 chromatin was extensively oligomeric, whereas activator-dependent p300 acetylation effectively disrupted the tertiary interactions that stabilize this higher order state. Interestingly, oligomerization of HTLV-1 chromatin acetylated by p300 in the absence of activators was roughly comparable with unacetylated chromatin (supplemental Fig. S2). Thus, effective disruption of chromatin oligomerization required p300 acetylation mediated by activators.
The folding of unacetylated HTLV-1 chromatin could not be determined under transcription conditions because almost of the sample was oligomeric and rapidly pelleted during ultracentrifugation. However, because only ∼25% of the p300-acetylated HTLV-1 chromatin pelleted after microcentrifugation (Fig. 2C), the extent of folding of the chromatin in the supernatant could be determined by sedimentation velocity in the analytical ultracentrifuge. HTLV-1 chromatin was assembled with activators and p300, exposed to acetyl-CoA, and analyzed in either transcription buffer or low salt TEN buffer (which serves as a reference by causing the fibers to completely unfold). Samples were characterized by sedimentation velocity, and data were analyzed to obtain the integral distribution of diffusion-corrected sedimentation coefficients, G(s) (40) (Fig. 2D). Roughly 25% of the sample in transcription buffer pelleted within the first few scans, consistent with the results of oligomerization assay. For a monomeric 12-mer nucleosomal array, the completely unfolded beads-on-a-string conformation sediments at ∼29 S and an extensively folded species (equivalent in compaction to the “30-nm fiber”) sediments at ∼55 S (5). The results in Fig. 2D show that the G(s) profiles of the monomeric p300-acetylated fibers were shifted to slightly higher sedimentation coefficients in transcription buffer compared with TEN. However, there was no evidence for extensively folded ∼55 S fibers. Thus, activator-dependent p300-acetylated HTLV-1 chromatin was mostly unfolded under transcription conditions. In an important complimentary folding experiment, the G(s) profiles of unacetylated and p300-acetylated HTLV-1 chromatin were determined in 1.4 mm MgCl2. At this salt concentration folding of unacetylated and p300-acetylated chromatin could be directly compared because nearly 100% of both samples remained in the supernatant after microcentrifugation. In 1.4 mm MgCl2, unacetylated chromatin was highly folded (s20,w = 40–55 S), with ∼35% of material beginning to form oligomers (s20,w > 60 S) (Fig. 2E). In contrast, the p300-acetylated chromatin was entirely monomeric and exhibited mostly extended conformations (s20,w = 30–40 S). Thus, our data demonstrate that activator-dependent p300 histone acetylation potently disrupted the stability of secondary and tertiary higher order chromatin structures under two different sets of ionic conditions, including those of the transcription experiments
Acetylation Density Is Heterogeneous
An important question is the extent to which p300 HAT activity targets nucleosomes along the HTLV-1 template. In other words, does p300 preferentially acetylate nucleosomes at the promoter region or equally throughout the entire ∼2500-bp chromatin fiber? To address this question, model nucleosomal arrays in the absence or presence of Tax and pCREB were acetylated with p300 and [14C]acetyl-CoA. The samples were then digested with EcoRI to liberate the 5 S-positioned nucleosomes (flanking the promoter) from the internal region that carries the promoter and G-less cassette (Fig. 3A). The digests were resolved by electrophoretic mobility shift assays (EMSA) using mixed native gels (1% agarose, 2% polyacrylamide). The EMSAs were analyzed by Coomassie staining followed by autoradiography. A representative result of three experiments is shown in Fig. 3B. Digestion of HTLV-1 nucleosomal arrays and chromatin yielded multiple complexes, 5 S mono- and dinucleosomes, the free HTLV-1 promoter region lacking activators and p300 (Fig. 3B, lane 2), and the super-shifted HTLV-1 promoter region presumably bound by Tax/pCREB and p300, which is formed in a concentration-dependent manner (Fig. 3B, lanes 4–6). Without added acetyl-CoA, no acetylation was observed (Fig. 3B, lanes 7 and 8). In the absence of activators, added p300 and [14C]acetyl-CoA produced moderate levels of acetylation throughout all regions of the template without preference for the promoter region or the 5 S nucleosomes (Fig. 3B, lane 9). In the presence of activators the level of 14C-labeled 5 S mononucleosomes remained largely unaffected (compare −/+ activators, lanes 9–12). In contrast, the addition of activators converted the promoter region into a more slowly migrating complex and resulted in an ∼3-fold increase in [14C]acetate incorporation (compare HTLV-1 promoter signal to HTLV-1 promoter-p300-activator complex signal in lanes 9 and 12). These data suggest that activator-dependent p300 acetylation targets mainly nucleosomal histones within the HTLV-1 promoter region. This is consistent with the previous observations of promoter-proximal nucleosome acetylation by p300 (48, 53). Collectively, our data indicate that p300 HAT activity can occur by two distinct mechanisms; that is, non-targeted, ubiquitous HAT activity in the absence of activators and activator-dependent HAT activity targeted primarily to promoter-proximal nucleosomes.
FIGURE 3.
Activator-dependent p300 acetylation preferentially targets the promoter region of HTLV-1 chromatin. A, shown is a schematic depicting physical separation of 208-bp 5 S-positioned mononucleosomes on the extremities from the internal promoter region of HTLV-1 chromatin. E represents an EcoRI digestion site. B, [14C]acetyl-CoA labeling and electrophoretic mobility shift assay of p300-acetylated promoter and distal nucleosomes is shown. HTLV-1 arrays (0.08 μm) were incubated with (+; 0.16 μm) and without (−) p300 and [14C]acetyl-CoA and with (0.46, 0.92, or 1.84 μm) and without (−) activators (pCREB/Tax) for 60 min at 30 °C in HAT buffer. [14C]Acetyl-CoA was removed, and HAT buffer was exchanged for EcoRI digestion buffer using a Bio-Rad spin 6 column before digestion with EcoRI for 120 min at 30 °C. Reaction products were then resolved on a native mixed 1% agarose, 2% polyacrylamide gel and analyzed by Coomassie staining on the left and phosphorimaging on the right. Migration of undigested HTLV-1 chromatin (*), digested 5 S mono- and di-nucleosomes, HTLV-1 promoter, and HTLV-1 promoter-p300-activator complexes are indicated. Relative 14C incorporation was determined by measured density (using ImageQuant 5.1) of HTLV-1 promoter and HTLV-1 promoter-p300-activator complexes normalized to 5 S mononucleosome signal in the same reaction.
Determination of Activator-dependent p300 Acetylation Sites on Chromatin
Last, we determined the identity of p300-acetylated lysines on the promoter nucleosomal histones. Previous reports using mononucleosomes showed that activator-independent p300 acetylation occurs on specific lysines located on all four core histone NTDs (54, 55). To map activator-dependent p300 histone acetylation sites on HTLV-1 promoter nucleosomes, we utilized a 588-bp fragment that carried the natural HTLV-1 promoter without the flanking 5 S-positioning sequences (Fig. 4A). This DNA was biotinylated at the upstream end of the fragment, assembled into nucleosomal arrays with recombinant Xenopus histone octamers, and bound to streptavidin-coated magnetic beads. The immobilized HTLV-1 templates were used for HAT assays and peptide mass spectrometry (Fig. 4B). The histone octamers used are shown in Figs. 4, C and D. We first performed a time-course HAT assay using the immobilized templates p300 and [14C]acetyl-CoA with and without activators. HAT reactions were analyzed by polyacrylamide gel electrophoresis, and acetylation levels were determined by Coomassie staining followed by autoradiography. The relative amount of 14C-labeled histones (H3, H4, H2A, and/or H2B) in the reactions containing activators compared with reactions lacking activators demonstrate activator-dependent stimulation of p300 HAT activity (Figs. 4, E and F), as expected from previous results (44, 50). Kinetic analysis of HAT reactions revealed that steady-state levels of acetylation were reached within 10–20 min, and acetylation levels plateaued for the remaining reaction time tested (60 min) (Fig. 4F), suggesting that p300 acetylation is limited to a finite set of nucleosomal lysines in this system.
FIGURE 4.
Kinetics and specificity of activator-dependent p300 acetylation of HTLV-1 chromatin. A, shown is an immobilized HTLV-1 fiber consisting of a biotinylated 588-bp DNA fragment containing the HTLV-1 promoter and G-less cassette bound to streptavidin-coated beads, assembled into nucleosomal arrays, exposed to activators and p300, and used for the acetylation mapping studies. The positions of the HTLV-1 promoter (three vCRE sites) and TATA box are indicated relative to the transcription start site (+1) linked to the G-less cassette. The dashed triangles indicate this template can assemble a maximum of three nucleosomes on the promoter region. B, shown is a schematic of the experimental design. C, purity of the recombinant X. laevis wild type (WT) core histone octamers (H3, H2A, H2B, H4) and Lys to Gln (KQ) mutant histone octamers (with multiple lysine-to-glutamine substitutions at Lys-5 and -9 for H2A; Lys-5, -12, -15, -20, -24, and -27 for H2B; Lys-4, -9, -14, -18, -23, and -27 for H3; Lys-5, -8, -12, and -16 for H4 (23) that were used in these experiments is shown. Histone samples were resolved by SDS-PAGE (15% polyacrylamide) and Coomassie-stained. D, sites of Lys to Gln (KQ) amino acid substitutions on recombinant X. laevis core histones are shown. The ovals represent the histone fold domains. E and F, kinetics of activator-dependent p300 HAT activity (with [14C]acetyl-CoA) was determined for WT immobilized HTLV-1 chromatin templates in the presence (+) and absence (−) of activators (pCREB/Tax). Reaction products were resolved by SDS-PAGE (15% polyacrylamide) and analyzed by Coomassie staining (E) followed by phosphorimaging (F). G, the kinetics of activator-dependent p300 HAT activity (with [14C]acetyl-CoA) was determined for WT and KQ-immobilized HTLV-1 chromatin templates in the presence (+) of activators (pCREB/Tax). Reaction products were resolved by SDS-PAGE (15% polyacrylamide) and analyzed by Coomassie staining (upper panel) followed sequentially by phosphorimaging (lower panel).
p300 HAT activity on wild type HTLV-1 nucleosomal arrays compared with HTLV-1 nucleosomal arrays carrying lysine to glutamine (Lys to Gln) mutant histones is shown in Fig. 4G (23). We reasoned that any reduction in histone acetylation due to Lys to Gln substitution would provide a preliminary indication of p300 site usage. In other words, because p300 HAT activity is specific for lysine, a glutamine substitution at a p300 target site will block acetylation and result in a reduction of 14C incorporation for that histone. Results indicated an almost complete abolishment of 14C labeling of Lys to Gln H4, H2A, and/or H2B ([14C]H2A and [14C]H2B migrated similarly and could not be distinguished) (Fig. 4G). Reduction of 14C-label suggests that p300 acetylation occurs at lysines corresponding to Lys to Gln-substituted sites on H4, H2A, and/or H2B (Fig. 4D). Notably, Lys to Gln H3 showed partial 14C incorporation, suggesting additional p300 target sites on H3 other than Lys-4, -9, -14, -18, -23, and -27 (locations of the Lys to Gln substitutions).
Direct identification of acetylation sites on histones H3 and H4 was determined by MALDI-TOF/TOF mass spectrometry (inadequate detection by MALDI-TOF of peptides corresponding to the NTDs of H2A/H2B prevented further analysis of these histones). Acetylated nucleosomal histones H4 and H3 from HAT reactions containing reconstituted immobilized chromatin, activators (pCREB/Tax), p300 and acetyl-CoA (Fig. 4A) were resolved by SDS-PAGE, digested with Arg-C endoproteinase, and analyzed by MALDI-TOF/TOF mass spectrometry (Figs. 5, supplemental Figs. S3 and S4). Protein identification was confirmed via data base search using the Mascot search engine, and acetylation sites were confirmed with manual analysis of the raw MS/MS spectra. Fig. 5 represents the peptide mass fingerprints derived from Arg-C digestion of acetylated histones H4 (panel A) and H3 (panel B). From a single MALDI-TOF run of Arg-C-digested H4, we obtained a total of 2 acetylated peptides and a total of 10 peptides corresponding to H4 (∼74% coverage, Fig. 5A, Table 1). The MS spectra indicate that lysines 5, 8, and 12 were acetylated in the peptide 4GKACGGKACGLGKACGGAKR17 at m/z 1396.8, and lysines 5, 8, 12, and 16 were acetylated in the peptide 4GKACGGKACGLGKACGGAKACR17 at m/z 1438.9. The MS/MS spectrum for each peptide confirms the location of acetylation based on the coverage of corresponding fragment ions. Lysine acetylation was additionally confirmed by the presence of the diagnostic immonium ion at m/z 126.1, corresponding to acetylated lysine-NH3 (supplemental Fig. S3). MALDI-TOF analysis of Arg-C digests of H3 revealed a total of 4 acetylated peptides out of a total of 13 peptides corresponding to H3 (∼63% coverage, (Fig. 5B, Table 1). The MS spectra indicate that lysines 9 and 14 were acetylated in the peptide 9KACSTGGKACAPR17 at m/z 985.5; lysines 18 and 23 were acetylated in the peptide 18KACQLATKACAAR26 at m/z 1070.6; lysines 27 and 36 were acetylated in the peptide 27KACSAPATGGVKACKPHR40 at m/z 1517.9; lysines 27, 36, and 37 were acetylated in peptide 27KACSAPATGGVKACKACPHR40 at m/z 1559.9. The MS/MS spectrum for each peptide confirmed the specific location of the acetylation sites (Figs. 5C, supplemental Fig. S4). Unacetylated peptides were detected for H4 (containing lysines 31, 79, and 91) and H3 (containing lysines 56, 64, 79, and 122) (Table 1). As an important control, MALDI-TOF was carried out as described above, except the reactions were performed in the absence of the activators (pCREB/Tax). This approach allowed us to assess whether p300 recruitment by activators conferred lysine targeting specificity. Results indicate that a similar pattern of acetylation was obtained with and without activators (supplemental Table S1). Taken together with the kinetic data in Fig. 4, E and F, these results indicate that the activators stimulate the efficiency of acetylation by p300 rather than determining the specific sites of modification. In summary, p300 acetylation of immobilized HTLV-1 chromatin occurred on histone sites previously identified for mononucleosomes (55) and for free histone H3 (56). In addition, several novel p300 acetylation sites on nucleosomal H3 were identified in our studies, including H3K9, H3K27, H3K36, and H3K37. The p300 histone acetylation sites identified in the HTLV-1 system in vitro are consistent with naturally occurring acetylation sites identified in vivo (18, 56, 57) with the exception of H3K37. This residue, although highly conserved in histone H3, has not yet been previously reported as a physiological acetylation site.
FIGURE 5.
Identification of lysines acetylated by p300. A, MALDI-TOF MS spectrum of histone H4 peptides from activator-dependent p300-acetylated HTLV-1 chromatin templates is shown. Starred peaks (*) indicate matches to predicted H4 peptides; acetylated peptides are labeled as indicated. B, shown is the MALDI-TOF MS spectrum of histone H3 peptides from activator-dependent p300-acetylated HTLV-1 chromatin templates. Starred peaks (*) indicate matches to predicted H3 peptides; acetylated peptides are labeled as indicated. C, the MS/MS spectrum of peptide 27KACSAPATGGVKACKACPHR40 (amino acids 27–40 of X. laevis H3 sequence) confirm acetylation on lysines 27, 36, and 37. Confirmation was based on the Mascot ion score of 104 and coverage of theoretical y and b ions including the sequence region of acetylated Lys-27, -36, and -37. The peak at m/z 126 corresponds to an acetylated lysine immonium ion, NH3, which further confirms the presence of acetylated lysine. Peaks corresponding to internal fragmentation are unlabeled shown in light gray.
TABLE 1.
Summary of p300-acetylated histones by MALDI-TOF
Identification of H3 and H4 was confirmed by a database search (Mascot ion score (identity threshold) 339 (48) and 192 (48), respectively). Ion scores (identity threshold) for individual MS/MS spectra are listed with the corresponding peptides. ND, not determined.
| Histone | Measured (MH+) | Calculated mass | Peptide sequence (Mascot ion score (identity threshold)) |
|---|---|---|---|
| H3 | |||
| Acetylated | |||
| Lys-9, Lys-14 | 985.547 | 984.5352 | 9KACSTGGKACAPR17 (50 (18)) |
| Lys-18, Lys-23 | 1070.6397 | 1069.6243 | 18KACQLATKACAAR26 (56 (18)) |
| Lys-27, Lys-36 | 1517.8715 | 1516.8474 | 27KACSAPATGGVKACKPHR40 (118 (18)) |
| Lys-27, Lys-36, Lys-37 | 1559.8842 | 1558.8579 | 27KACSAPATGGVKACKACPHR40 (104 (18)) |
| Unmodified | |||
| Lys-56 | 1250.7232 | 1249.703 | 54YQKSTELLIR63 |
| Lys-64 | 788.4926 | 787.4704 | 64KLPFQR69 |
| Lys-79 | 1335.6887 | 1334.683 | 73EIAQDFKTDLR83 |
| Lys-122 | 1384.7994 | 1383.7908 | 117VTIMPKDIQLAR128 |
| Not determined | |||
| Lys-4 | ND | 704.4 | 3TKQTAR8 |
| Lys-115 | ND | 3598.8 | 84FQSSAVMALQEASEAYLVGLFEDTNLCGIHAKR116 |
| H4 | |||
| Acetylated | |||
| Lys-5, Lys-8, Lys-12 | 1396.8068 | 1395.7946 | 4GKACGGKACGLGKACGGAKR17 (107 (18)) |
| Lys-5, Lys-8, Lys-12, Lys-16 | 1438.8546 | 1437.8052 | 4GKACGGKACGLGKACGGAKACR17 (117 (18)) |
| Unmodified | |||
| Lys-31 | 1325.78 | 1324.7463 | 24DNIQGITKPAIR35 |
| Lys-79,Lys-91 | 1594.921 | 1594.8912 | 79KTVTAMDVVYALKR92 |
| Not determined | |||
| Lys-20 | ND | 808.5 | 18HRKVLR23 |
| Lys-59 | ND | 1385.84 | 56GVLKVFLENVIR67 |
DISCUSSION
Unacetylated HTLV-1 chromatin is extensively condensed under the ionic conditions required for optimal RNAPII transcriptional activity even in the absence of linker histones (Fig. 2, C and D). This raises the possibility that activator-dependent p300 histone acetylation functions to activate transcription in part by disrupting the intrinsic condensation of H1-depleted chromatin fibers. Our results indicate that activator-dependent p300 histone acetylation was effective at destabilizing two types of nucleosome-nucleosome interactions under transcription conditions; that is, tertiary interactions that lead to interfiber oligomerization (Fig. 2C, supplemental Fig. S2) and secondary interactions that lead to intrafiber folding (Figs. 2, D and E). Concomitantly, a substantial increase in the level of transcription was observed from the decondensed p300-acetylated HTLV-1 chromatin relative to the same chromatin exposed to activators and p300 but no acetyl-CoA (Fig. 2B). Thus, our structure/function studies demonstrate that p300 acetylation-induced destabilization of higher order chromatin structures is strongly correlated with activation of transcription from the HTLV-1 model chromatin. Although the repressive nature of condensed chromatin is widely perceived (6–9), the present work is novel in that it uses a highly defined chromatin model system to define the biochemical relationships between activator-dependent recruitment of p300, targeted p300 histone tail acetylation, the stability of higher order chromatin architecture, and RNAPII transcription levels. Repression of transcription in vitro due to chromatin generally has focused on nucleosomes acting as barriers to RNAPII at the primary structure level. Our results suggest a more complex activation pathway that reflects the hierarchical nature of the chromatin fiber itself (Fig. 6). Subsequent to linker histone dissociation, activator-dependent p300 histone acetylation causes nearly complete decondensation of promoter chromatin. The promoter nucleosomes can then more easily undergo primary structure changes such as additional histone post-translational modification (18, 19), remodeling (58, 59), or eviction (46, 60). Thus, we see the process of transcriptional activation as involving mechanistic steps at the tertiary, secondary, and primary structure levels, each of which makes its own contribution to the overall level of activation achieved. From a biological perspective, such layered control allows for very fine-tuned regulation of RNAPII transcription.
FIGURE 6.
Model for functional effects of activator-dependent p300 histone acetylation. Based on our data, promoter-proximal p300 histone acetylation is proposed to perform two functions. One antagonizes histone methylation-dependent transduction pathways via acetylation of H3K9, H3K27, and H3K36, whereas the other destabilizes higher order structure via acetylation-dependent disruption of both secondary and tertiary chromatin structures leading to subsequent primary structure changes of the chromatin fiber and fine-tuned regulation of gene expression.
Our results also have several important implications relating to higher order chromatin structure. Previous studies have shown that all four core histone NTDs contribute additively to chromatin oligomerization (17). Thus, the accumulation of acetylation events on each of the core histone NTDs is likely to be responsible for the potent disruption of oligomerization seen in our experiments. Extensive charge neutralization by acetylation presumably plays a key mechanistic role given the importance of charge density in stabilizing oligomeric chromatin structures (61). An important conclusion is that activator-dependent acetylation of promoter-proximal nucleosomes by p300 is sufficient to disrupt oligomerization of ∼2500-bp chromatin molecules. Thus, localized hyperacetylation of promoter nucleosomes is able to disrupt higher order structure over at least several kilobases of surrounding chromatin, perhaps much more. A likely explanation for this observation is that because oligomerization is cooperative (5, 14), hyperacetylation of only a subset of nucleosomes could have a disproportionate affect on disrupting tertiary nucleosome-nucleosome interactions over a long range. In terms of folding, we note that acetylation of H4K16 leads to disruption of intrafiber nucleosome-nucleosome interactions (21, 24). H4 Lys-16 is one of the residues acetylated by p300 in our system (Table 1, Fig. 5A), and this acetylation event presumably is responsible for much of the unfolding of the HTLV-1 chromatin that occurs under transcription conditions. Whether acetylation at one or more other sites also contributes to fiber unfolding remains to be determined.
The mapping studies have identified four novel p300 acetylation sites on the H3 NTDs of promoter-proximal nucleosomes: H3K9, H3K27, H3K36, and H3K37. The other mapped sites are listed in Table 1. Interestingly, the novel sites of lysine acetylation (with exception to H3K37) are also sites of methylation (for review, see Ref. 18)). It has recently been proposed that HATs can indirectly regulate histone methylation at loci where HATs and methyltransferases compete for a common lysine (56, 57, 62–64). Acetylation of H3K27 by CBP (a highly related p300 paralog) has been proposed to obstruct H3K27 methylation and antagonize Polycomb silencing at target genes during early embryogenesis in Drosophila (56). Acetylation of H3K36 (by yeast Gcn5) localizes to gene promoters (57), whereas methylation of H3K36 localizes to open-reading frames and correlates with transcription elongation (65, 66). Therefore, the modification status of H3K36 with respect to acetylation or methylation may coordinate transcription initiation or elongation, respectively (57, 67–70). Our identification of H3K9, H3K27, and H3K36 acetylation suggests a role for human p300 to counteract the methylation-dependent silencing pathways of HP1 (through H3K9 acetylation) or Polycomb (through H3K27 acetylation) and regulates transcription initiation (through H3K36 acetylation) at p300-targeted loci.
In closing, we have recently defined the “chromogenome” as the structural and functional status of the genome at any given moment within a eukaryotic cell (60). Our studies suggest that two of the important functions of p300 histone acetylation are to decondense promoter chromatin for the purposes of enhancing gene expression via primary chromatin structure alterations and to act as a temporal or spatial dynamic “switch” to antagonize methylation-dependent transduction pathways (Fig. 6). In this regard, p300 acetylation is poised to have profound effects on the fluidity of the chromogenome acting through these and other (60, 71) pathways.
Supplementary Material
Acknowledgments
We are grateful to Drs. Karolin Luger, Paul Laybourn, and Steven McBryant for helpful discussions and Dinaida Lopez and Jeanne Mick for purified Tax and pCREB.
This work was supported, in whole or in part, by National Institutes of Health Grants GM45916 (to J. C. H.) and GM088409 and CA055035 (to J. K. N.). This work was also supported by an American Heart Association postdoctoral fellowship (to H. J. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- RNAPII
- RNA polymerase II
- CREB
- cAMP-response element-binding protein
- NTD
- amino-terminal domain
- HAT
- histone acetyltransferase
- HTLV-1
- human T-cell leukemia virus type-1
- vCRE
- viral cAMP-response DNA element
- pCREB
- phosphorylated CREB.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2.Tse C., Sera T., Wolffe A. P., Hansen J. C. (1998) Mol. Cell. Biol. 18, 4629–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen J. C., Wolffe A. P. (1992) Biochemistry 31, 7977–7988 [DOI] [PubMed] [Google Scholar]
- 4.Li G., Margueron R., Hu G., Stokes D., Wang Y. H., Reinberg D. (2010) Mol. Cell. 38, 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen J. C. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 361–392 [DOI] [PubMed] [Google Scholar]
- 6.Choi J. K., Howe L. J. (2009) Biochem. Cell. Biol. 87, 139–150 [DOI] [PubMed] [Google Scholar]
- 7.Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 8.Felsenfeld G. (1992) Nature 355, 219–224 [DOI] [PubMed] [Google Scholar]
- 9.Woodcock C. L., Dimitrov S. (2001) Curr. Opin. Genet. Dev. 11, 130–135 [DOI] [PubMed] [Google Scholar]
- 10.Horowitz-Scherer R. A., Woodcock C. L. (2006) Chromosoma 115, 1–14 [DOI] [PubMed] [Google Scholar]
- 11.Zheng C., Hayes J. J. (2003) Biopolymers 68, 539–546 [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Ramirez M., Dong F., Ausio J. (1992) J. Biol. Chem. 267, 19587–19595 [PubMed] [Google Scholar]
- 13.Fletcher T. M., Hansen J. C. (1995) J. Biol. Chem. 270, 25359–25362 [DOI] [PubMed] [Google Scholar]
- 14.Schwarz P. M., Felthauser A., Fletcher T. M., Hansen J. C. (1996) Biochemistry 35, 4009–4015 [DOI] [PubMed] [Google Scholar]
- 15.Tse C., Hansen J. C. (1997) Biochemistry 36, 11381–11388 [DOI] [PubMed] [Google Scholar]
- 16.Dorigo B., Schalch T., Bystricky K., Richmond T. J. (2003) J. Mol. Biol. 327, 85–96 [DOI] [PubMed] [Google Scholar]
- 17.Gordon F., Luger K., Hansen J. C. (2005) J. Biol. Chem. 280, 33701–33706 [DOI] [PubMed] [Google Scholar]
- 18.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 19.Roth S. Y., Denu J. M., Allis C. D. (2001) Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Ramirez M., Rocchini C., Ausio J. (1995) J. Biol. Chem. 270, 17923–17928 [DOI] [PubMed] [Google Scholar]
- 21.Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S., Resch M., Lilyestrom W., Clark N., Hansen J. C., Peterson C., Luger K. (2010) Biochim. Biophys. Acta 1799, 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X., Hayes J. J. (2008) Mol. Cell. Biol. 28, 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson P. J., An W., Routh A., Martino F., Chapman L., Roeder R. G., Rhodes D. (2008) J. Mol. Biol. 381, 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumann H., Hancock S. M., Buning R., Routh A., Chapman L., Somers J., Owen-Hughes T., van Noort J., Rhodes D., Chin J. W. (2009) Mol. Cell 36, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus W. L., Kadonaga J. T. (1998) Genes Dev. 12, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez D. I., Mick J. E., Nyborg J. K. (2007) Protein Expr. Purif. 55, 406–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma N., Nyborg J. K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7959–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao L. J., Giam C. Z. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11445–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giebler H. A., Loring J. E., van Orden K., Colgin M. A., Garrus J. E., Escudero K. W., Brauweiler A., Nyborg J. K. (1997) Mol. Cell. Biol. 17, 5156–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dynan W. S. (1987) Genet. Eng. 9, 75–87 [Google Scholar]
- 32.Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Enzymol. 304, 3–19 [DOI] [PubMed] [Google Scholar]
- 33.Luger K., Rechsteiner T. J., Richmond T. J. (1999) Methods Mol. Biol. 119, 1–16 [DOI] [PubMed] [Google Scholar]
- 34.Simpson R. T., Thoma F., Brubaker J. M. (1985) Cell 42, 799–808 [DOI] [PubMed] [Google Scholar]
- 35.Hansen J. C., Ausio J., Stanik V. H., van Holde K. E. (1989) Biochemistry 28, 9129–9136 [DOI] [PubMed] [Google Scholar]
- 36.Hansen J. C., Lohr D. (1993) J. Biol. Chem. 268, 5840–5848 [PubMed] [Google Scholar]
- 37.Geiger T. R., Sharma N., Kim Y. M., Nyborg J. K. (2008) Mol. Cell. Biol. 28, 1383–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konesky K. L., Nyborg J. K., Laybourn P. J. (2006) J. Virol. 80, 10542–10553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwarz P. M., Hansen J. C. (1994) J. Biol. Chem. 269, 16284–16289 [PubMed] [Google Scholar]
- 40.Demeler B., van Holde K. E. (2004) Anal. Biochem. 335, 279–288 [DOI] [PubMed] [Google Scholar]
- 41.van Holde K. E., Weischet W. O. (1978) Biopolymers 17, 1387–1403 [Google Scholar]
- 42.Carruthers L. M., Bednar J., Woodcock C. L., Hansen J. C. (1998) Biochemistry 37, 14776–14787 [DOI] [PubMed] [Google Scholar]
- 43.Harrod R., Tang Y., Nicot C., Lu H. S., Vassilev A., Nakatani Y., Giam C. Z. (1998) Mol. Cell. Biol. 18, 5052–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georges S. A., Giebler H. A., Cole P. A., Luger K., Laybourn P. J., Nyborg J. K. (2003) Mol. Cell. Biol. 23, 3392–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwok R. P., Laurance M. E., Lundblad J. R., Goldman P. S., Shih H., Connor L. M., Marriott S. J., Goodman R. H. (1996) Nature 380, 642–646 [DOI] [PubMed] [Google Scholar]
- 46.Nyborg J. K., Egan D., Sharma N. (2010) Biochim. Biophys. Acta 1799, 266–274 [DOI] [PubMed] [Google Scholar]
- 47.Lenzmeier B. A., Nyborg J. K. (1997) J. Virol. 71, 2577–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu H., Pise-Masison C. A., Fletcher T. M., Schiltz R. L., Nagaich A. K., Radonovich M., Hager G., Cole P. A., Brady J. N. (2002) Mol. Cell. Biol. 22, 4450–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemasson I., Polakowski N. J., Laybourn P. J., Nyborg J. K. (2002) J. Biol. Chem. 277, 49459–49465 [DOI] [PubMed] [Google Scholar]
- 50.Georges S. A., Kraus W. L., Luger K., Nyborg J. K., Laybourn P. J. (2002) Mol. Cell. Biol. 22, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lemasson I., Polakowski N. J., Laybourn P. J., Nyborg J. K. (2006) J. Biol. Chem. 281, 13075–13082 [DOI] [PubMed] [Google Scholar]
- 52.Widom J. (1998) Annu. Rev. Biophys. Biomol. Struct. 27, 285–327 [DOI] [PubMed] [Google Scholar]
- 53.Kundu T. K., Palhan V. B., Wang Z., An W., Cole P. A., Roeder R. G. (2000) Mol. Cell 6, 551–561 [DOI] [PubMed] [Google Scholar]
- 54.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 55.Schiltz R. L., Mizzen C. A., Vassilev A., Cook R. G., Allis C. D., Nakatani Y. (1999) J. Biol. Chem. 274, 1189–1192 [DOI] [PubMed] [Google Scholar]
- 56.Tie F., Banerjee R., Stratton C. A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M. O., Scacheri P. C., Harte P. J. (2009) Development 136, 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris S. A., Rao B., Garcia B. A., Hake S. B., Diaz R. L., Shabanowitz J., Hunt D. F., Allis C. D., Lieb J. D., Strahl B. D. (2007) J. Biol. Chem. 282, 7632–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clapier C. R., Cairns B. R. (2009) Annu. Rev. Biochem. 78, 273–304 [DOI] [PubMed] [Google Scholar]
- 59.Cairns B. R. (2009) Nature 461, 193–198 [DOI] [PubMed] [Google Scholar]
- 60.Hansen J. C., Nyborg J. K., Luger K., Stargell L. A. (2010) J. Cell. Physiol. 224, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McBryant S. J., Klonoski J., Sorensen T. C., Norskog S. S., Williams S., Resch M. G., Toombs J. A., 3rd, Hobdey S. E., Hansen J. C. (2009) J. Biol. Chem. 284, 16716–16722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Y., Li T., Vu T. H., Ulaner G. A., Hu J. F., Hoffman A. R. (2003) Endocrinology 144, 5658–5670 [DOI] [PubMed] [Google Scholar]
- 63.Garcia B. A., Hake S. B., Diaz R. L., Kauer M., Morris S. A., Recht J., Shabanowitz J., Mishra N., Strahl B. D., Allis C. D., Hunt D. F. (2007) J. Biol. Chem. 282, 7641–7655 [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y., Reinberg D. (2001) Genes Dev. 15, 2343–2360 [DOI] [PubMed] [Google Scholar]
- 65.Shilatifard A. (2004) Biochim. Biophys. Acta 1677, 79–86 [DOI] [PubMed] [Google Scholar]
- 66.Sims R. J., 3rd, Belotserkovskaya R., Reinberg D. (2004) Genes Dev. 18, 2437–2468 [DOI] [PubMed] [Google Scholar]
- 67.Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 68.Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 69.Joshi A. A., Struhl K. (2005) Mol. Cell. 20, 971–978 [DOI] [PubMed] [Google Scholar]
- 70.Rao B., Shibata Y., Strahl B. D., Lieb J. D. (2005) Mol. Cell. Biol. 25, 9447–9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L., Tang Y., Cole P. A., Marmorstein R. (2008) Curr. Opin. Struct. Biol. 18, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.