Abstract
Osterix, a zinc finger transcription factor, is specifically expressed in osteoblasts and osteocytes of all developing bones. Because no bone formation occurs in Osx-null mice, Osterix is thought to be an essential regulator of osteoblast differentiation. We report that, in several mesenchymal and osteoblastic cell types, BMP-2 induces an increase in expression of the two isoforms of Osterix arising from two alternative promoters. We identified a consensus Sp1 sequence (GGGCGG) as Osterix binding regions in the fibromodulin and the bone sialoprotein promoters in vitro and in vivo. Furthermore, we show that Osterix is a novel substrate for p38 MAPK in vitro and in vivo and that Ser-73 and Ser-77 are the regulatory sites phosphorylated by p38. Our data also demonstrate that Osterix is able to increase recruitment of p300 and Brg1 to the promoters of its target genes fibromodulin and bone sialoprotein in vivo and that it directly associates with these cofactors through protein-protein interactions. Phosphorylation of Osterix at Ser-73/77 increased its ability to recruit p300 and SWI/SNF to either fibromodulin or bone sialoprotein promoters. We therefore propose that Osterix binds to Sp1 sequences on target gene promoters and that its phosphorylation by p38 enhances recruitment of coactivators to form transcriptionally active complexes.
Keywords: Bone, Bone Morphogenetic Protein (BMP), Cell Differentiation, Gene Transcription, p38 MAPK
Introduction
Bone is a highly dynamic tissue that is constantly remodeled throughout life. Bone remodeling activity is dependent on a delicate balance between osteoclast resorption and osteoblast new bone formation. Deregulation of these two activities unleashes pathological states such as osteoporosis and osteosclerosis. Both endochondral and intramembranous ossification depends on osteoblasts that are derived from pluripotent mesenchymal stem cells that, in response to various cellular and environmental signals, commit to the osteoblast phenotype. Among them, BMPs5 are essential for commitment and differentiation to the osteoblast lineage; they promote osteoblast differentiation in vitro and in vivo, bone regeneration, and ectopic bone formation in vivo (1–3). The BMP signal is transduced through the binding to its heteromeric cell membrane receptors (4, 5). BMP binding to receptors results in the activation of the Smad family of transcription factors, which directly regulate target gene expression (6).
BMP target genes include a growing number of osteoblast-determining transcription factors. For instance, in vivo genetic evidence as well as osteogenic induction of bone marrow mesenchymal stem cells in vitro has identified several types of transcription factors such as Id1, homeodomain proteins such as Dlx3 and Dlx5, ATF4, Runx2, and Osterix (Osx) (7–9). Runx2 and Osx have been widely accepted as master osteogenic factors because neither Runx2- nor Osx-null mice form mature osteoblasts (10, 11). Osx contains a proline- and serine-rich transactivation domain located in the N-terminal part of the protein and three zinc fingers with homology to the Sp1/Kruppel transcription factor family. Osx expression is specifically restricted to osteoblasts and osteocytes of all developing bones. In Osx-null mice, no bone formation takes place, although Runx2 is expressed, suggesting that Osx acts downstream of Runx2 during bone development (11). Moreover, it has been suggested that Runx2 may function from the commitment step to the point where osteochondroprogenitors appear, whereas Osx may have a role in the segregation of osteoblasts from osteochondroprogenitors (8). In addition, it has been shown that genetic polymorphisms in the Osx gene locus are associated with low bone mineral density (12, 13). This essential role of Osx relies on its ability to regulate the expression of a number of osteoblast markers such as osteopontin, osteocalcin, Dkk1, and collagen type I. In addition, transcriptional regulators such as NFATc or NO66 have been shown to interact with Osx and regulate its transcriptional responses (14, 15).
Runx2 and Osx transcription is stimulated by BMP treatment in vitro (11, 16, 17). Pretreatment with cycloheximide blocks Osx induction, suggesting that Osx is not a direct target of the BMP signaling cascade but requires the expression of newly synthesized intermediates (18). Interestingly, although expression of Osx in vivo requires Runx2, BMP-2 is still able to stimulate alkaline phosphatase activity and Osx expression in Runx2-deficient cells (18). Recent data indicate that BMP-2 activates expression of Osx through Runx2-dependent as well as -independent mechanisms involving Dlx5 and Msx2 (17, 19). These studies also showed that BMP-2 induction of Osx required the transcriptional activation of Dlx5 by p38 MAPK-mediated phosphorylation (17, 20).
Activation of p38 MAPK signaling by BMP-2, insulin-like growth factor I, or mechanical stress has been shown to be relevant in the induction of their osteogenic effects (20–25). Thus, although these data suggest that osteoblast-specific transcription factors and p38 are involved in BMP-induced Osx expression, little is known about the transcriptional mechanisms by which Osx promotes osteoblast-specific gene expression. In this study, we identified a consensus Sp1 sequence (GGGCGG) as the Osx binding regions in the fibromodulin (Fmod) and the bone sialoprotein (Ibsp) promoters in vitro and in vivo. We also show that Osx is a novel substrate for p38 MAPK in vitro and in vivo and that Ser-73 and Ser-77 are the regulatory sites phosphorylated by p38. The transactivation potential of Osx was increased by p38 phosphorylation at least in part through increased interaction of Osx with the transcriptional cofactors p300 and Brg1. We propose a regulatory network in which BMP-2 activates expression of Osx and enhances its transcriptional activation by p38-mediated phosphorylation.
EXPERIMENTAL PROCEDURES
Plasmids, Reagents, and Antibodies
cDNA fragments encoding both long and short Osterix-pCDNA3 expression vectors were kindly provided by Dr. K. Watanabe and Dr. B. de Crombrugghe, respectively. Osx cDNA was used as a parental plasmid to generate the phosphorylation mutants using PCR approaches or subcloned into pGEX vector to generate bacterial GST-Osx fusion constructs. Activated MKK6 expression vector (MKK6EE) and recombinant p38 were provided by Dr. P. Muñoz-Cánoves. Fmod promoter-reporter construct from −2032 to +100 (2-kb pFmod-Luc) was kindly provided by Dr. F. Cimino. The pFmod300-Luc promoter-reporter was generated by PCR. The Ibsp promoter-reporter (pIbsp-Luc) construction encoding a conserved 300-bp enhancer of human IBSP gene, located at −12400 from the transcriptional start site, was cloned in the minimal promoter of c-fos gene. BMP-2 was generously provided by Wyeth and was used at a final concentration of 2 nm. SB203580 (Calbiochem) was used at a final concentration of 10 μm. Antibodies against Osx (Abcam), phosphoserine (Abcam), MKK6 (Abcam), Brg1 (Upstate), p300 (Santa Cruz Biotechnology), and α-tubulin (Sigma) were used at 1:1000.
Cell Culture and Transfection
C2C12, Saos-2, and mouse embryonic fibroblast (MEF) cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen). MC3T3-E1 cell line was maintained in the α-modified minimum Eagle's medium (LabClinics) supplemented with 10% fetal bovine serum (FBS). Human primary osteoblasts were obtained from Lonza and cultured in DMEM/F-12 supplemented with 15% FBS, 10 μg/ml ascorbic acid, and 1.25 μg/ml amphotericin B. Bone marrow mesenchymal stem cells were maintained in DMEM supplemented with 15% FBS. All media were supplemented with 0.2 mm glutamine, 0.1 mm pyruvate, and 100 units/ml penicillin-streptomycin. C2C12, MC3T3-E1, MEF, and Saos-2 cell differentiation was induced with medium lacking serum and BMP-2 at a final concentration of 2 nm. For human primary osteoblasts, differentiation was induced with medium lacking serum, 50 μg/ml ascorbic acid, and 2 nm BMP-2, and for bone marrow mesenchymal stem cells, differentiation was induced with 10% FBS, 50 μm ascorbic acid, 5 mm β-glycerophosphate, and 2 nm BMP-2. C2C12, MC3T3-E1, and MEF cells were transiently transfected using Lipofectamine LTX (Invitrogen).
Western Blot Analysis
Cells were washed twice in cold PBS and lysed with 50 mm Tris, pH 7.5, 150 mm NaCl, 0.2% Igepal, 10% glycerol supplemented with protease and phosphatase inhibitors. Protein extracts were resolved on 10% SDS-polyacrylamide gels, transferred to nitrocellulose membrane (Millipore), and subjected to Western blot using the above indicated antibodies at 1:1000 dilution. Immunocomplexes were visualized with a horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG antibody (1:10,000) followed by incubation with ECL Western blot reagent (GE Healthcare).
Immunofluorescence Assay
24 h after either transfection with Osx or treatment with BMP-2, C2C12 cells were fixed as described previously (17). Cells were stained with rabbit anti-Osx (Abcam) at 1:500 followed by goat anti-rabbit IgG conjugated with Alexa Fluor 488 at 1:400. Labeling was detected using a Leica TCS SL inverted laser scanning confocal microscope.
Quantitative PCR (RT-qPCR) Analysis
Total RNA was isolated from C2C12, MC3T3-E1, Saos-2, and bone marrow mesenchymal stem cells using the Ultraspec RNA Isolation System (Biotecx). 5 μg of total RNA were reverse transcribed using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative PCRs were carried out using the ABI Prism 7900 HT Fast Real Time PCR System and a TaqMan 5′-nuclease probe method (Applied Biosystems). All transcripts were normalized to GAPDH, and transfection efficiency was assessed by GFP expression. Designed human TaqMan assays (Applied Biosystems) were used to quantify gene expression of Osx (Mm00504574_m1), Fmod (Mm00491215_m1), Ibsp (Mm00492555_m1), and GAPDH (Mm99999915_g1).
Reporter Assays
Luciferase activities were quantified using the Luciferase Assay System (Promega). Luciferase values were normalized using β-galactosidase activity measured with the Luminescent β-Galactosidase Detection kit II (Clontech).
Biotinylated Oligonucleotide Precipitation Assays
C2C12 cells were lysed as described previously (26). Biotinylated oligonucleotides were incubated overnight with cell extracts in the presence of 1 μg of poly(dI-dC) (GE Healthcare) and collected with streptavidin-agarose beads (GE Healthcare). Bound Osx was detected by immunoblotting.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assays were performed using the Chromatin Immunoprecipitation Assay kit (Upstate) according to the manufacturer's instructions but repeating all the washes twice. Antibodies used were anti-Osx (Abcam), anti-p300 (N-15, Santa Cruz Biotechnology), Brg1 (Upstate), anti-RNA polymerase II, and anti-IgG (Upstate). Primer sequences used for the PCR were as follows: for human primary osteoblasts and Saos-2 cells, HsIbspF, 5′-cttctttctcatgtggccaacactcg; HsIbspR, 5′-tggcatcaggagatgtcctctctt; HsFmodF, 5′-ggacccagggctgccaat; and HsFmodR, 5′-cgtccctctgtctggcctccttgggtt; and for C2C12 cells, MmIbspF, 5′-ttcaggctgccaatgctccagggaa; MmFmodR, 5′-tgaccacgtccttctgtctggtct; MmIbspF, 5′-ccagttttcaaacatccaaatccatagg; and MmIbspR, 5′-ttggcactgggagatgtcctccctt.
Immunoprecipitation
Cells were lysed as above. The supernatant fraction was incubated with 1 μg of anti-FLAG-M2 antibody (Sigma) overnight followed by incubation with protein G-Sepharose beads for 1 h. Bound proteins were washed four times in lysis buffer. Bound proteins were detected by immunoblotting using the antibodies described above.
In Vitro Phosphorylation by p38 MAPK
GST-p38 MAPK was activated by MBP-MKK6-DD (5:1 ratio) in 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 2 mm DTT, 200 μm ATP. Activated p38 (200 ng for each condition) was incubated with recombinant Osx proteins in the same buffer supplemented with 1 μm ATP and 3 pmol of [γ-32P]ATP at 30 °C for 20 min.
RESULTS
Expression of Two Isoforms of Osterix Is Stimulated by BMP-2 in Mesenchymal and Osteogenic Cells
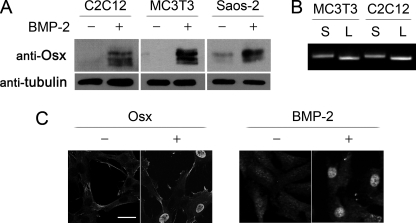
Previous studies showed that Osx mRNA levels increase after BMP-2 addition in mesenchymal cell types, including primary osteoblasts, mesenchymal stem cells, mesenchymal C2C12 cells, and osteoblast MC3T3-E1 cells (11, 17, 20, 27). Analysis of protein expression by immunoblotting indicated that Osx was not significantly expressed in undifferentiated C2C12 cells or MC3T3-E1 cells and showed some expression in the osteoblastic cell line Saos-2 (Fig. 1A). However, Osx protein levels were strongly increased in all cell types at 24 h after addition of 2 nm BMP-2. In all cell types, Osx appeared as multiple bands of different mobility (Fig. 1A). Previous work showed that both the human and mouse Osx mRNAs are expressed as two isoforms arising from two alternative promoters (28–30). RT-PCR using unique 5′-primers and a common 3′-primer yielded different species that corresponded to the long (MASSLL) and the short isoforms (MLTAAC) of Osx. These results indicated that both isoforms are induced at significant levels by BMP-2 in these cell lines (Fig. 1B). We also analyzed the subcellular localization of Osx after BMP-2 induction. Immunofluorescence analysis of C2C12 cells, either transfected with an Osx expression vector or 24 h after BMP-2 addition, showed that Osx is a constitutively nuclear transcription factor (Fig. 1C).
FIGURE 1.
Expression of two isoforms of Osterix by BMP-2 in mesenchymal and osteogenic cells. A and B, cells were treated with BMP-2 in medium without serum for 24 h. A, Osx and tubulin were detected by immunoblotting. B, both short (S) and long (L) Osx mRNA were detected by PCR from cells treated with 2 nm BMP-2 for 24 h using specific forward primers (short, 5′-cacccattgccagtaatcttcaagcca; long, 5′-ctcgaggatggcgtcctctctgcttg) and a common reverse primer (5′-ggactgcctgcaggagagagga). C, subcellular localization of Osx in C2C12 cells was analyzed by immunofluorescence with anti-Osx antibody in cells either transiently transfected with short Osx construct or incubated in medium without serum and treated with BMP-2 for 24 h. Scale bar, 10 μm.
Osx-responsive Regions in Promoters of Fibromodulin and Bone Sialoprotein
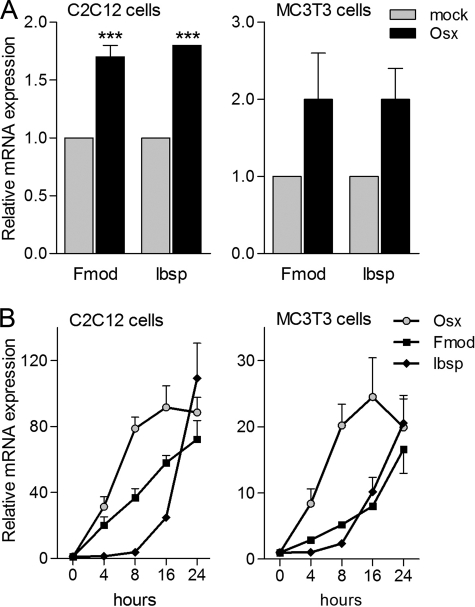
To analyze the mechanisms of transcriptional activation by Osx, we used microarrays to search for Osx target genes by comparing RNA expression of C2C12 cells transfected with a mock construct or transfected with an Osx expression vector. Among the differentially expressed genes, we focused on Fmod, whose expression increased 2-fold in response to Osx. We also focused on Ibsp as a late osteoblast marker; its expression has been shown to be abolished in Osx-deficient cells (11, 31). RT-qPCR confirmed that Osx overexpression increased the expression levels of both Fmod and Ibsp in C2C12. Moreover, we analyzed the expression levels of Fmod and Ibsp in MC3T3-E1 cells. Similarly to C2C12 cells, Osx induced expression of both genes (Fig. 2A).
FIGURE 2.
Fmod and Ibsp expression is induced by Osx. A, cells were transiently transfected with mock or Osx construct overnight and incubated in medium without serum for 24 h. Fmod and Ibsp mRNAs were measured by RT-qPCR, normalized to GAPDH, and expressed as relative expression ± S.E. of four independent experiments. Expression of the different constructs was analyzed by immunoblotting (data not shown). B, confluent cells were incubated in medium without serum overnight and treated with BMP-2 for the indicated times. Osx, Fmod, and Ibsp mRNAs were measured by RT-qPCR, normalized to GAPDH, and plotted as relative expression to time 0 ± S.E. of three independent experiments (***, p < 0.001 compared with mock condition, two-way ANOVA followed by Bonferroni's test).
We also analyzed the temporal expression pattern of both Fmod and Ibsp in response to BMP-2 compared with the induction of Osx. As shown in Fig. 2B, induction of Fmod and Ibsp gene expression in either C2C12 or MC3T3-E1 cells strongly correlated with that of Osx. All genes required at least 8 h to increase their mRNA levels significantly after BMP-2 addition.
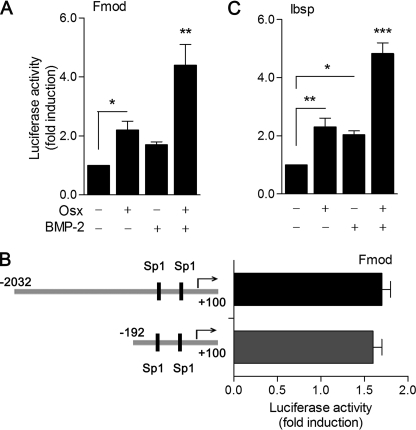
Because these findings suggested that either Osx or BMP-2 is able to increase Fmod expression, we decided to determine whether Osx and BMP-2 stimulation was transcriptional by analysis of Fmod promoter activity. Using a 2-kb upstream regulatory sequence of the murine Fmod gene (2-kb pFmod-Luc) in C2C12 cells, we observed a 2.3-fold increase in activation in cells overexpressing Osx and a 1.7-fold increase in activation after treatment with 2 nm BMP-2 (Fig. 3A). More interestingly, BMP-2 promoted an additive effect to Osx overexpression in C2C12 cells. We then used reporter assays to analyze deletions of the 2-kb promoter. The deletion of the sequence from −2032 to −192 provoked only a slight decreases in Osx or BMP-2 reporter induction (Fig. 3B). Homology analysis using the ECR Browser showed that the −297/+100 region on the Fmod promoter is highly conserved among mammalian species and detected two putative Sp1 transcription factor binding sites (supplemental Fig. 1 and Ref. 32). Similar analysis on the Ibsp promoter region detected a highly conserved 300-bp enhancer located at −12400 bp from the Ibsp transcription start site that contains a Runx2 binding site, a palindromic homeobox binding site (TAATTA), and an Sp1 binding site (supplemental Fig. 1). To test whether this region alone has the ability to render a minimal promoter responsive to Osx and/or BMP-2, we further assayed the responses of a reporter construct containing this region upstream of a heterologous c-fos minimal promoter. Whereas the minimal c-fos promoter showed no response at all to Osx or BMP-2, the Ibsp enhancer region was increased 2- and 3-fold by overexpression of Osx and addition of 2 nm BMP-2, respectively (Fig. 3C). Similarly to Fmod reporter assays, BMP-2 promoted an additive effect to Osx overexpression on this enhancer in C2C12 cells. The reporter assays of Fmod and Ibsp indicate that, in addition to the induction of Osx expression, BMP-2 activates alternative pathways that result in further Fmod and Ibsp transcriptional activation by Osx.
FIGURE 3.
Fmod and Ibsp transcriptional activation by Osx and BMP-2. A, cells were co-transfected with 2-kb pFmod-Luc reporter vector and either mock or Osx construct overnight and treated with or without BMP-2 in medium without serum for 24 h. B, cells were transfected with 2-kb pFmod-Luc or pFmod300-Luc reporter vectors overnight and treated with BMP-2 in medium without serum for 24 h. C, cells were co-transfected with pIbsp-Luc reporter vector and either mock or Osx construct overnight and treated with or without BMP-2 in medium without serum for 24 h. A, B, and C, luciferase activity was measured and normalized against β-galactosidase activity. Relative luciferase activities were expressed as mean ± S.E. for triplicates from five independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001; one-way ANOVA followed by Bonferroni's multiple comparison test).
Osx Interacts with Sp1 Consensus Regions in Regulatory Sites of Fmod and Ibsp
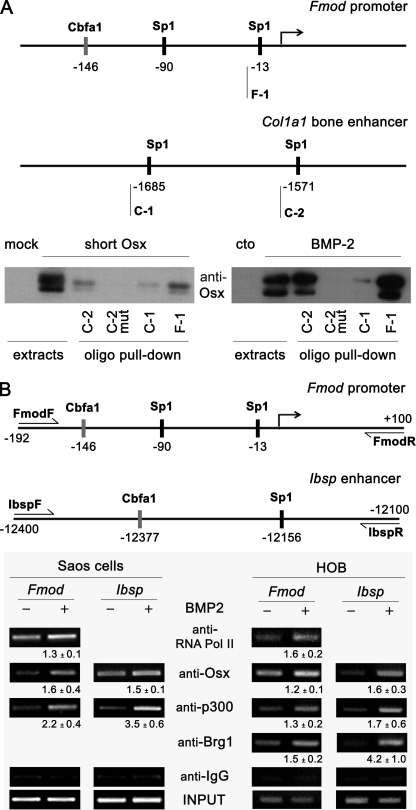
It has been shown that Osx interacts with the promoters of osteogenic genes such as Col1a1, Dkk1, or Bglap in vivo (14, 15, 31). In all cases, a GC-rich canonical Sp1 sequence has been shown to be the specific Osx binding site. Therefore, we analyzed whether Osx bound to the Sp1 sequence of Fmod (GGGGCGG) and studied the Osx binding preferences by comparing them with Sp1 sequences located in the bone enhancer of the Col1a1 promoter (AGGGCGG or GGGGAGG, respectively) that have been shown to be bound by Osx (Fig. 4A) (33). We performed oligonucleotide pulldown assays from either C2C12 cells overexpressing Osx or cells after induction of endogenous Osx by BMP-2 addition. Osx required a functional Sp1 site because no binding at all to the DNA was observed when extracts were incubated with the Sp1-mutated oligonucleotide (Fig. 4A). The results also suggest that the sequence GGGCGG is preferred for Osx binding at least in vitro because it bound better than the sequence GGGAGG.
FIGURE 4.
Osx interaction with Sp1 regions in regulatory sites of Fmod and Ibsp genes. A, upper panel, Sp1 regions analyzed by oligo pulldown. Biotinylated (biot) oligonucleotide sequences used were as follows: F-1, 5′-taggaatttggggcgggaccctggt-biot; C-1, 5′-ggaacagaaggggagggagc-biot; C-2, 5′-cagggagtcccgcctcctccaaac-biot; and C-2 mut, 5′-cagggagtattgtttcctccaaac-biot. Lower panel, the indicated double-stranded biotinylated oligonucleotides were incubated with equal aliquots of extracts from C2C12 cells either transfected with an Osx construct using mock as control or treated without (cto) or with BMP-2 in medium without serum for 24 h. Precipitated complexes were analyzed by immunoblotting using Osx antibody. B, upper panel, promoter regions analyzed by chromatin immunoprecipitation. Lower panels, Saos-2 cells or human primary osteoblasts (HOB) were treated with BMP-2 for 24 h in medium without serum. Cells were fixed with formaldehyde, and chromatin immunoprecipitation analysis was performed by incubating DNA-protein complexes using the indicated antibodies and IgG as a control. Quantification of the results of two independent experiments is shown below each panel as mean ± S.E. Pol, polymerase.
To investigate binding of Osterix in both Fmod promoter and Ibsp enhancer in vivo, we performed chromatin immunoprecipitation assays in Saos-2 cells or human primary osteoblasts treated with 2 nm BMP-2 for 24 h. As shown in Fig. 4B, binding of Osx to the proximal Fmod promoter or Ibsp enhancer in vivo was increased after treatment with BMP-2. The increased binding of Osx to these promoter regions correlated with increased recruitment of RNA polymerase II to the start site of Fmod gene. It has been reported previously that p300 histone acetyltransferase is required for the expression of several osteogenic genes in response to BMP-2 (17, 34–36). Results of the chromatin immunoprecipitation assay indicated that p300 was recruited onto both Fmod proximal promoter and Ibsp enhancer sequences in response to BMP-2. Similarly, binding of the SWI/SNF subunit Brg1 was also increased in human primary osteoblasts after BMP-2 addition (Fig. 4B). Altogether, these findings suggest that Osx and transcriptional coactivators are recruited onto these promoter regions and involved in the transcriptional program of induction of both Fmod and Ibsp expression by BMP-2.
Osx Is Phosphorylated by p38
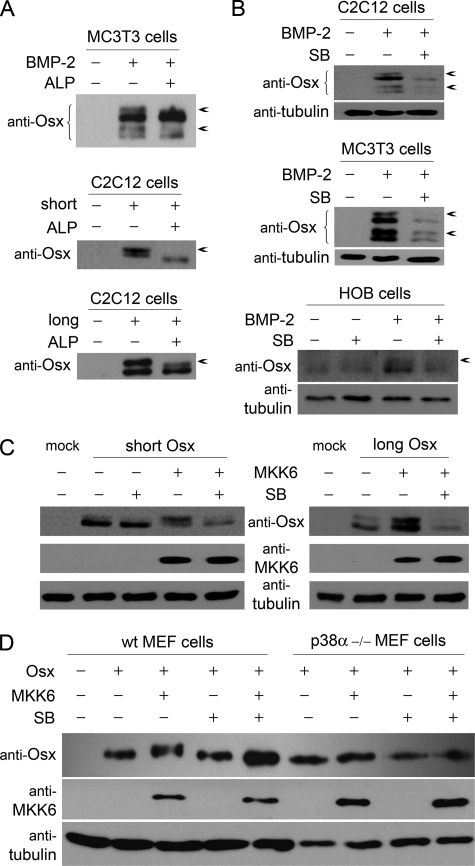
The fact that addition of BMP-2 was able to further increase the responsiveness to Osx of both the proximal Fmod and the Ibsp enhancer regions led us to investigate the additional transcriptional effects of BMP-2 in the expression of Fmod and Ibsp target genes. Interestingly, SDS-PAGE analysis of cell extracts showed that both long and short forms of Osx appeared as two species. To determine whether the respective retarded bands were phosphorylated forms, cell extracts from MC3T3-E1 cells after BMP-2 induction were treated with alkaline phosphatase. Phosphatase treatment abrogated slow migrating forms, indicating that those were due to phosphorylation events (Fig. 5A). Similar results were obtained by exogenously overexpressing either long or short forms of Osx, indicating that both forms were substrates of cellular kinase(s) (Fig. 5A).
FIGURE 5.
Osx phosphorylation by p38. A, MC3T3-E1 cells were treated with or without BMP-2 in medium without serum. C2C12 cells were transfected with empty vector, short Osx, or long Osx constructs. After 24 h, cell extracts were incubated with alkaline phosphatase (ALP) for 1 h. Osx forms were detected by immunoblotting. B, cells were treated with or without BMP-2 in medium without serum for 24 h and pretreated with SB203580 (SB) for 30 min where indicated. Osx and tubulin were detected by immunoblotting. C, C2C12 cells were co-transfected with constitutively active MKK6 (MKK6EE) and short or long Osx constructs. After 24 h, cells were treated with SB203580 for 3 h. Osx, MKK6, and tubulin were detected by immunoblotting. D, wild type (wt) and p38α−/− MEF cells were co-transfected with MKK6 construct and the short Osx construct. After 24 h, cells were treated with SB203580 for 3 h. Osx, MKK6, and tubulin were detected by immunoblotting. Arrows indicate phosphorylated forms.
Activation of p38 MAPK by BMP-2 has been shown to play a relevant role in the osteogenic effects of this cytokine (21–23). More specifically, the p38 pathway has been directly implicated in the BMP-induced sequential recruitment of the transcriptional coactivators p300 and the SWI/SNF complex onto osteogenic genes (34, 36). We therefore analyzed whether Osx could be a target of the p38 pathway. In agreement with previous reports (17), treatment of C2C12 cells, MC3T3-E1 cells, or human primary osteoblasts with the specific p38 inhibitor SB203580 inhibited the BMP-2-induced expression of Osx (Fig. 5B). In addition to the decrease in total expression, SB203580 reduced the phosphorylated Osx bands. To further confirm the ability of p38 to phosphorylate Osx, we used a constitutively active form of MKK6 (MKK6EE), which phosphorylates and activates p38, and analyzed cell extracts from C2C12 cells expressing either short or long forms of Osx. The amount of the upper band corresponding to phosphorylated Osx was increased in the presence of MKK6EE compared with extracts from cells expressing Osx alone (Fig. 5C). Moreover, treatment of cells with SB203580 significantly decreased the amount of retarded bands. To further examine the requirement of distinct p38 isoforms for Osx phosphorylation, we analyzed MKK6EE-induced Osx phosphorylation in p38α−/− MEFs. MKK6EE induced phosphorylation of Osx in wild type but not in p38α−/− MEFs (Fig. 5D). Taken together, these results demonstrate that p38 MAPK is able to phosphorylate Osx.
Osx Is Phosphorylated by p38 at Ser-73 and Ser-77
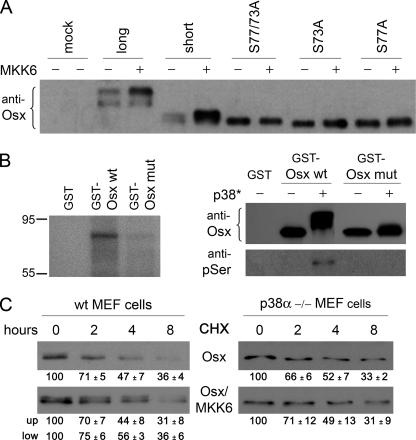
Sequence analysis of Osx revealed four distinct serines corresponding to consensus sites for MAPKs conserved in Osx orthologs, and one of these was only present in the long form of Osx. We generated site-specific mutants corresponding to the two sites present in both long and short forms of Osx where Ser was changed to Ala. Expression of MKK6EE induced the appearance of retarded phosphorylated bands when co-transfected with either the short or long forms of Osx (Fig. 6A). However, both of the single point mutants S73A and S77A as well as the double mutant S73A/S77A lost p38-dependent phosphorylation and were detected exclusively as a single fast migrating band irrespective of expression of MKK6EE (Fig. 6A). The fact that neither single mutant was phosphorylated by p38 may indicate that both of them are phosphorylated, or because both sites are located very close to each other in a region of Osx with a predicted coiled structure, Ser to Ala mutation in one site might also hinder phosphorylation at the other site.
FIGURE 6.
Phosphorylation of Osx at Ser-73 and Ser-77 by p38. A, C2C12 cells were co-transfected with constitutively active MKK6 (MKK6EE) and the different Osx constructs as indicated using an empty vector as control. B, 5 μg of GST-Osx wild type or the S73A/S77A mutant (mut) were incubated with activated p38 MAPK (200 ng for each condition) in the presence of γ-[32P]ATP and visualized by SDS-PAGE and autoradiography (left panel) or visualized by immunoblotting against Osx or phosphoserine (pSer) (right panel). Addition of MKK6-activated recombinant p38 is indicated by an asterisk (p38*). C, wild type (wt) and p38α−/− MEF cells were co-transfected with MKK6EE and Osx constructs as indicated. After 24 h, cells were treated with cycloheximide for the indicated times. Osx was detected by immunoblotting. Quantification of the results of three independent experiments is shown below each panel as mean ± S.E.
We then examined whether Osx was a direct substrate of p38 by in vitro phosphorylation assays using recombinant proteins. Recombinant wild type Osx was phosphorylated in vitro by activated recombinant p38, whereas the double mutant S73A/S77A showed impaired phosphorylation by p38 (Fig. 6B). Analysis of the efficiency of this phosphorylation indicates that addition of recombinant p38 induced almost a complete phosphorylation of wild type Osx, whereas the double mutant S73A/S77A only showed minor effects on either phosphorylation on serines or in its electrophoretic mobility (Fig. 6B).
To examine the functional consequences of Osx phosphorylation, we first analyzed whether phosphorylation changed the Osx protein turnover. Wild type and p38α-defective MEFs were transfected with Osx and MKK6EE and treated with the protein synthesis inhibitor cycloheximide for different periods of time. Quantification of the kinetics of Osx disappearance displayed no significant differences between the phosphorylated and unphosphorylated forms of Osx. Similarly, the profile of degradation of Osx was similar irrespective of the presence or absence of MKK6EE or co-transfection of wild type Osx with MKK6EE in p38α−/− MEFs (Fig. 6C).
p38 Phosphorylation Enhances Osx Transcriptional Activity and Increases Its Interaction with Coactivators
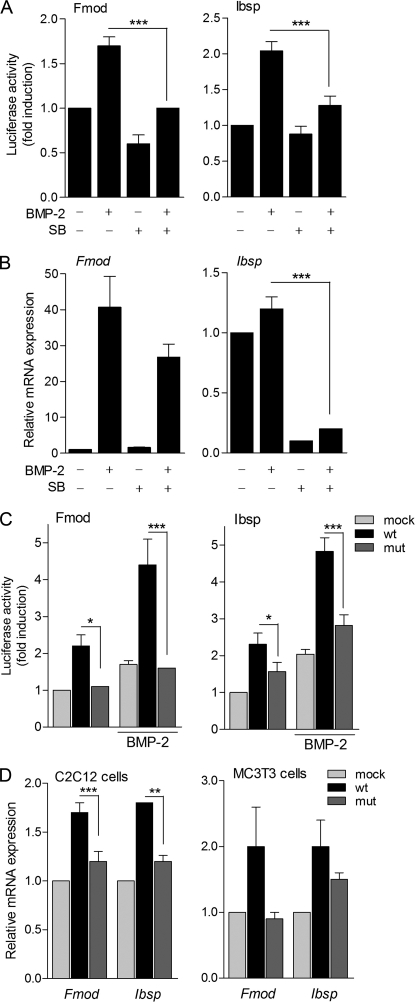
To investigate whether phosphorylation by p38 has a functional effect on Osx transcriptional activity, C2C12 cells were transfected with either Fmod or Ibsp promoter constructs, and cells were treated with BMP-2 in the presence or absence of the p38 inhibitor SB203580. Increases in reporter activity induced by BMP-2 were significantly inhibited by addition of SB203580 in both cases, suggesting a functional role for p38-induced Osx phosphorylation (Fig. 7A). We further analyzed the endogenous expression of these target genes by RT-qPCR upon differentiation of bone marrow mesenchymal stem cell cultures. Induction of Fmod and Ibsp gene expression by BMP-2 was reduced in the presence of the p38 inhibitor SB203580 (Fig. 7B). In view of these results, we assessed the relevance of Ser-73 and Ser-77 phosphorylation to the transcriptional activity of Osx. Whereas overexpression of Osx wild type led to a 2–3-fold induction of the activity of Fmod and Ibsp reporter constructs, induction of promoter activity by the S73A/S77A mutant construct was significantly lower (Fig. 7C). Similarly, addition of BMP-2 further increased the transcriptional activity of wild type Osx on both reporters, whereas the mutant form showed impaired responses to the cytokine addition (Fig. 7C). Likewise, we further analyzed the endogenous expression of these target genes by RT-qPCR. As expected, induction of Fmod and Ibsp endogenous genes by S73A/S77A Osx was reduced in comparison with induction by wild type Osx in both C2C12 and MC3T3-E1 cells (Fig. 7D).
FIGURE 7.
Induction of Osx transcriptional activity by p38 phosphorylation. A, C2C12 cells were transfected with either 2-kb pFmod-Luc or pIbsp-Luc reporter vectors overnight, treated with or without BMP-2 in medium without serum for 24 h, and pretreated with SB203580 (SB) for 30 min when indicated. Luciferase activity was measured and normalized against β-galactosidase activity. Relative luciferase activities were expressed as mean ± S.E. for triplicates from four independent experiments (***, p < 0.001; one-way ANOVA followed by Bonferroni's multiple comparison test). B, bone marrow mesenchymal stem cells were cultured for 4 days and treated at a confluent state at days 0 and 3 with BMP-2 (2 nm) and/or SB203580 during the last 24 h in osteogenic differentiating medium. Fmod and Ibsp mRNAs were measured by RT-qPCR, normalized to GAPDH, and expressed as relative expression ± S.E. of three independent experiments (***, p < 0.001; two-way ANOVA followed by Bonferroni's multiple comparison test). C, C2C12 cells were co-transfected with 2-kb pFmod-Luc or pIbsp-Luc vectors and mock, wild type (wt), or double mutant (mut) Osx constructs and treated with or without BMP-2 in medium without serum for 24 h. Luciferase activity was measured and expressed as above (*, p < 0.05; ***, p < 0.001; two-way ANOVA followed by Bonferroni's multiple comparison test). D, cells were transfected with mock, wild type (wt), or double mutant (mut) Osx constructs for 24 h. Fmod and Ibsp mRNAs were measured by RT-qPCR, normalized to GAPDH, and expressed as relative expression ± S.E. of three independent experiments (**, p < 0.01; ***, p < 0.001; two-way ANOVA followed by Bonferroni's multiple comparison test).
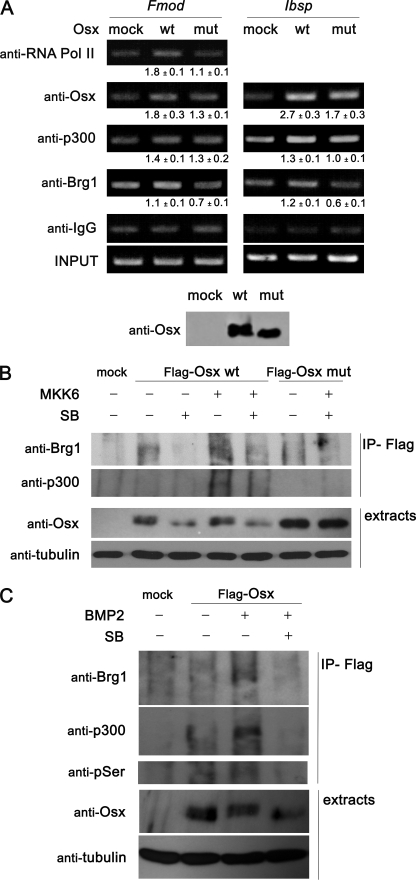
To investigate the relevance of the phosphorylation of Osx to its binding and transcriptional activation of the Fmod and Ibsp genes in vivo, we performed chromatin immunoprecipitation assays in C2C12 cells transfected with wild type or double mutant Osx. As shown in Fig. 8A, Osx bound to Fmod promoter and Ibsp enhancer. Increased Osx binding was sufficient to recruit p300 and the RNA polymerase II to the Fmod promoter and p300 to the Ibsp enhancer. More importantly, although transfection efficiency and expression levels of either wild type or mutant Osx were similar, binding of mutant Osx and its ability to recruit RNA polymerase II, p300, or Brg1 to the Fmod or Ibsp regulatory regions were lower, suggesting that phosphorylation of Osx enhances assembly of transcriptionally active complexes in Osx target genes.
FIGURE 8.
p300 and Brg1 recruitment induced by Osx phosphorylation. A, C2C12 cells were transfected with mock, wild type (wt), or double mutant (mut) Osx constructs for 24 h. Cells were fixed with formaldehyde, and chromatin immunoprecipitation analysis was performed by incubating DNA-protein complexes using the indicated antibodies. IgG was used as a control. Quantification of the results of three independent experiments is shown below each panel as mean ± S.E. Expression of the constructs was analyzed by immunoblotting. B, C2C12 cells were transfected with mock, FLAG-tagged wild type (wt), or double mutant (mut) Osx constructs and constitutively active MKK6 (MKK6EE) as indicated. After 24 h, cells were treated with SB203580 (SB) for 3 h as indicated. Expression of the constructs and anti-FLAG-co-immunoprecipitated proteins (IP-Flag) were analyzed by immunoblotting C, C2C12 cells were transfected with mock or FLAG-tagged wild type Osx. After 24 h, cells were treated with BMP and/or SB203580 (SB) for 3 h as indicated. Expression of the constructs and anti-FLAG-co-immunoprecipitated proteins were analyzed by immunoblotting. Pol, polymerase.
Because overexpression of Osx is sufficient to induce recruitment of p300 and Brg1 to the responsive promoters, we analyzed whether these factors might interact with each other and studied the role of Osx phosphorylation in this interaction. We analyzed the ability of Osx to interact with endogenous p300 and Brg1. Thus, we expressed FLAG-tagged wild type and the S73A/S77A forms of Osx in C2C12 cells either alone or together with MKK6EE. After immunoprecipitation of Osx complexes, we found p300 and Brg1 to be associated to Osx by Western blot analysis (Fig. 8B). In addition, overexpression of MKK6EE, which led to phosphorylated Osx, increased the amounts of interacting p300 and Brg1, whereas addition of p38 inhibitor SB203580 decreased the binding of both transcriptional coactivators. These interactions were mostly abrogated when the Osx mutated form S73A/S77A was analyzed. Moreover, addition of BMP-2 to cells expressing FLAG-tagged wild type Osx also increased interaction of Osx with p300 and Brg1. These BMP-induced effects were partially abolished by addition of SB203580 (Fig. 8C). Taken together, our results strongly suggest that Osx binds to Sp1 sequences on target gene promoters and that phosphorylation of Osx by p38 may enhance recruitment of coactivators.
DISCUSSION
Osterix and Runx2 are widely considered to be master osteogenic factors because neither Runx2- nor Osx-null mice form mature osteoblasts (11). Here, we show that exposure of distinct mesenchymal and osteoblastic cells to BMP-2 induces the appearance of both the long and short Osx forms, which also become phosphorylated by p38 MAPK. Our results also emphasize the essential role of p38 and chromatin remodeling in the control of Osx expression and function. Osx, by virtue of its binding to Sp1 sequences, activates transcription of several target genes in osteoblasts and mesenchymal cell lines such as Fmod and Ibsp. More importantly, we identified that phosphorylation of Osx by p38 at Ser-73/77 promotes assembly of stable, transcriptionally active complexes containing Osx, p300, and Brg1.
The essential role of Osx in osteogenesis relies on its ability to regulate the expression of a number of osteoblast markers such as Osteopontin, Dkk1, and collagen type I. We identified Fmod and Ibsp as novel osteoblastic genes regulated by Osx and characterized Sp1 binding regions in their promoters that are able to mediate BMP-2 and Osx activation. Our data constitute the first evidence of the mechanisms of osteogenic regulation of Fmod expression by Osx and BMP-2. In the case of Ibsp, functional cooperativity was previously shown between the osteogenic factors Dlx5 and Runx2 in the proximal promoter of Ibsp where Runx2 and Dlx5 bind to adjacent sites and physically interact to drive promoter activation (37). There is some evidence that Osx binding activity also plays an essential role in mediating these effects on Ibsp expression. Previous studies using 2.5- and 2.7-kb Ibsp promoters failed to show absolute tissue-specific expression in transgenic mice (38) and required at least a 9-kb promoter to show osteoblast-specific expression (39). We identified an Ibsp enhancer region with an 80% sequence homology across human, mouse, and dog genes (supplemental Fig. 1). This enhancer contains fully conserved binding sites for Osx, Runx2, and homeodomain transcription factors, suggesting that it could be critical for osteoblast-specific expression. Induction of Ibsp and Fmod by BMP-2 correlates with that of Osx and requires at least 16–24 h to reach maximal induction, whereas the induction of Runx2 and Dlx5 by BMP-2 requires much less time (17, 40). Similarly, the temporal expression pattern in other models of differentiation of osteoblast precursors indicates that the expression of Osx always precedes that of Fmod, Ibsp, and other markers of terminal differentiation such as osteocalcin or alkaline phosphatase. Finally, blocking the Osx function either in mice or cell cultures leads to loss of expression of both Ibsp and Fmod (11, 15, 31, 41, 42). Altogether, our findings are consistent with the idea that BMP-dependent induction of expression of late osteogenic markers depends on Osx and its cooperative action with other osteogenic master genes.
Osx belong to the Sp1/Kruppel family of transcription factors that presents a marked preference for binding to GC-rich motifs (43). However, because only a few mammalian Osx-responsive promoters have been characterized, there is no evidence for preferences to bind to specific motifs. Our results suggest that Osx displays the ability to bind either the GGGCGG or the GGGAGG sequences with, at least in vitro, a higher DNA binding affinity to GGGCGG compared with GGGAGG. Interestingly, our sequence analysis of the Osx-responsive Sp1 boxes in both Fmod and Ibsp promoters is consistent with other Osx-responsive genes characterized like Col1a1, Col11a2, and Dkk1. In all cases, the responsive motifs in vitro and in vivo contain one of these two types of sequences (14, 31, 41).
p38 MAPK is known to play an important role in several steps of the osteoblast lineage progression and is necessary but not sufficient for the BMP-induced acquisition of the osteoblast phenotype (21, 44). Several evidences also indicate that p38 activity modulates Osx expression induced by BMP in calvarial as well as bone marrow-derived mesenchymal stem cells (17, 20, 24, 27). Similarly, other osteogenic stimuli such as mechanical stress, drugs, or cytokines exert their osteogenic effects through activation of p38 followed by an increase in Osx expression and function (17, 45–47). Our results provide a mechanism by which BMP-2-activated p38 could enhance transcriptional activity of Osx through direct phosphorylation on Ser-73 and Ser-77. First, activated p38 phosphorylates Osx in vitro and in vivo. Second, phosphorylation of Osx is induced by the expression of constitutively active MKK6, whereas it is blocked by the pharmacological inhibition of p38. Finally, p38-mediated phosphorylation of Osx appears to increase its interaction with the transcriptional coactivators p300 and Brg1. Indeed, the p38 inhibitor SB203580 inhibited BMP-2- and Osx-driven Fmod or Ibsp promoter activation, and the mutant Osx S73A/S77A displayed reduced transcriptional activity and BMP-2 responsiveness. The existence of regulatory phosphorylations affecting the activity of Sp1 is well known (32), and some reports indicate that this also could be the case for Osx because it has been shown to interact and become dephosphorylated by calcineurin (48). Regulatory effects of p38 phosphorylation were also observed previously in Dlx5 transcriptional activity on the promoter of Osx itself (17). We therefore propose a positive osteogenic regulatory network where p38 activation regulates the expression of Osx through Dlx5 phosphorylation and further transcriptional activation of Osx through its phosphorylation on Ser-73/77.
Ser-73 and Ser-77 are located in a solvent-exposed coiled region of the proline- and serine-rich transactivation domain of Osx. This region has been shown to interact with cofactors such as transcription factor IIB, a Jumonji family histone demethylase (NO66), and Brg1 through its zinc finger domain (15, 49). Functionally equivalent regions of Sp1 have been shown to be phosphorylated by distinct kinases, resulting in activity changes based on their ability to recruit transcriptional cofactors (for a review, see Ref. 32). Thus, it is likely that phosphorylation of Osx in the transactivation domain might modulate its ability to recruit transcriptional cofactors.
Acetylation and/or methylation at specific sites in histones in chromatin is commonly associated with transcriptionally active genes, and p300, histone deacetylases (HDACs), and the NO66 methylase have been shown to bind and regulate the acetylation or methylation of the core histones H3 and H4 in several osteogenic gene promoters. For instance, p300 is recruited to the osteocalcin promoter in response to osteogenic signals (35, 50), and JunB recruits p300 to activate dentin matrix protein 1 (Dmp1) (34), whereas HDAC4 inhibits Runx2 activity, and HDAC4-null mice exhibit strong skeletal defects (51). BMPs have been shown to switch recruitment from HDAC1 to p300 in osteocalcin, Osx, or Fgfr3 gene promoters and to promote hypomethylation of CpG sites in Dlx5 and Osx promoter regions (35–36, 52). Interestingly, BMP-2 has been shown to increase binding of Osx to Ibsp or osteocalcin promoters with a concomitant decrease of NO66 binding and activity (15). Similarly, BMPs increase the expression and function of the ATP-dependent chromatin-remodeling complex SWI/SNF, which has been shown to be absolutely essential for osteogenesis, especially the complexes containing Brg1 as a catalytic core (53–55). Our study demonstrates that Osx is able to increase recruitment of p300 and Brg1 to the promoters of its target genes Fmod and Ibsp in vivo and that Osx directly associates to these cofactors through protein-protein interactions that are further enhanced by BMP signaling. Furthermore, phosphorylation of Osx at Ser-73/77 increases its ability to recruit p300 and SWI/SNF to Fmod or Ibsp promoter as has been shown for p38-dependent recruitment of p300 by JunB on Dmp1 promoter or by Sp1 to Fgfr3 promoter (34, 36). Recruitment of both cofactors is likely to be cooperative because, in addition to its histone acetyltransferase activity, the p300 protein may act as a bridging factor to connect sequence-specific transcription factors to other cofactors and to the basal transcriptional machinery, and acetylated chromatin is the preferred substrate for SWI/SNF recruitment (56). In conclusion, Osx is able to up-regulate Fmod and Ibsp expression levels through its ability to recruit p300 and Brg1. Furthermore, phosphorylation of Osx by p38 is able to enhance the recruitment of these transcriptional cofactors. Formation of this complex may initiate chromatin remodeling responsible for the initiation and maintenance of high transcription rates of their target genes.
Supplementary Material
Acknowledgments
We thank Wyeth for providing BMP-2 and Drs. F. Cimino, B. de Crombrugghe, P. Muñoz-Cánoves, and K. Watanabe for reagents. We also thank E. Adanero, E. Castaño, and B. Torrejón for technical assistance.
This work was supported in part by Ministerio de Educación y Ciencia (MEC) Grant BFU2008-02010 and Instituto de Salud Carlos III Redes Temáticas de Investigación Cooperativa en Salud Grant RD06/0020.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- BMP
- bone morphogenetic protein
- Osx
- Osterix
- Fmod
- fibromodulin
- Ibsp
- bone sialoprotein
- MEF
- mouse embryonic fibroblast
- qPCR
- quantitative PCR
- HDAC
- histone deacetylase
- ANOVA
- analysis of variance.
REFERENCES
- 1.Chen D., Zhao M., Mundy G. R. (2004) Growth Factors 22, 233–241 [DOI] [PubMed] [Google Scholar]
- 2.Tsumaki N., Yoshikawa H. (2005) Cytokine Growth Factor Rev. 16, 279–285 [DOI] [PubMed] [Google Scholar]
- 3.Wan M., Cao X. (2005) Biochem. Biophys. Res. Commun. 328, 651–657 [DOI] [PubMed] [Google Scholar]
- 4.Moustakas A., Heldin C. H. (2009) Development 136, 3699–3714 [DOI] [PubMed] [Google Scholar]
- 5.Miyazono K., Maeda S., Imamura T. (2005) Cytokine Growth Factor Rev. 16, 251–263 [DOI] [PubMed] [Google Scholar]
- 6.Massagué J., Seoane J., Wotton D. (2005) Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 7.Karsenty G. (2008) Annu. Rev. Genomics Hum. Genet. 9, 183–196 [DOI] [PubMed] [Google Scholar]
- 8.Nakashima K., de Crombrugghe B. (2003) Trends Genet. 19, 458–466 [DOI] [PubMed] [Google Scholar]
- 9.Lian J. B., Stein G. S., Javed A., van Wijnen A. J., Stein J. L., Montecino M., Hassan M. Q., Gaur T., Lengner C. J., Young D. W. (2006) Rev. Endocr. Metab. Disord. 7, 1–16 [DOI] [PubMed] [Google Scholar]
- 10.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 11.Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J. M., Behringer R. R., de Crombrugghe B. (2002) Cell 108, 17–29 [DOI] [PubMed] [Google Scholar]
- 12.Styrkarsdottir U., Halldorsson B. V., Gretarsdottir S., Gudbjartsson D. F., Walters G. B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Center J. R., Nguyen T. V., Bagger Y., Gulcher J. R., Eisman J. A., Christiansen C., Sigurdsson G., Kong A., Thorsteinsdottir U., Stefansson K. (2008) N. Engl. J. Med. 358, 2355–2365 [DOI] [PubMed] [Google Scholar]
- 13.Timpson N. J., Tobias J. H., Richards J. B., Soranzo N., Duncan E. L., Sims A. M., Whittaker P., Kumanduri V., Zhai G., Glaser B., Eisman J., Jones G., Nicholson G., Prince R., Seeman E., Spector T. D., Brown M. A., Peltonen L., Smith G. D., Deloukas P., Evans D. M. (2009) Hum. Mol. Genet. 18, 1510–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koga T., Matsui Y., Asagiri M., Kodama T., de Crombrugghe B., Nakashima K., Takayanagi H. (2005) Nat. Med. 11, 880–885 [DOI] [PubMed] [Google Scholar]
- 15.Sinha K. M., Yasuda H., Coombes M. M., Dent S. Y., de Crombrugghe B. (2010) EMBO J. 29, 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M. H., Javed A., Kim H. J., Shin H. I., Gutierrez S., Choi J. Y., Rosen V., Stein J. L., van Wijnen A. J., Stein G. S., Lian J. B., Ryoo H. M. (1999) J. Cell. Biochem. 73, 114–125 [PubMed] [Google Scholar]
- 17.Ulsamer A., Ortuño M. J., Ruiz S., Susperregui A. R., Osses N., Rosa J. L., Ventura F. (2008) J. Biol. Chem. 283, 3816–3826 [DOI] [PubMed] [Google Scholar]
- 18.Lee M. H., Kwon T. G., Park H. S., Wozney J. M., Ryoo H. M. (2003) Biochem. Biophys. Res. Commun. 309, 689–694 [DOI] [PubMed] [Google Scholar]
- 19.Matsubara T., Kida K., Yamaguchi A., Hata K., Ichida F., Meguro H., Aburatani H., Nishimura R., Yoneda T. (2008) J. Biol. Chem. 283, 29119–29125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X., Goh C. H., Li B. (2007) Endocrinology 148, 1629–1637 [DOI] [PubMed] [Google Scholar]
- 21.Guicheux J., Lemonnier J., Ghayor C., Suzuki A., Palmer G., Caverzasio J. (2003) J. Bone Miner. Res. 18, 2060–2068 [DOI] [PubMed] [Google Scholar]
- 22.Nöth U., Tuli R., Seghatoleslami R., Howard M., Shah A., Hall D. J., Hickok N. J., Tuan R. S. (2003) Exp. Cell. Res. 291, 201–211 [DOI] [PubMed] [Google Scholar]
- 23.Viñals F., López-Rovira T., Rosa J. L., Ventura F. (2002) FEBS Lett. 510, 99–104 [DOI] [PubMed] [Google Scholar]
- 24.Celil A. B., Hollinger J. O., Campbell P. G. (2005) J. Cell. Biochem. 95, 518–528 [DOI] [PubMed] [Google Scholar]
- 25.Fan D., Chen Z., Wang D., Guo Z., Qiang Q., Shang Y. (2007) J. Cell. Physiol. 211, 577–584 [DOI] [PubMed] [Google Scholar]
- 26.López-Rovira T., Chalaux E., Massagué J., Rosa J. L., Ventura F. (2002) J. Biol. Chem. 277, 3176–3185 [DOI] [PubMed] [Google Scholar]
- 27.Celil A. B., Campbell P. G. (2005) J. Biol. Chem. 280, 31353–31359 [DOI] [PubMed] [Google Scholar]
- 28.Lu X., Gilbert L., He X., Rubin J., Nanes M. S. (2006) J. Biol. Chem. 281, 6297–6306 [DOI] [PubMed] [Google Scholar]
- 29.Milona M. A., Gough J. E., Edgar A. J. (2003) BMC Genomics 4, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishio Y., Dong Y., Paris M., O'Keefe R. J., Schwarz E. M., Drissi H. (2006) Gene 372, 62–70 [DOI] [PubMed] [Google Scholar]
- 31.Zhang C., Cho K., Huang Y., Lyons J. P., Zhou X., Sinha K., McCrea P. D., de Crombrugghe B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6936–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan N. Y., Khachigian L. M. (2009) Mol. Cell. Biol. 29, 2483–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin H., van't Hof R. J., Albagha O. M., Ralston S. H. (2009) Hum. Mol. Genet. 18, 2729–2738 [DOI] [PubMed] [Google Scholar]
- 34.Narayanan K., Srinivas R., Peterson M. C., Ramachandran A., Hao J., Thimmapaya B., Scherer P. E., George A. (2004) J. Biol. Chem. 279, 44294–44302 [DOI] [PubMed] [Google Scholar]
- 35.Lee H. W., Suh J. H., Kim A. Y., Lee Y. S., Park S. Y., Kim J. B. (2006) Mol. Endocrinol. 20, 2432–2443 [DOI] [PubMed] [Google Scholar]
- 36.Sun F., Chen Q., Yang S., Pan Q., Ma J., Wan Y., Chang C. H., Hong A. (2009) Nucleic Acids Res. 37, 3897–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roca H., Phimphilai M., Gopalakrishnan R., Xiao G., Franceschi R. T. (2005) J. Biol. Chem. 280, 30845–30855 [DOI] [PubMed] [Google Scholar]
- 38.Chen J., Thomas H. F., Jin H., Jiang H., Sodek J. (1996) J. Bone Miner. Res. 11, 654–664 [DOI] [PubMed] [Google Scholar]
- 39.Paz J., Wade K., Kiyoshima T., Sodek J., Tang J., Tu Q., Yamauchi M., Chen J. (2005) Matrix Biol. 24, 341–352 [DOI] [PubMed] [Google Scholar]
- 40.Hassan M. Q., Tare R. S., Lee S. H., Mandeville M., Morasso M. I., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2006) J. Biol. Chem. 281, 40515–40526 [DOI] [PubMed] [Google Scholar]
- 41.Goto T., Matsui Y., Fernandes R. J., Hanson D. A., Kubo T., Yukata K., Michigami T., Komori T., Fujita T., Yang L., Eyre D. R., Yasui N. (2006) J. Bone Miner. Res. 21, 661–673 [DOI] [PubMed] [Google Scholar]
- 42.Baek W. Y., de Crombrugghe B., Kim J. E. (2010) Bone 46, 920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wierstra I. (2008) Biochem. Biophys. Res. Commun. 372, 1–13 [DOI] [PubMed] [Google Scholar]
- 44.Caverzasio J., Higgins L., Ammann P. (2008) J. Bone Miner. Res. 23, 1389–1397 [DOI] [PubMed] [Google Scholar]
- 45.Chang J., Sonoyama W., Wang Z., Jin Q., Zhang C., Krebsbach P. H., Giannobile W., Shi S., Wang C. Y. (2007) J. Biol. Chem. 282, 30938–30948 [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y., Wang C., Li S., Song H., Wei F., Pan K., Zhu K., Yang P., Tu Q., Chen J. (2008) Eur. J. Oral Sci. 116, 199–206 [DOI] [PubMed] [Google Scholar]
- 47.Lee H. W., Suh J. H., Kim H. N., Kim A. Y., Park S. Y., Shin C. S., Choi J. Y., Kim J. B. (2008) J. Bone Miner. Res. 23, 1227–1237 [DOI] [PubMed] [Google Scholar]
- 48.Okamura H., Amorim B. R., Wang J., Yoshida K., Haneji T. (2009) Biochem. Biophys. Res. Commun. 379, 440–444 [DOI] [PubMed] [Google Scholar]
- 49.Hatta M., Yoshimura Y., Deyama Y., Fukamizu A., Suzuki K. (2006) Int. J. Mol. Med. 17, 425–430 [PubMed] [Google Scholar]
- 50.Sierra J., Villagra A., Paredes R., Cruzat F., Gutierrez S., Javed A., Arriagada G., Olate J., Imschenetzky M., Van Wijnen A. J., Lian J. B., Stein G. S., Stein J. L., Montecino M. (2003) Mol. Cell. Biol. 23, 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vega R. B., Matsuda K., Oh J., Barbosa A. C., Yang X., Meadows E., McAnally J., Pomajzl C., Shelton J. M., Richardson J. A., Karsenty G., Olson E. N. (2004) Cell 119, 555–566 [DOI] [PubMed] [Google Scholar]
- 52.Lee J. Y., Lee Y. M., Kim M. J., Choi J. Y., Park E. K., Kim S. Y., Lee S. P., Yang J. S., Kim D. S. (2006) Mol. Cells 22, 182–188 [PubMed] [Google Scholar]
- 53.Young D. W., Pratap J., Javed A., Weiner B., Ohkawa Y., van Wijnen A., Montecino M., Stein G. S., Stein J. L., Imbalzano A. N., Lian J. B. (2005) J. Cell. Biochem. 94, 720–730 [DOI] [PubMed] [Google Scholar]
- 54.Villagra A., Cruzat F., Carvallo L., Paredes R., Olate J., van Wijnen A. J., Stein G. S., Lian J. B., Stein J. L., Imbalzano A. N., Montecino M. (2006) J. Biol. Chem. 281, 22695–22706 [DOI] [PubMed] [Google Scholar]
- 55.Flowers S., Nagl N. G., Jr, Beck G. R., Jr, Moran E. (2009) J. Biol. Chem. 284, 10067–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mujtaba S., Zeng L., Zhou M. M. (2007) Oncogene 26, 5521–5527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.