Abstract
Hsp90 is a ubiquitous, ATP-dependent chaperone, essential for eukaryotes. It possesses a broad spectrum of substrates, among which is the p53 transcription factor, encoded by a tumor-suppressor gene. Here, we elucidate the role of the adenine nucleotide in the Hsp90 chaperone cycle, by taking advantage of a unique in vitro assay measuring Hsp90-dependent p53 binding to the promoter sequence. E42A and D88N Hsp90β variants bind but do not hydrolyze ATP, whereas E42A has increased and D88N decreased ATP affinity, compared with WT Hsp90β. Nevertheless, both of these mutants interact with WT p53 with a similar affinity. Surprisingly, in the case of WT, but also E42A Hsp90β, the presence of ATP stimulates dissociation of Hsp90-p53 complexes and results in p53 binding to the promoter sequence. D88N Hsp90β is not efficient in both of these reactions. Using a trap version of the chaperonin GroEL, which irreversibly captures unfolded proteins, we show that Hsp90 chaperone action on WT p53 results in a partial unfolding of the substrate. The ATP-dependent dissociation of p53-Hsp90 complex allows further folding of p53 protein to an active conformation, able to bind to the promoter sequence. Furthermore, in support of these results, the overproduction of WT or E42A Hsp90β stimulates transcription from the WAF1 gene promoter in H1299 cells. Altogether, our research indicates that ATP binding to Hsp90β is a sufficient step for effective WT p53 client protein chaperoning.
Keywords: ATPases, Chaperone Chaperonin, Heat Shock Protein, Protein-Protein Interactions, Transcription Factors, Hsp90
Introduction
Hsp90 is an abundant protein in cells of all known organisms, with the exception of the Archea kingdom. Although in bacteria its presence is not required for survival (1), yeast and higher eukaryotes are fully dependent on its activity (2, 3). In multicellular organisms Hsp90 plays a key role in the activation and stabilization of various protein substrates. Among these are kinases (Raf1, Akt, and Src), telomerase, Rab GDP dissociation inhibitors, glucocorticoid hormone receptors (GR),3 and transcription factors such as the p53 tumor suppressor protein (4–6). These Hsp90 substrates belong to different protein families and do not share common sequence or structural motifs. Hence, modes of interaction and chaperoning may possess both common features and specific differences.
Hsp90 is functional as a dimer, each monomer consisting of three domains connected with flexible linkers of different length and sequence depending on organism and isoform. The main substrate binding region is proposed to be localized in the middle domain (7), however structural (8) and biochemical studies (9–11) suggest that at least two distinct surfaces of interaction should exist on Hsp90 chaperone while binding its protein substrate. Repositioning of the domains of the Hsp90 may be translated into conformational rearrangements of a protein substrate, changing its tertiary structure and exposing buried residues, thus enabling interaction with other proteins and ligands, such as hormones or nucleic acids.
Despite the initial controversy on the ATP dependence of Hsp90 (12), it was unambiguously shown that yeast and human Hsp90 possess an adenine nucleotide binding site localized in the N-terminal part of the protein (13, 14). Further studies have revealed that yeast Hsp82 is able to hydrolyze ATP and that changes of amino acids engaged directly in ATP binding (D79N) or Mg2+ ion binding and ATP hydrolysis (E33A) in Hsp82 are lethal (15). Finally, a low intrinsic ATPase activity of human Hsp90β was confirmed (16).
More recently, crystal structures of full-length Hsp90 from yeast and bacteria were published (17, 18) as well as mammalian Grp96 Hsp90 endoplasmic reticulum homologue (19). Based on these findings a model for chaperoning cycle of Hsp90 was proposed for a bacterial HptG protein (18). The model assumes that the ATP hydrolysis is the crucial step in the Hsp90 chaperoning cycle, providing energy for most prominent rearrangements in the Hsp90 protein. In contrast, new studies on conformation dynamics of both HptG (20) and yeast Hsp82 (21, 22) demonstrate that severe structural changes of Hsp90 dimers could occur upon ATP binding and are rate-limiting for the nucleotide hydrolysis. This suggests that at least partial influence on the substrate may be evoked before the hydrolysis step. Additionally, the crystallographic data indicate that, although Grp94 is an ATP-hydrolyzing enzyme (23), the decisive step in Grp94 client protein chaperoning cycle is possibly the ATP binding, rather than hydrolysis (19).
In the best studied Hsp90-dependent reaction, where GR is activated for hormone binding, Hsp90 does not work alone. Hsp70 machinery as well as co-chaperones are required for the efficient receptor activation (24). In our previous reports we have established that Hsp90 rescues the WT p53 tumor suppressor protein activity at physiological temperature in a single chaperone reaction. We demonstrated that WT p53 binding to the specific DNA promoter sequence in vitro at physiological temperature is Hsp90- and ATP-dependent and that the p53 transcriptional activity in cells requires Hsp90 (25, 26). WT p53 was shown to bind to Hsp90 in native or nearly native conformation (27). Muller and co-workers detected the WT p53 DNA binding domain as a minimal region of the protein responsible for Hsp90 interaction, whereas Hsp90 middle and C-terminal domains were proposed to be engaged in p53 binding (11).
In the current work we took advantage of the simplicity of the Hsp90-dependent reaction of p53 chaperoning and tested the role of adenine nucleotide in this reaction. We used Hsp90β proteins with changes in single residues, which disturb ATP binding or hydrolysis, to demonstrate that protein substrate binds stably to the nucleotide-free Hsp90β. Binding of ATP, but not its hydrolysis, was required for dissociation of p53-Hsp90β complex, and such a reaction was sufficient to rescue the p53 capability to bind to the promoter sequence in vitro. In accordance, the ability to hydrolyze ATP by an overproduced Hsp90β was not required to increase the p53 activity in cultured cells.
EXPERIMENTAL PROCEDURES
Protein Purification
Human full-length Hsp90β isoform was cloned from human cDNA library and inserted into pBAD24 plasmid. E42A and D88N changes were introduced by site-directed mutagenesis. Hsp90β variants were purified as described for WT Hsp90β (26). Human MBP-Hsp90α was purified, and the tag was removed as described previously (25). GroEL D87K trap was overproduced in the BL21(DE3) Escherichia coli strain and purified using ion-exchange chromatography on Q-Sepharose beads following heparin and ResQ FPLC columns (Amersham Biosciences).
Full-length human p23 protein was overproduced from plasmid pT7p23 in BL21(DE3) E. coli strain by 5-h induction with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside. Bacterial cells were harvested by centrifugation and lysed in buffer: 25 mm Tris, pH 7.5, 50 mm KCl, 0.1% β-mercaptoethanol, 1 mg/ml lysozyme (Sigma) followed by short sonication. The lysate was purified using phenyl-Sepharose and ResQ FPLC columns and dialyzed against the storage buffer: 25 mm Hepes, pH 7.6, 150 mm KCl, 20% glycerol, and 5 mm DTT.
Hsp90β ATP Binding Assay
Hsp90β variants were incubated at 1 μm concentrations on ice for 30 min in a buffer A (50-μl total): 25 mm HEPES, pH 7.9, 1 mm MgCl2, 15 mm NaCl, 0.5 mm DTT, 10% glycerol, 0.01% Triton X-100, 20 μm BSA (20-fold excess over Hsp90) and 14 μCi of [α-32P]ATP. Afterward, the whole volume of the mixture was applied onto the nitrocellulose 0.44-μm filter and washed with 30 ml of buffer A. Dry nitrocellulose filters were put into scintillation vials, and the radioactivity was measured using a scintillation counter. The experiment was independently repeated three times.
Hsp90β ATPase Assay
ATPase activity of Hsp90β chaperones was measured as described earlier (25).
Binding of Hsp90β to p23 Affinity Column
A p23 affinity column was prepared by covalent cross-linking of the purified proteins to the CNBr-activated Sepharose (Amersham Biosciences) according to the producer's protocol. Roughly 1 mg of the protein was conjugated to 0.5 g of the resin. Free uncoupled groups were blocked by addition of glycine. Pull-down experiments were performed by incubation of 10 μl of the prepared affinity resin with 5 μg of the purified Hsp90 WT or mutant in the buffer: 25 mm Tris, pH 7.6, 50 mm KCl, 5 mm DTT, 10 mm MgCl2, 5 mm ATP. Beads were washed three times in buffer containing 150 mm KCl and eluted with Laemmli SDS-PAGE buffer. Samples were loaded on 10% SDS-PAGE gel, after electrophoresis proteins were visualized by a silver stain.
Hsp90β-p53 ELISA
Binding of WT p53 to Hsp90β variants was tested by ELISA, as described previously for p53-Hsp90 interaction in the presence or absence of a nucleotide (25).
In Vitro p53 DNA Binding Assay
The p53 electrophoretic mobility shift assay (EMSA) was performed as described previously (25), with the modified reaction buffer composition: 25 mm Hepes, pH 7.5, 5% glycerol, 100 mm KCl, 10 mm MgCl2, and 3 mm DTT. GroEL trap protein was added to selected reactions as indicated, together with the promoter DNA, during the stage of p53 activation for DNA binding.
Hsp90β Co-immunoprecipitation
H1299 human p53-null cells were grown to 90% confluence in 6-well plates and transfected by using Lipofectamine 2000 reagent (Invitrogen) with 5 μg of pcDNA3.1 plasmids carrying WT or E42A/D88N FLAG-HA-tagged Hsp90β coding sequences and with 100 ng of pCMV plasmid carrying the WT p53 gene as indicated. At 24 h post-transfection cells were washed with PBS and scraped from plates in ice-cold IP buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 5 mm EGTA, 1.5 mm EDTA, 0.05% Triton X-100, 1 mm PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin). Cells were lysed by brief sonication, and lysates were clarified by centrifugation at 14,000 rpm for 10 min at 4 °C. Equal amounts of clarified lysates, as determined by Bradford assay, were incubated with 7 μl of anti-FLAG-agarose beads (Sigma) for 3 h at 4 °C, and subsequently washed three times with IP buffer. Bound proteins were eluted from beads with 30 μl of FLAG peptide (100 ng/μl) in IP buffer. Eluates were resolved in SDS-PAGE system and blotted, and proteins were detected with anti-Hsp90α/β (SPA-846, Stressgen), anti-p53 (rabbit polyclonal, Moravian Biotechnology), and anti-p23 (JJ3, Affinity Bioreagents) primary antibodies and suitable secondary antibodies coupled to HRP.
Luciferase Reporter Assay, p53 mRNA, and Protein Level Measurements
H1299 cells were grown and transfected as described for Hsp90β co-immunoprecipitation with the following vectors: p21-luc, containing Firefly luciferase under WAF1 p53-inducible promoter (350 ng), pCMV-luc with Firefly luciferase under the CMV promoter (350 ng), and pCMV-RL with Renilla luciferase under the CMV promoter (150 ng), along with other vectors required in the experiment: FLAG-HA-pcDNA/FLAG-HA-Hsp90β (5 μg) and pCMV-WTp53 (50 ng). 18-h post transfection cells were detached using trypsin, washed 2× with PBS, and 20% of the cell number was lysed to measure luciferase activity, 80% for the RNA isolation. The luminescence of both luciferases was measured using Dual-Luciferase Reporter Assay System (Promega) on the LumiStar Galaxy luminometer (BMG). Promoter activity was calculated by the division of a measured value of the Firefly luciferase activity by the value of Renilla luciferase activity. RNA was isolated, cDNA synthesized, and Real-Time PCR performed for TP53 and GAPDH cDNA as described previously (25). The following antibodies were used to detect proteins on Western blot: for Hsp90β, PA-012 (Affinity Bioreagents); for p53, DO-1 (Moravian Biotechnology); and for GAPDH, SC-25778 (Santa Cruz Biotechnology).
RESULTS
Characterization of Hsp90β Variants
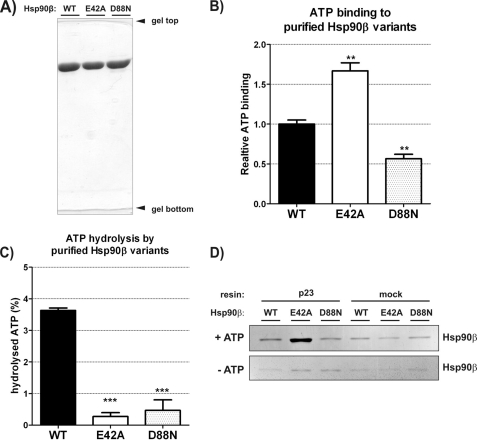
To elucidate the role of the adenine nucleotides in the Hsp90β chaperoning cycle we constructed point mutants in human HSP90β coding sequence, which resulted, on the protein level, in amino acid changes: E42A and D88N. The changed residues are strictly conserved in the whole Hsp90 family and correspond to positions Glu-33 and Asp-79 in yeast Hsp82 and Glu-46 and Asp-93 in human Hsp90α. Yeast and human Hsp90α protein variants were described previously to have impaired ATP cycling properties (14, 28). By analogy to these results, the E42A variant of human Hsp90β protein was supposed to bind but not hydrolyze ATP, whereas the D88N residue change, to abolish the binding of ATP to Hsp90β. To verify these assumptions the WT and changed Hsp90β proteins were overproduced in E. coli and purified to homogeneity (Fig. 1A). They were properly folded as judged by CD spectrum and mostly dimeric, as judged by size-exclusion chromatography (data not shown). All purified proteins were tested for ATP binding and hydrolysis activities. As shown in Fig. 1B, the E42A variant of Hsp90β binds ATP more efficiently than WT Hsp90β, whereas the binding of ATP by D88N Hsp90β is weaker. The D88N Hsp90β variant still binds ATP more efficiently than BSA (background) probably due to the existence of the second ATP binding site located in the C-terminal domain of Hsp90 (29, 30). Both variants, E42A and D88N, do not catalyze the ATP hydrolysis (Fig. 1C).
FIGURE 1.
In vitro ATP binding, hydrolysis, and interaction with p23 by human Hsp90β variants: WT, E42A, and D88N. A, 10% SDS-PAGE result demonstrating the purity of Hsp90β variants used in studies in vitro. B, ATP binding efficiency by WT, E42A, and D88N Hsp90β, detected by α-radiolabeled ATP (described under “Experimental Procedures”), shown as a fraction of WT Hsp90β. Each bar is a mean of three experiments, standard errors are shown. **, indicates a statistical significance (p < 0.01) in one-way ANOVA test (Dunnett post-test against the WT Hsp90β result). Bars have been corrected for background binding of ATP to 20-fold excess of BSA, present also in the reactions containing Hsp90β variants. Background was 16% of the WT Hsp90β result. C, ATP hydrolysis efficiency by WT, E42A, and D88N Hsp90β, shown as a fraction of a hydrolyzed γ-radiolabeled ATP, during a 3-h reaction, at 37 °C. Results were corrected for the background ATPase activity, remaining in control samples of all Hsp90β variants after inhibition by radicicol (see “Experimental Procedures”). Each bar is a mean of three experiments, and standard errors are shown. ***, indicates a statistical significance (p < 0.001) in one-way ANOVA test (Dunnett post-test against the WT Hsp90β result). D, silver-stained SDS-PAGE gel showing indicated Hsp90β variants eluted from the complex with p23-conjugated or mock resin, in the presence or absence of 5 mm ATP (see “Experimental Procedures”). Mock controls contained no p23 protein conjugated to the resin. The interaction with p23 co-chaperone is the most efficient in the case of E42A Hsp90β in the presence of ATP.
To confirm that the full-length WT or mutant variants of the Hsp90β chaperone interact with the p23 co-chaperone (31, 32) in a manner previously described, we prepared affinity column, conjugating p23 to the CNBr-activated resin. We performed an in vitro pulldown experiment to check whether the full-length WT Hsp90β and studied E42A or D88N variants were capable of interacting with p23. In the presence of ATP, E42A Hsp90β interacted with p23 affinity resin more efficiently than WT and D88N Hsp90β. In control experiments, in the absence of ATP, the affinity of all Hsp90 variants to p23 was at a background level (Fig. 1D). These results are in agreement with earlier reports of yeast p23 affinity to the yeast Hsp82 WT/E33A/D79N variants (14).
Influence of ATP on Interaction of Hsp90β Variants with WT p53
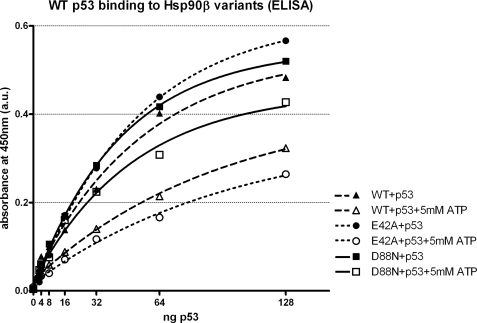
To monitor the effect of ATP on binding of Hsp90 to the p53 tumor suppressor protein in vitro, we have used a modified ELISA. In the absence of ATP all three proteins, namely WT Hsp90β, E42A Hsp90β, and D88N Hsp90β possessed a similar affinity toward WT p53 (Fig. 2). In all three variants, the presence of ATP shifted the equilibrium toward dissociation of Hsp90β-p53 complex (Fig. 2). The strongest effect was achieved in the case of E42A Hsp90β, suggesting that ATP binding, rather than hydrolysis, was sufficient for releasing of p53 from the Hsp90β complex (Fig. 2). Supporting this hypothesis, the presence of ATP only mildly influenced the binding of p53 to D88N Hsp90β (Fig. 2). In control experiments we confirmed that the preincubation of p53 with Hsp90 variants does not change the results presented in Fig. 2, suggesting that ATP can also disrupt pre-formed p53-Hsp90 complexes (result not shown).
FIGURE 2.
Influence of ATP on the interaction of Hsp90β variants and p53 in vitro. The ELISA test (performed as described under “Experimental Procedures”) was used to study the binding of Hsp90β to increasing amounts of WT p53, with or without 5 mm ATP, at 25 °C. ATP presence dissociates Hsp90β-WT p53 complexes, and the effect is strongest on E42A and weakest on D88N Hsp90β. Results are means of two measurements at each point and are corrected for the background binding of WT p53 to BSA, in the presence or absence of ATP, respectively. Curves are fitted with a one-phase exponential association model, with all R2 values above 0.99. The result is a representative of three independent experiments.
The efficient binding of WT p53 to D88N Hsp90β independently of the presence of ATP was additionally supported by results of a glycerol gradient centrifugation (supplemental data) and glutaraldehyde cross-linking experiments (result not shown).
Nucleotide Dependence of Hsp90β Variants in WT p53 Chaperoning in Vitro
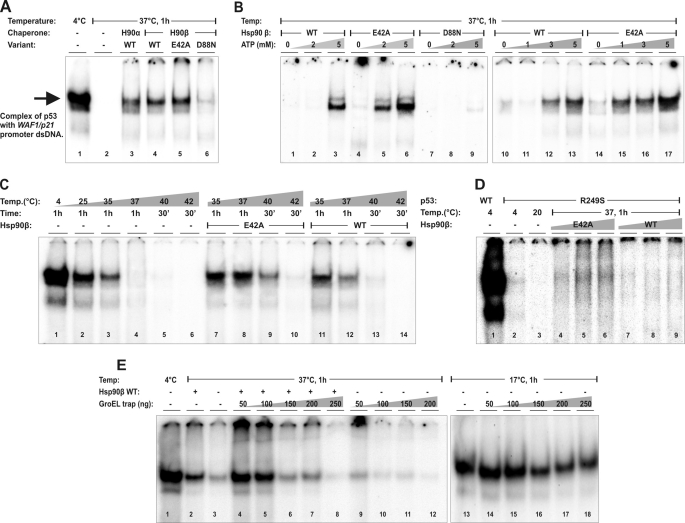
To answer the question whether the ATPase of Hsp90β is required for rescue of the in vitro WT p53 activity at the physiological temperature, we performed an EMSA of p53 binding to a WAF1 (p21 encoding gene) promoter sequence, as published before (25). p53 was able to bind the WAF1 promoter-derived sequence in the absence or presence of Hsp90β at a temperature <35 °C (Fig. 3, A (lane 1) and C (lanes 1–3)). After 1-h incubation at 37 °C, p53 was not able to bind to the promoter sequence (Fig. 3A, lane 2) unless WT Hsp90 α or β and at least 3 mm ATP concentration was present (Fig. 3, A (lanes 3 and 4) and B (lanes 1–3 and 10–13)). In the case of E42A Hsp90β, which can efficiently bind, but not hydrolyze ATP, a lower concentration of ATP (1–2 mm) was sufficient to allow chaperoning of the binding of p53 to the promoter-derived sequence (Fig. 3B, lanes 4–6 and 14–17). In contrast, the D88N Hsp90β protein, in the presence or absence of ATP, retained only traces of chaperoning activity toward WT p53 (Fig. 3, A (lane 6) and B (lanes 7–9)). Surprisingly, at heat-shock conditions (40 °C) and with a presence of 5 mm ATP, the E42A Hsp90β variant exhibited more pronounced chaperone activity toward WT p53 than WT Hsp90β (Fig. 3C). In accordance, the R249S oncogenic p53 variant, which is unable to bind WAF1 sequence already at >20 °C (26), was partially reactivated in vitro by E42A and not by WT Hsp90β at 37 °C (Fig. 3D).
FIGURE 3.
Hsp90β variants possess a different efficiency in the in vitro chaperoning of WT and mutant p53. A, Hsp90α and variants of Hsp90β (WT, E42A, and D88N) were used at 10 μm to rescue 0.5 μm WT p53 (concentrations estimated for monomers) from 1-h inactivation at 37 °C, in the presence of 5 mm ATP. Following the reaction, a p53-specific DNA-binding activity to the WAF1-promoter-derived sequence was tested by EMSA assay as described under “Experimental Procedures.” The main, marked band represents the p53-specific DNA complex. The D88N (lane 6) is the only Hsp90 tested with a decreased ability to rescue WT p53. B, ATP dependence of the WT p53 rescue reactions by Hsp90β variants. Reaction conditions, protein concentrations, and EMSA procedures were as in A. ATP was titrated as indicated. The E42A Hsp90β has an increased sensitivity to ATP in this reaction (lanes 4–6 and 14–17) compared with WT Hsp90β (lanes 1–3 and 10–13), whereas the D88N variant is inactive regardless of the presence or absence of ATP (lanes 7–9). C, temperature dependence of the WT p53 rescue reactions by Hsp90β variants. Protein, ATP concentrations, and EMSA procedures were as in A. WT p53 rescue reactions were carried out as indicated, at 35/37 °C for 1 h or 40/42 °C for 30 min. The E42A Hsp90β has the increased ability to rescue WT p53 at heat-shock conditions (40 °C). D, a restoration of the specific DNA-binding activity of R249S p53 by WT or E42A Hsp90β. p53, ATP concentrations, and EMSA procedures were as in A. Rescue reactions were carried out at indicated temperatures, for 1 h. Hsp90β variants were titrated at 5 μm, 10 μm, and 20 μm monomer concentrations of each protein variant. E42A but not WT Hsp90β was capable of a limited restoration of the specific DNA-binding activity by the otherwise inactive R249S p53 oncogenic variant. E, effect of GroEL trap variant on WT p53-specific DNA-binding activity in the presence or absence of WT Hsp90β at 37 and 17 °C. In marked reactions, indicated amounts of purified GroEL trap were added during the DNA binding stage, before being applied to the EMSA gel. The GroEL trap irreversibly bound partially unfolded WT p53 and decreased its activity at 37 °C, also in the presence of Hsp90. At 17 °C the effect of the GroEL trap was not pronounced.
These data suggest that the in vitro rescue of p53 activity from thermal inactivation by Hsp90β is dependent mainly on the ATP binding to Hsp90β and may be increased by reducing the Hsp90β ATPase activity. Notably, we do not observe any additional supershift when Hsp90β is present in the reaction mixture, suggesting that p53 dissociates completely from Hsp90β complex upon interacting with the promoter sequence. In control experiments we found that the presence of anti-Hsp90 IgGs (SPA-842 and SPA-846, Stressgen) does not supershift the p53-promoter complex (result not shown). As shown in Fig. 2, the presence of ATP is sufficient to shift the equilibrium toward dissociation of p53-Hsp90β complex that allows formation of the specific p53-DNA complex.
Hsp90 Is Not Only a Passive WT p53 Chaperone
A passive, “holdase,” mode of Hsp90 action should be diminished in the presence of ATP, which stimulates the dissociation of p53-Hsp90 complexes (Fig. 2). However, the ATP binding is required for efficient chaperone activity of Hsp90, which facilitates p53 binding to the promoter sequence at 37 °C (Fig. 3). This potential discrepancy needs to be resolved. An attractive hypothesis is that Hsp90 is also an active chaperone, that similarly to Hsp100 binds to client proteins, partially unfolds them, and, in the presence of ATP, dissociates, allowing for a spontaneous folding of the client protein to its active conformation (33). To test this hypothesis we used a GroEL D87K trap protein (34), which irreversibly binds to unfolded protein intermediates. Increasing concentration of the GroEL trap, added during the specific DNA binding by WT p53, efficiently inhibits Hsp90-dependent binding of p53 to the promoter sequence at 37 °C (Fig. 3E). This suggests that partially unfolded WT p53 intermediates are released from Hsp90. The effect of the GroEL trap is not pronounced when the reaction is carried out in the absence of Hsp90 at lower temperatures, such as 17 °C (Fig. 3F). A moderate inhibition of p53 binding to the promoter sequence at 17 °C by the GroEL trap is likely due to the presence of intrinsically unfolded regions of p53 (35).
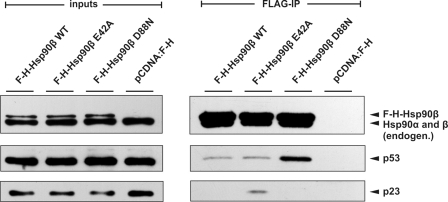
Interaction of Hsp90 Variants with WT p53 in Cells
We used a p53-null, H1299 human lung carcinoma cell line to verify the interaction of p53 with Hsp90 variants in a cell system. We have transiently overproduced FLAG-HA tagged variants of Hsp90β together with WT p53. The suitability of cell-transfected system for this study was ensured by showing the interaction of p23 protein exclusively with E42A Hsp90β mutant (Fig. 4). This is in line with in vitro experiments (Fig. 1) and previously published data on yeast Hsp82 (36). Results presented in Fig. 4 show that p53 co-immunoprecipitates most efficiently with Hsp90β D88N and to a lesser extent with WT and E42A variants. Enhanced binding of p53 to Hsp90β D88N most probably reflects a diminished dynamics of such interaction in an ATP-containing lysate as compared with p53-WT Hsp90β or E42A interactions. It is in agreement with in vitro WT p53-Hsp90β interaction results in the presence of ATP (Fig. 2).
FIGURE 4.
Immunoprecipitation of WT p53 with Hsp90β variants. FLAG-HA-Hsp90β was immunoprecipitated with anti-FLAG agarose beads and p53 and p23 proteins detected as described under “Experimental Procedures.” Equal protein amounts were loaded as inputs as determined by the Bradford assay. Hsp90β D88N interacted preferably with the client protein WT p53, and Hsp90β E42A with the p23 co-chaperone.
Influence of Hsp90β on WT p53 Activity in Cells
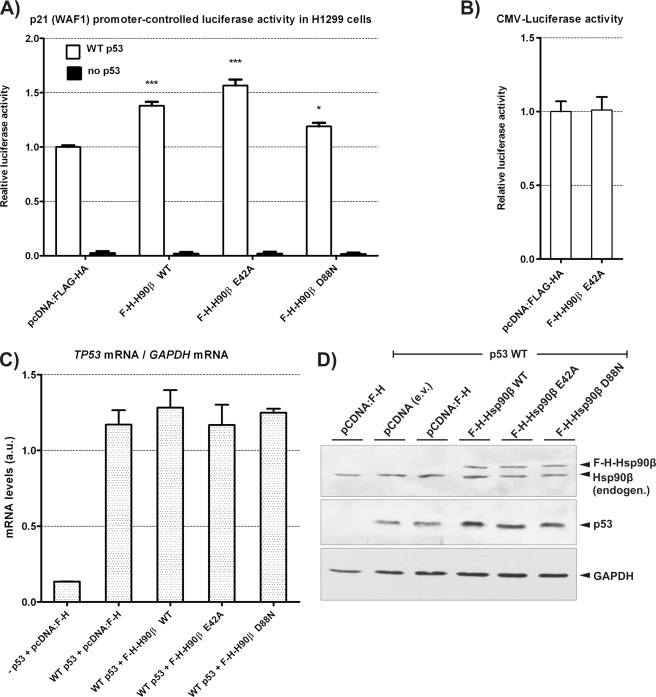
The p53 transcriptional activity in H1299 cells was monitored by a co-transfection of a limiting amount of WT p53 vector (2× less WT p53-encoding vector than in co-immunoprecipitation experiments, see “Experimental Procedures”) and with the vector containing Firefly luciferase gene under the control of the WAF1 promoter. E42A Hsp90β overexpression up-regulated the WT p53 transcriptional activity more efficiently than WT Hsp90β (Fig. 5A). D88N Hsp90β, similarly to in vitro experiments, retained a positive p53 activity regulation when compared with controls, suggesting that a part of the influence of Hsp90 on WT p53 may be exerted by a passive, ATP-independent chaperone mechanism. The increase in the Firefly luciferase activity by E42A Hsp90β was not caused by the direct chaperoning of luciferase (Fig. 5B). Described effects were also not caused by p53 mRNA level differences as tested by real-time PCR (Fig. 5C), or difference in Hsp90β level after overexpression, as shown by Western blots (Fig. 5D). However, the limiting amount of overexpressed WT p53, which allowed us to observe p53 activity differences, revealed that Hsp90β variants increased the amount of WT p53 protein (Fig. 5D). This suggested that Hsp90β may act on p53 stability. It has been demonstrated earlier that a transient overproduction of Hsp90 increases WT p53 level in U2OS cells (37).
FIGURE 5.
Hsp90β variants differentially influence the WT p53 transcriptional activity in H1299 cells. A, the transcriptional activity of WT p53, 18-h post transient transfection of H1299 cells, monitored by the luciferase reporter assay. Cells were transfected with p21-luc vector (containing Firefly luciferase coding sequence under the control of the WAF1 gene promoter), pCMV-RL vector (Renilla luciferase coding sequence under the CMV promoter), pcDNA vectors indicated below the graph bars encoding N-terminally FLAG-HA-tagged Hsp90β variants (F-H-Hsp90β WT, E42A, or D88N), or empty pcDNA-FLAG-HA. The pCMV-wtp53 vector was transfected optionally, as indicated (WT p53 coding sequence under the CMV promoter). Graph bars indicate averages with standard errors of Firefly luciferase activity ratios to Renilla luciferase, from three separate experiments, normalized to results of pcDNA-FLAG-HA. *** and *, indicate a statistical significance (p < 0.001 and p < 0.05, respectively) in a one-way ANOVA test (Dunnett post-test against the control pcDNA-FLAG-HA result). B, the influence of the Hsp90β E42A variant on constitutively expressed Firefly luciferase. pCMV-luc (CMV-controlled Firefly luciferase coding sequence), pCMV-RL, and pCMV-p53 vectors were co-transfected with either pcDNA-FLAG-HA (pcDNA-F-H) or pcDNA-FLAG-HA-Hsp90β E42A (F-H-Hsp90β E42A) as indicated. Firefly luciferase activity was measured and calculated as described in A. C, p53 mRNA levels in cells used in A measured by real-time PCR. Total RNA was extracted from selected, representative transfection experiments from A (80% of cells were used for mRNA isolation, 20% for luciferase reporter assay). The result in the case of the first bar (no p53 vector transfected) is above zero in p53-null cells, as in a total lack of the specific RT-PCR product for p53 mRNA, otherwise absent unspecific products appear with limited overlap of melting temperatures to the specific product detection range. D, p53 and Hsp90β protein level changes in transfected H1299 cells observed by Western blots. Selected protein extracts used in luciferase reporter assay in A were subjected to Western blot: Hsp90β, p53, and GAPDH were detected as indicated under “Experimental Procedures.”
DISCUSSION
The Hsp90 ATPase cycle has been extensively studied (15, 32, 38, 39). It was shown that functional coupling between ATP turnover and conformational changes of Hsp90 is a general feature conserved among Hsp90 proteins (40). Recent research on HptG protein (20) and yeast Hsp82 (21, 22) indicated that a majority of structural transitions of these chaperones are not tightly coupled to the ATP hydrolysis. However, the coupling of these events to a chaperone action of Hsp90 on a client protein has remained unsolved. The situation is complicated by the fact that, in several systems studied, Hsp90 is not working alone. Such is the case of the GR (24) and kinases, where the kinase-specific co-chaperone p50 (cdc37) is required for Hsp90-dependent activation reaction (8, 41).
We decided to couple the ATP binding of human Hsp90β with its chaperone activity using the p53 client protein. This system is unique, because at the physiological temperature of 37 °C no other chaperones or co-chaperones are required for rescue of WT p53 binding to the promoter sequence in vitro (25). Recently, we have found that only under heat shock conditions other chaperones, like Hsp70-Hsp40 machine and co-chaperone Hop, are required for folding p53 to the conformation capable of binding to the promoter sequence (26).
The purified Hsp90β variants used in this work, E42A and D88N, were both initially designed as negative controls in the p53 chaperoning tests. Although D88N indeed retained only a little activity in the in vitro p53 rescue reactions, E42A surprisingly possessed the activity at least comparable to WT Hsp90β with a higher ATP sensitivity and efficiency at heat-shock temperature (Fig. 3). In contrast to the WT Hsp90β, it was also able to partially reactivate the specific DNA-binding activity of the R249S p53 oncogenic variant. This suggests that its chaperone activity is increased to the range of Hsp70-Hsp40 system, and it is more effective than WT Hsp90β on heat-shocked WT and mutant p53 (26). The additional activity of E42A on p53 arises most likely due to an increased affinity to the nucleotide (Fig. 1), together with the fact that nucleotide hydrolysis seems dispensable for the effective release of the substrate (Fig. 2 and supplemental data) and completion of the p53 chaperoning cycle (Fig. 3). At the same time, diminished, but not abolished, binding of ATP by D88N Hsp90β (Fig. 1) is sufficient to inhibit ATP-dependent WT p53 release (Fig. 2) and the chaperone activity toward WT p53 (Fig. 3). In other tests, such as a nucleotide hydrolysis and binding to p23, purified E42A and D88N Hsp90β behaved as expected from previously published data on yeast Hsp82 and the human α isoform (14, 36, 42). These studies focus on trapping co-chaperones and substrates with E33A/E47A variants of yeast Hsp82/human Hsp90α, bringing little information on D79N/D93N Hsp90 variants interactions with various client proteins. We may therefore only speculate whether the stable binding of WT p53 to D88N Hsp90β variant, in the presence of ATP in vitro (Fig. 2) and in vivo (Fig. 4), is a unique feature of p53 or a more general trait of Hsp90 substrates. In the well studied case of a ligand binding domain of the GR there are similarities (the in vitro GR binding to D93N Hsp90α is not decreased upon ATP treatment) but also differences (ATP-dependent release of the GR is weaker in E47A than in WT Hsp90α) (28). Our results may, however, help to explain why WT p53 has not been previously found in stable complexes with WT Hsp90 in cells (43).
The data presented in this paper also address a crucial question in the chaperone field: Does the passive, holdase mode of a molecular chaperone action, a protection of client protein by binding, by itself explain the behavior of Hsp90? Here we show that binding of ATP to Hsp90-p53 complex triggers its dissociation. If only the passive mode of Hsp90 action occurred, the increased dissociation upon ATP binding should inhibit Hsp90-dependent binding of p53 to the promoter sequence. However, the situation is quite opposite: the Hsp90 variant that displays the most efficient ATP-mediated p53 release also acts as the most potent activator of p53 binding to the promoter sequence. The efficient inhibition of binding of p53 to the promoter sequence by GroEL trap variant indicates that, after ATP-dependent dissociation of Hsp90-p53 complex, p53 is in a partially unfolded state, exposing regions irreversibly trapped by GroEL. In the absence of GroEL trap, p53 spontaneously refolds and reaches the conformation capable of specific binding to the promoter sequence. Such an active, “unfoldase” mode of action, previously proposed (33) and confirmed (34) for Hsp100, is also the most likely mechanism of action of Hsp90 on WT p53.
Recently, using a chromatin immunoprecipitation, we demonstrated that Hsp90 was required in vivo for binding of WT p53 to the promoter sequence (26). In agreement with those results, here we show that overproduction of Hsp90 stimulates the WAF1 promoter. Surprisingly, the overproduction of Hsp90 also partially stabilizes WT p53. It had been previously reported that the Hsp90 overproduction increased the cellular level of WT p53 (37), but the molecular mechanism of those reactions is under further investigation. Our preliminary results suggest that, in H1299 cells, where the level of MDM2 E3 ligase is limited, WT p53 is primarily ubiquitinated by E3-ubiquitin ligase CHIP (C-terminus of Hsc70 Interacting Protein), and in this case the overproduction of Hsp90 variants inhibit the in vivo ubiquitination of p53.4 These findings are in agreement with recently published results, that Hsp90 inhibits a CHIP-dependent ubiquitination of neuronal nitric-oxide synthase (44, 45).
In general, there is a correlation between the in vitro and in vivo WT p53 chaperoning by the studied Hsp90β variants, suggesting that in the presence of ATP the same interaction and mechanism are responsible for effects under both conditions. On the other hand, using E42A Hsp90β, it was not possible to reactivate the p53 oncogenic R249S variant in the same cellular background, in which WT p53 was additionally activated (supplemental data), even though a traceable reactivation was possible in vitro (Fig. 3). Also, a knockdown of Hsp90 ATPase-stimulating Aha1 co-chaperone (38) increases neither WT nor mutant p53 activity in H1299 cells.5 Aha1 inhibition is advantageous in folding of a cystic fibrosis-causing CFTR ΔF508 protein (46). Therefore, similar to p53, rescue of this protein activity may depend on a lowered Hsp90 ATPase activity. The differences between both substrates remain to be discovered, and the effect of E42A Hsp90β on CFTR folding is under further investigation. Another Hsp90 client is an IRF-1 transcription factor. It is stabilized in human cells by the WT Hsp90α overproduction, whereas E46A Hsp90α is less effective and D93N acts in a dominant-negative manner, destabilizing IRF-1 (47). The dominant negative effect of D88N Hsp90β was also shown on migration of endothelial cells (48). These results together underscore the fact that there is a variety of Hsp90-chaperoning mechanisms, depending on a particular substrate. Distinguishing these differences may help to explain functional reasons behind a low intrinsic ATPase activity of eukaryotic Hsp90 and its extensive regulation by co-chaperones. Although some substrates require the Hsp90 ATPase (e.g. IRF-1), others, like p53 and CFTR ΔF508, may depend mostly on the nucleotide binding to Hsp90, and yet another group of protein clients could rely on the passive chaperoning (49). It is tempting to speculate that Hsp90 affinity toward different substrates and interaction with other chaperones are important parameters in these reactions. If the affinity is limited and reaction is single-chaperone (like in the case of p53 at 37 °C), ATP binding to Hsp90 is sufficient to trigger the chaperone activity and substrate release. If the affinity is higher and Hsp70 chaperones are engaged in the chaperoning reactions (like in the case of IRF-1 or GR), ATP hydrolysis is required for efficient client release and chaperone activity of Hsp90.
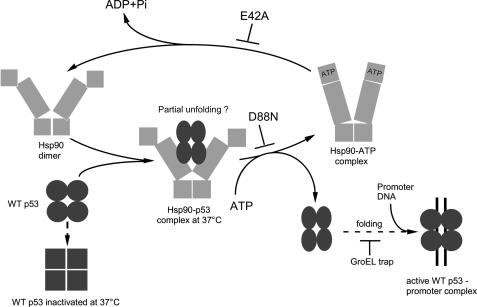
To summarize our findings on the p53 substrate, we propose a model of Hsp90-dependent chaperoning of p53, fueled with the ATP nucleotide (Fig. 6). At the first stage, when Hsp90 dimer is most likely in the “open” (“apo”) conformation (50), p53 enters the cycle and interacts with Hsp90 in a native or near-native state (27), without a requirement for a nucleotide. Binding of Hsp90 induces the conformational changes of p53 resulting in its partial unfolding. The D88N change in Hsp90β stops the cycle at this stage, because this variant is unable to bind ATP but able to trap a latent form of the client protein. In cases of WT or E42A Hsp90β, binding of ATP causes structural rearrangements of the Hsp90 domains sufficient for dissociation of Hsp90-p53 complex. Partially unfolded p53, free from Hsp90, can spontaneously further refold to the active conformation. This step is efficiently inhibited by the GroEL trap, which irreversibly binds to unfolded proteins. The model does not exclude the possibility that, in the case of other substrates or/and stress conditions, ATP hydrolysis is required for efficient dissociation of the Hsp90-client protein complexes.
FIGURE 6.
A proposed pathway of p53 chaperoning by Hsp90. The p53 client protein is captured by a nucleotide-free Hsp90, in a native or near-native conformation. Otherwise WT p53 becomes irreversibly inactivated at 37 °C. The Hsp90β D88N variant binds p53, not being able to proceed with further steps. Hsp90β WT and E42A are both able to chaperone and release the client, indicating that necessary structural rearrangements of Hsp90β must occur prior to the ATP hydrolysis (see “Discussion” for more details and references).
Supplementary Material
Acknowledgments
We thank Peter Csermely for the plasmid encoding human Hsp90α, Ted Hupp for plasmid constructs for p53 overexpression, Christomos Prodromou for the vector with human p23, and F. Ulrich Hartl for the GroEL trap-overproducing E. coli strain.
This work was supported, in part, by a grant from the Polish Ministry of Science and Higher Education.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, references, and Figs. 1 and 2.
D. Walerych, M. Gutkowska, M. P. Klejman, B. Wawrzynow, Z. Tracz, M. Wiech, M. Zylicz, and A. Zylicz, unpublished results.
- GR
- glucocorticoid receptor
- ANOVA
- analysis of variance.
REFERENCES
- 1.Bardwell J. C., Craig E. A. (1988) J. Bacteriol. 170, 2977–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang H. C., Lindquist S. (1994) J. Biol. Chem. 269, 24983–24988 [PubMed] [Google Scholar]
- 3.Voss A. K., Thomas T., Gruss P. (2000) Development 127, 1–11 [DOI] [PubMed] [Google Scholar]
- 4.Whitesell L., Lindquist S. L. (2005) Nat. Rev. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 5.Picard D. (2002) Cell Mol. Life Sci. 59, 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wegele H., Müller L., Buchner J. (2004) Rev. Physiol. Biochem. Pharmacol. 151, 1–44 [DOI] [PubMed] [Google Scholar]
- 7.Meyer P., Prodromou C., Hu B., Vaughan C., Roe S. M., Panaretou B., Piper P. W., Pearl L. H. (2003) Mol. Cell 11, 647–658 [DOI] [PubMed] [Google Scholar]
- 8.Vaughan C. K., Gohlke U., Sobott F., Good V. M., Ali M. M., Prodromou C., Robinson C. V., Saibil H. R., Pearl L. H. (2006) Mol. Cell 23, 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang L., Ricketson D., Getubig L., Darimont B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18487–18492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheibel T., Weikl T., Rimerman R., Smith D., Lindquist S., Buchner J. (1999) Mol. Microbiol. 34, 701–713 [DOI] [PubMed] [Google Scholar]
- 11.Müller L., Schaupp A., Walerych D., Wegele H., Buchner J. (2004) J. Biol. Chem. 279, 48846–48854 [DOI] [PubMed] [Google Scholar]
- 12.Jakob U., Scheibel T., Bose S., Reinstein J., Buchner J. (1996) J. Biol. Chem. 271, 10035–10041 [DOI] [PubMed] [Google Scholar]
- 13.Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1997) Cell 90, 65–75 [DOI] [PubMed] [Google Scholar]
- 14.Obermann W. M., Sondermann H., Russo A. A., Pavletich N. P., Hartl F. U. (1998) J. Cell Biol. 143, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panaretou B., Prodromou C., Roe S. M., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1998) EMBO J. 17, 4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLaughlin S. H., Ventouras L. A., Lobbezoo B., Jackson S. E. (2004) J. Mol. Biol. 344, 813–826 [DOI] [PubMed] [Google Scholar]
- 17.Ali M. M., Roe S. M., Vaughan C. K., Meyer P., Panaretou B., Piper P. W., Prodromou C., Pearl L. H. (2006) Nature 440, 1013–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiau A. K., Harris S. F., Southworth D. R., Agard D. A. (2006) Cell 127, 329–340 [DOI] [PubMed] [Google Scholar]
- 19.Dollins D. E., Warren J. J., Immormino R. M., Gewirth D. T. (2007) Mol. Cell 28, 41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graf C., Stankiewicz M., Kramer G., Mayer M. P. (2009) EMBO. J. 28, 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mickler M., Hessling M., Ratzke C., Buchner J., Hugel T. (2009) Nat. Struct. Mol. Biol. 16, 281–286 [DOI] [PubMed] [Google Scholar]
- 22.Hessling M., Richter K., Buchner J. (2009) Nat. Struct. Mol. Biol. 16, 287–293 [DOI] [PubMed] [Google Scholar]
- 23.Frey S., Leskovar A., Reinstein J., Buchner J. (2007) J. Biol. Chem. 282, 35612–35620 [DOI] [PubMed] [Google Scholar]
- 24.Pratt W. B., Toft D. O. (2003) Exp. Biol. Med. (Maywood) 228, 111–133 [DOI] [PubMed] [Google Scholar]
- 25.Walerych D., Kudla G., Gutkowska M., Wawrzynow B., Muller L., King F. W., Helwak A., Boros J., Zylicz A., Zylicz M. (2004) J. Biol. Chem. 279, 48836–48845 [DOI] [PubMed] [Google Scholar]
- 26.Walerych D., Olszewski M. B., Gutkowska M., Helwak A., Zylicz M., Zylicz A. (2009) Oncogene 28, 4284–4294 [DOI] [PubMed] [Google Scholar]
- 27.King F. W., Wawrzynow A., Höhfeld J., Zylicz M. (2001) EMBO J. 20, 6297–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young J. C., Hartl F. U. (2000) EMBO J. 19, 5930–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Söti C., Rácz A., Csermely P. (2002) J. Biol. Chem. 277, 7066–7075 [DOI] [PubMed] [Google Scholar]
- 30.Garnier C., Lafitte D., Tsvetkov P. O., Barbier P., Leclerc-Devin J., Millot J. M., Briand C., Makarov A. A., Catelli M. G., Peyrot V. (2002) J. Biol. Chem. 277, 12208–12214 [DOI] [PubMed] [Google Scholar]
- 31.Richter K., Walter S., Buchner J. (2004) J. Mol. Biol. 342, 1403–1413 [DOI] [PubMed] [Google Scholar]
- 32.McLaughlin S. H., Sobott F., Yao Z. P., Zhang W., Nielsen P. R., Grossmann J. G., Laue E. D., Robinson C. V., Jackson S. E. (2006) J. Mol. Biol. 356, 746–758 [DOI] [PubMed] [Google Scholar]
- 33.Wawrzynow A., Banecki B., Zylicz M. (1996) Mol. Microbiol. 21, 895–899 [DOI] [PubMed] [Google Scholar]
- 34.Weber-Ban E. U., Reid B. G., Miranker A. D., Horwich A. L. (1999) Nature 401, 90–93 [DOI] [PubMed] [Google Scholar]
- 35.Bell S., Klein C., Müller L., Hansen S., Buchner J. (2002) J. Mol. Biol. 322, 917–927 [DOI] [PubMed] [Google Scholar]
- 36.Millson S. H., Truman A. W., King V., Prodromou C., Pearl L. H., Piper P. W. (2005) Eukaryot Cell 4, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki M., Nie L., Maki C. G. (2007) J. Biol. Chem. 282, 14626–14634 [DOI] [PubMed] [Google Scholar]
- 38.Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J. K., Singh S., Millson S. H., Clarke P. A., Naaby-Hansen S., Stein R., Cramer R., Mollapour M., Workman P., Piper P. W., Pearl L. H., Prodromou C. (2002) Mol. Cell 10, 1307–1318 [DOI] [PubMed] [Google Scholar]
- 39.McLaughlin S. H., Smith H. W., Jackson S. E. (2002) J. Mol. Biol. 315, 787–798 [DOI] [PubMed] [Google Scholar]
- 40.Richter K., Soroka J., Skalniak L., Leskovar A., Hessling M., Reinstein J., Buchner J. (2008) J. Biol. Chem. 283, 17757–17765 [DOI] [PubMed] [Google Scholar]
- 41.Arlander S. J., Felts S. J., Wagner J. M., Stensgard B., Toft D. O., Karnitz L. M. (2006) J. Biol. Chem. 281, 2989–2998 [DOI] [PubMed] [Google Scholar]
- 42.Gano J. J., Simon J. A. (2010) Mol. Cell Proteomics 9, 255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sepehrnia B., Paz I. B., Dasgupta G., Momand J. (1996) J. Biol. Chem. 271, 15084–15090 [DOI] [PubMed] [Google Scholar]
- 44.Peng H. M., Morishima Y., Clapp K. M., Lau M., Pratt W. B., Osawa Y. (2009) Biochemistry 48, 8483–8490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pratt W. B., Morishima Y., Peng H. M., Osawa Y. (2010) Exp. Biol. Med. 235, 278–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 47.Narayan V., Eckert M., Zylicz A., Zylicz M., Ball K. L. (2009) J. Biol. Chem. 284, 25889–25899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao R. Q., Fontana J., Fulton D., Lin M. I., Harrison K. D., Sessa W. C. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 105–111 [DOI] [PubMed] [Google Scholar]
- 49.Minami Y., Minami M. (1999) Genes Cells 4, 721–729 [DOI] [PubMed] [Google Scholar]
- 50.Bron P., Giudice E., Rolland J. P., Buey R. M., Barbier P., Díaz J. F., Peyrot V., Thomas D., Garnier C. (2008) Biol. Cell 100, 413–425 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.