Abstract
Innate immune recognition of flagellin is shared by transmembrane TLR5 and cytosolic Nlrc4 (NOD-like receptor family CARD (caspase activation recruitment domain) domain containing 4)/Naip5 (neuronal apoptosis inhibitory protein 5). TLR5 activates inflammatory genes through MYD88 pathway, whereas Nlrc4 and Naip5 assemble multiprotein complexes called inflammasomes, culminating in caspase-1 activation, IL-1β/IL-18 secretion, and pyroptosis. Although both TLR5 and Naip5/Nlrc4 pathways cooperate to clear infections, little is known about the relative anti-pathogen effector mechanisms operating through each of them. Here we show that the cytosolic flagellin (FLA-BSDot) was able to activate iNOS, an enzyme previously associated with TLR5 pathway. Using Nlrc4- or Naip5-deficient macrophages, we found that both receptors are involved in iNOS activation by FLA-BSDot. Moreover, distinct from extracellular flagellin (FLA-BS), iNOS activation by intracellular flagellin is completely abrogated in the absence of caspase-1. Interestingly, IL-1β and IL-18 do not seem to be important for FLA-BSDot-mediated iNOS production. Together, our data defined an additional anti-pathogen effector mechanism operated through Naip5 and Nlrc4 inflammasomes and illustrated a novel signaling transduction pathway that activates iNOS.
Keywords: Innate Immunity, Macrophage, MyD88, Nitric-oxide Synthase, Pattern Recognition Receptor, Inflammasomes, NOD-like Receptor (NLR), Caspase-1, Flagellin
Introduction
Innate immune system employs intracellular and extracellular pattern recognition receptors (PRRs)2 to detect microbial products through the identification of pathogen-associated molecular patterns (PAMPs). Among many families of PRRs, Toll-like receptors (TLRs) and NOD-like receptors (NLRs) have been described as central elements in triggering innate immune responses (1, 2).
TLRs and NLRs have been shown to activate distinct biochemical pathways, resulting in diverse mechanisms of resistance to pathogens. These mechanisms may act independently or may cooperate to the resolution of infections. TLRs primarily activate transcriptional factors such as NF-κB and IFN-responsive factors (IRF) (3, 4), culminating in the production of effector molecules such as inflammatory cytokines and type I interferons. The contribution of these pathways to resistance to infection is manifold: recruitment of other inflammatory cells, production of cytotoxic and cytostatic molecules, and activation of adaptive immune response. NLRs, on the other hand, although also capable of activating NF-κB, have a peculiar ability of initiating an inflammatory response through caspase-1 activation via the assembly of a multiprotein complex called inflammasome (5–7). Besides the well known role of caspase-1 on IL-1β and IL-18 maturation and secretion, this protease has also been described to induce macrophages death by a process known as pyroptosis (6, 7). Importantly, pyroptosis has been ascribed as a mechanism of host immune response against certain pathogens, such as Salmonella, Legionella, Pseudomonas, and Shigella, although it is still not clear how pyroptosis effectively contributes for pathogen clearance (7).
The recognition of PAMPs by TLRs and NLRs occur independently. However, there are cases where a certain PAMP can be recognized by both TLRs and NLRs. Flagellin, the monomeric subunit of flagella present in several Gram-negative and Gram-positive bacteria is a good example of this redundancy. Extracellular flagellin is recognized by transmembrane TLR5 (8), whereas cytosolic sensing of flagellin requires Naip5 (neuronal apoptosis inhibitory protein 5) and Nlrc4 (NLR family CARD (caspase activation recruitment domain) domain-containing 4), also called Ipaf (ICE-(IL-1-converting enzyme) protease activating factor), both members of the NLR family (9–13). It was recently demonstrated that both Nlrc4 and Naip5 are required for signaling in response to the 35-amino acid of a peptide located on the C-terminal D0 domains of Legionella pneumophila and Salmonella typhimurium flagellins and that this region of flagellin is distinct from the D1 domain sensed by TLR5 (11). Importantly, TLR5 engagement by flagellin leads to NF-κB activation, whereas the recognition of flagellin by Naip5 and Nlrc4 induces the assembly of inflammasomes with the recruitment and activation of pro-caspase-1 (7).
Regardless the particularities between intracellular and extracellular receptor pathways, the relative contribution of TLR5 and Naip5/Nlrc4 inflammasomes to host resistance to infection is still not clear. Here, by using a purified flagellin, we defined an additional anti-pathogen effector mechanism operating through Naip5 and Nlrc4 inflammasomes that was previously associated with TLR pathway (14, 15), namely the activation of inducible nitric-oxide synthase (iNOS), the enzyme responsible for nitric oxide (NO) production (16). Interestingly, distinct from extracellular flagellin, iNOS activation by intracellular flagellin is completely abrogated in the absence of caspase-1 but not with neutralization of IL-1β and/or IL-18. Importantly, this effect is preserved in Myd88-deficient macrophage, ruling out the participation of TLR5. Together, our data indicated that by using distinct pathways, TLR and NLR systems can operate with a certain level of redundancy, which might improve the host resistance against infections.
EXPERIMENTAL PROCEDURES
Mice
6–8-Week-old wild-type (WT) C57BL/6 and Myd88-deficient female mice were bred in our animal facilities at the University of São Paulo. Nlrc4- and caspase-1-deficient mice were previously described (17) and backcrossed for six generations with C57BL/6. Mice with 99.2% A/J genome harboring the wild-type Lgn1 allele derived from C57BL/6 were constructed by backcrossing a progeny of C57BL/6 x A/J with A/J for 6 generations as described in supplemental Fig. 1. Briefly, each generation was screened by PCR using 2 pairs primers that amplify fragments within microsatellite regions flanking the Lgn1 locus, thus, differentiating C57BL/6 from A/J regions of chromosome 13 (13). The resultant mice (F7 AJxB6 Lgn1B6/AJ) are heterozygous for the dominant Lgn1 locus and show full Naip5 functions. In contrast, littermate controls homozygous for the A/J allele (F7 AJxB6 Lgn1AJ/AJ) harbors two copies of the defective Naip5 allele from A/J mice (13, 18). Naip5 functions of the F7 mice were monitored by measuring restriction of L. pneumophila multiplication in macrophages (supplemental Fig. 1). These mice were bred and constructed in the animal facilities at the University of São Paulo-Ribeirão Preto. Animal studies used protocols approved by the University of São Paulo Committee on Use and Care of Animals.
Preparation of Mouse Macrophages
Bone marrow-derived macrophages (BMDM) were prepared as described (19). Bone marrow cells were cultured for 7 days at 37 °C in 5% CO2 in RPMI 1640 medium supplemented with 30% L929 supernatant containing macrophage-stimulating factor, 10% heat-inactivated fetal calf serum (FCS), 10 mm HEPES, 1 mm sodium pyruvate, 2 mm l-glutamine, and 100 units/ml streptomycin and penicillin. All supplements were purchased from Invitrogen. Peritoneal macrophages (PM) were obtained as described (20). Briefly, peritoneal lavage was collected 4 days after injection of mice with 1% starch solution (Sigma). Adherent macrophages were prepared by overnight culture in RPMI supplemented with 10% heat inactivated FCS and antibiotics, resulting in a population that comprises 97–99% of F4/80+ cells (data not shown).
Stimulation of Macrophages with Flagellin
PM and BMDM (3 × 105 cells seeded in 96-well plates) were cultured for 5–24 h at 37 °C in 5% CO2 in RPMI 1640 medium supplemented with 3% FCS, amino acids, and antibiotics in the presence of purified flagellin from Bacillus subtilis (FLA-BS) or S. typhimurium (FliC) (Invivogen) (1–6 μg/ml) in its free form or inserted into DOTAP (Roche Diagnostics), a cationic lipid formulation that permits its delivery to cell cytosol (21). DOTAP was used accordingly to the manufacturer's instructions. Briefly, DOTAP (50 μl) was incubated for 15 min in serum-free media with 9 μg of purified flagellin. After incubation, 2.95 ml of RPMI 1640 medium was added, and an aliquot of 200 μl was added to 3 × 105 macrophages (for 3 μg/ml final concentration). In some cultures endogenous caspase-1 was blocked with specific inhibitor z-YVAD-fmk (Calbiochem) at a final concentration of 2–50 μm, and IL-1β and IL-18 were neutralized by adding anti-mouse IL-1β or IL-18 (AbD Serotec) at final concentration of 1 and 5 μg/ml, respectively.
Measurement of Cytokines and Nitric Oxide
IL-1β and IL-18 were measured in culture supernatants by enzyme-linked immunoabsorbent assay (ELISA) (kits from BD Biosciences) following the manufacturer's instructions. Culture supernatants were also assayed for NO by the Griess reaction. Briefly, 50 μl of supernatant was incubated with 50 μl of Griess reagent for 5 min at ambient temperature, and the NO2 concentration was determined by measuring the optical density at 550 nm in reference to a standard NaNO2 solution.
Western Blot
Western blot was performed as previously described (22). Cells were harvested, washed once in ice-cold PBS, lysed directly in SDS sample buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 2.5% β-mercaptoethanol), and boiled for 5 min. Samples were resolved under reducing conditions for 2 h at 15 V in SDS-polyacrylamide gels. Proteins were then transferred onto PVDF membranes for 1 h in a semi-dry system. Blots were blocked for 3 h in TBST (10 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.05% Tween) containing 0.1% sodium azide and 5% nonfat dried milk and then probed with polyclonal antibodies to iNOS (1:1000) and β-actin (1:20.000) overnight (Santa Cruz Biotechnology). Reactions were detected with suitable secondary antibody (1:2000) conjugated to horseradish peroxidase (The Jackson Laboratory and Amersham Biosciences) using enhanced chemiluminescence solution (Pierce).
Real-time Quantitative PCR
Total RNA was isolated using TRIzol RNA isolation. Concentration and purity of the RNA were verified by spectrophotometer NanoDROP nd-1000. The absorbance ratio at 260/280 nm of all samples ranged from 1.8 to 2.0, indicating they all were contaminant-free. The RNA integrity has been assayed on a 1% agarose gel stained with ethidium bromide. cDNA was generated from 3 μg of total RNA using oligo(dT) and the Moloney murine leukemia virus reverse transcriptase RNase H (Invitrogen) according to the manufacturer's protocol. Quantitative real-time PCR analysis was performed with the Taqman System (Applied Biosystems) (catalog no. Mm0130989_m1; Nos2). To standardize all the samples, the housekeeping gene glyceraldehydes 3-phosphate dehydrogenase (catalog no. 4352932) was used as an endogenous reference gene (TaqMan Endogenous Control concentration-limited primer, Applied Biosystems).
Macrophage Infection with L. pneumophila
BMDMs from C57BL/6- and Myd88-deficient mice (2 × 105 cells seeded in 24-well plates) were infected with wild-type or flagellin-deficient mutant FlaA L. pneumophila at a multiplicity of infection of 0.015 for 24, 48, and 72 h in the presence or absence of 0.1 mm aminoguanidine (selective inhibitor of iNOS (23); SIGMA). Macrophages were lysed in sterile H2O, and the cell lysates were combined with the tissue culture supernatant from that well, ensuring that all bacteria in the well would be counted. Serial dilutions from each well were plated on charcoal yeast extract agar plates. Colony-forming units were assessed by counting of bacterial colonies present after 96 h of incubation at 37 °C.
Statistical Analysis
Experiments were performed always in duplicate or triplicate at least three times. Data are presented as the mean values ± S.D. Statistical analysis of the data were carried out using one-way analysis of variance and Tukey as a post-test. Differences between experimental groups were considered significant for p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
RESULTS
Recognition of Extracellular or Cytosolic B. subtilis Flagellin Differentially Stimulates Macrophages
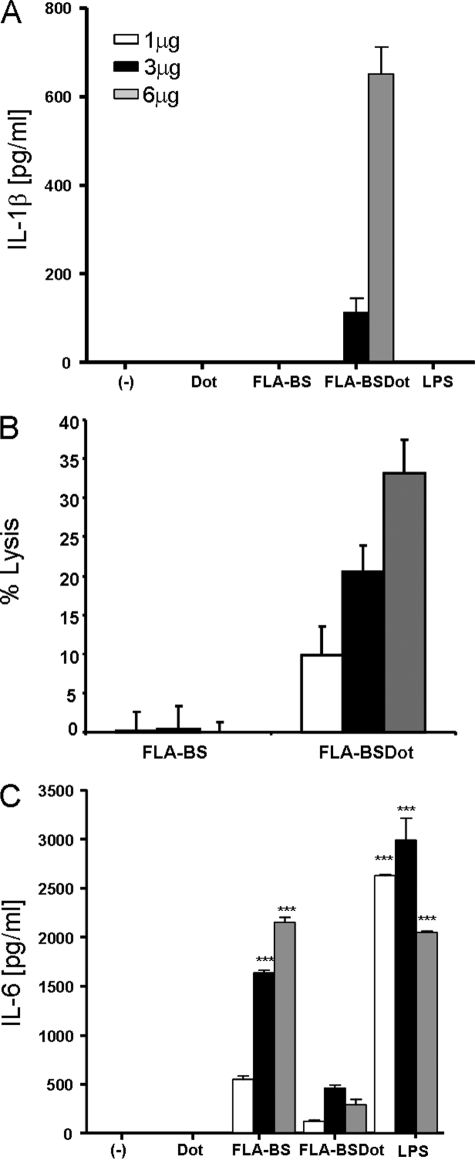
We first characterized the macrophage responses to extracellular (FLA-BS) or intracellular (FLA-BSDot) flagellin. PMs were stimulated with LPS, FLA-BS, or FLA-BSDot (1–6 μg/ml), and the secretion of IL-6, IL-1β, and macrophage lysis (monitored by LDH release) were determined respectively as surrogate markers of NF-κB and caspase-1 activation. As expected, only FLA-BSDot induced IL-1β maturation and secretion and caspase-1-dependent macrophage lysis in a dose-dependent manner (Fig. 1, A and B, and supplemental Fig. 2). Conversely, high levels of IL-6 were only observed in the presence of FLA-BS and LPS (Fig. 1C). DOTAP by itself did not induce either IL-1β or IL-6 secretion (Fig. 1). Also, as expected, LPS alone was not able to induce IL-1β secretion (Fig. 1). Moreover, as expected, the level of IL-1β secreted in response to FLA-BSDot stimulation alone is markedly lower than that observed after LPS priming (data not shown). IL-18 was not detected in these conditions (data not shown). Our results are in agreement with the literature showing that TLRs and NLRs drive macrophage activation in different ways.
FIGURE 1.
Differential activation of macrophages by extracellular and cytosolic B. subtilis. PM from C57BL/6 mice (3 × 105/200 μl) were stimulated with 1, 3, and 6 μg/ml LPS or purified flagellin from B. subtilis in its free form (FLA-BS) or inserted into DOTAP, a cationic lipid vesicules that permits its delivery to cell cytosol (FLA-BSDot). The release of IL-1β (A), IL-6 (C), and macrophage lysis (B) were determined in culture supernatants at 24 h by ELISA and at 6 h by LHD release, respectively. Numbers represent the mean ± S.D. of triplicate samples. Data are representative of three independent experiments.
Cytosolic Flagellin Induces iNOS Expression by Macrophages
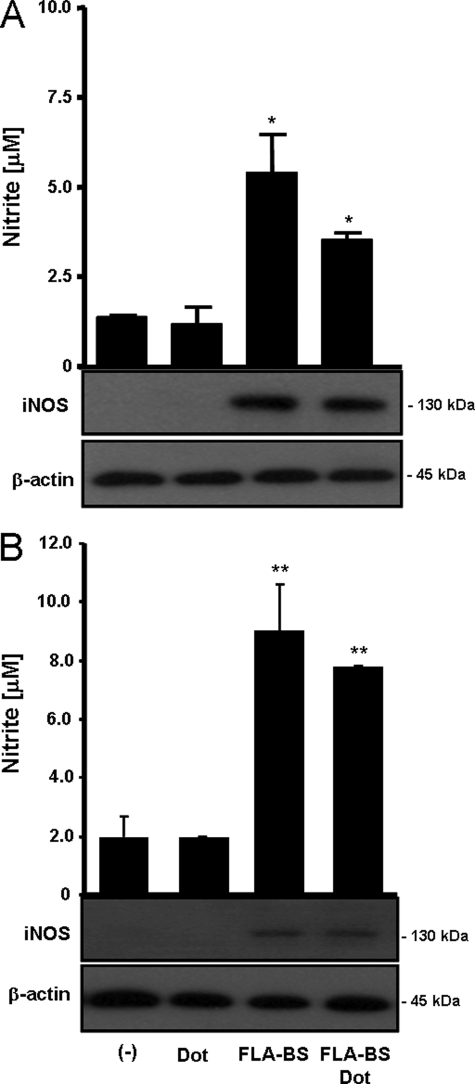
FLA-BS stimulation of PMs resulted in iNOS expression, which correlated with NO secretion (Fig. 2A). However, unexpectedly, iNOS expression and NO production were also observed when PMs were stimulated with FLA-BSDot. Similar results were also observed using adherent BMDM (Fig. 2B), indicating that FLA-BSDot up-regulates iNOS activity in different macrophage populations. Therefore, our data show for the first time that both extracellular and cytosolic flagellins are able to activate iNOS.
FIGURE 2.
Cytosolic flagellin induces iNOS activation. PM (A) or BMDM (B) from C57BL/6 mice (3 × 105/200 μl) were stimulated with 3 μg/ml purified flagellin from B. subtilis in its free form (FLA-BS) or inserted into DOTAP, a cationic lipid vesicules that permits its delivery to cell cytosol (FLA-BSDot). After 24 h iNOS expression was evaluated in cell extracts by Western blot, and nitrite concentrations in culture supernatants were determined by Griess method. Numbers represent the mean ± S.D. of triplicate samples. *, p < 0.05; **, p < 0.01 when compared with control group. Data are representative of three independent experiments.
Nlrc4 and Naip5 Participate in FLA-BSDot-induced iNOS Expression
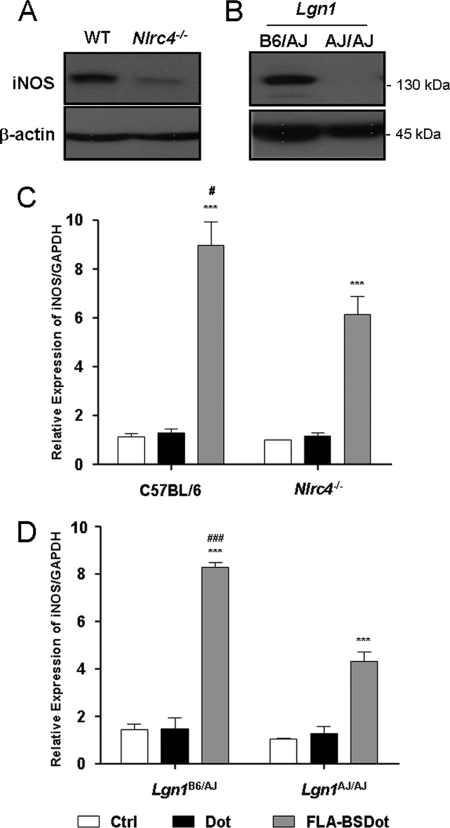
Because cytosolic flagellin has been shown to be sensed by Nlrc4 and Naip5 inflammasomes (9–13), we next determined their contribution to FLA-BSDot-induced iNOS activation by using BMDM from mice deficient in each of these molecules. As shown in Fig. 3, FLA-BSDot-induced iNOS expression in Nlrc4−/− BMDM was significantly lower than observed in WT C57BL/6 cells, but not absent, suggesting that Nlrc4 is important but not essential for iNOS expression in response to cytosolic flagellin (Fig. 3A). A similar pattern of results was observed at transcriptional levels by real-time RT-PCR, where a significant decrease in the iNOS expression was observed in Nlrc4−/− BMDM when compared with WT C57BL/6 cells (Fig. 3C). The Naip5 allele from A/J mouse strain is partially defective due to a series of amino acid substitutions (18, 24). Thus, we took advantage of these mutations and generated a pair of mice with a similar genetic background (A/J) that differentially expresses functional (Lgn1B6/AJ) or mutant (Lgn1AJ/AJ) Naip5 allele (see “Experimental Procedures” and supplemental Fig. 1). FLA-BSDot stimulates Naip5-competent (Lgn1B6/AJ) but not Naip5-deficient (Lgn1AJ/AJ) BMDM to up-regulate iNOS protein levels (Fig. 3B). Notwithstanding the absence of iNOS protein in Naip5-deficient (Naip5AJ/AJ) BMDM, low levels of iNOS transcripts were still observed by RT-PCR (Fig. 3D). These data indicate that functional Naip5 plays a major role for FLA-BSDot-induced iNOS. Similar results were obtained with flagellin purified from S. typhimurium (data not shown), a well characterized Naip5 and Nlrc4 agonist (11). Taken together, our results established that flagellin from B. subtilis is also a Naip5 and Nlrc4 agonist and that both receptors are involved in the up-regulation of iNOS.
FIGURE 3.
Nlrc4 and Naip5 are required for cytosolic flagellin-induced iNOS expression. BMDM from wild-type and Nlrc4−/− C57BL/6 mice (A and C) or Naip5-competent (Lgn1B6/AJ) and Naip5-deficient (Lgn1AJ/AJ) mice (B and D) were stimulated with 3 μg/ml purified flagellin from B. subtilis inserted into DOTAP, a cationic lipid vesicule that permits its delivery to cell cytosol (FLA-BSDot). After 24 or 5 h, iNOS expression was evaluated in cell extracts by Western blot (A and B) or at transcriptional levels by real-time RT-PCR (C and D), respectively. Data are representative of three independent experiments. Numbers represent the mean ± S.D. of triplicate samples. ***, p < 0.001 when compared with control group. #, p < 0.05; ###, p < 0.001 when compared with FLA-BSDot group from deficient mice. Ctrl, control.
FLA-BSDot-induced iNOS Expression Is Dependent on Caspase-1
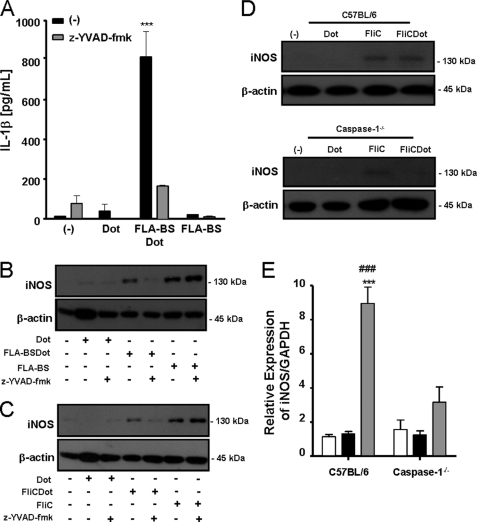
Because cytosolic flagellin recognition by inflammasomes results in caspase-1 activation (9–13), we asked whether caspase-1 was involved in FLA-BSDot-induced iNOS. Therefore, macrophages were cultured with FLA-BS or FLA-BSDot in the presence or absence of z-YVAD-fmk, a synthetic peptide that irreversibly inhibits caspase-1. As expected, z-YVAD-fmk inhibited IL-1β secretion in response to FLA-BSDot (Fig. 4A). Importantly, it also abrogated the effect of FLA-BSDot on iNOS expression (Fig. 4B). However, no effect of z-YVAD-fmk was observed in FLA-BS-induced iNOS expression (Fig. 4B). The same results were obtained with FliC and FliCDot from the Gram-negative S. typhimurium, showing that the effect was not restricted to flagellin from the Gram-positive B. subtilis (Fig. 4C). The requirement of caspase-1 for iNOS induction in response to cytosolic flagellin was confirmed at protein (Fig. 4D) and transcriptional levels (Fig. 4E) in caspase-1−/− BMDM, demonstrating that extracellular and intracellular flagellin use different pathways to activate iNOS.
FIGURE 4.
Caspase-1 is required for cytosolic flagellin-induced iNOS expression. PMs or BMDM from C57BL/6 or caspase-1−/− mice were stimulated with 3 μg/ml purified free (FLA-BS) or cytosolic (FLA-BSDot) flagellin from B. subtilis (A, B, and E) or flagellin from S. typhimurium (FliC or FliCDot, respectively) (C and D) in the presence or absence of z-YVAD-fmk (2 μm). After 24 or 5 h, iNOS expression was evaluated in cell extracts by Western blot (B, C, and D) or at transcriptional levels by real-time RT-PCR (E), respectively. IL-1β secretion was determined in culture supernatants by ELISA (A). Numbers represent the mean ± S.D. of triplicate samples. ***, p < 0.001 when compared with control group. ###, p < 0.001 when compared with FLA-BSDot group from caspase-1−/− BMDM. Data are representative of three independent experiments.
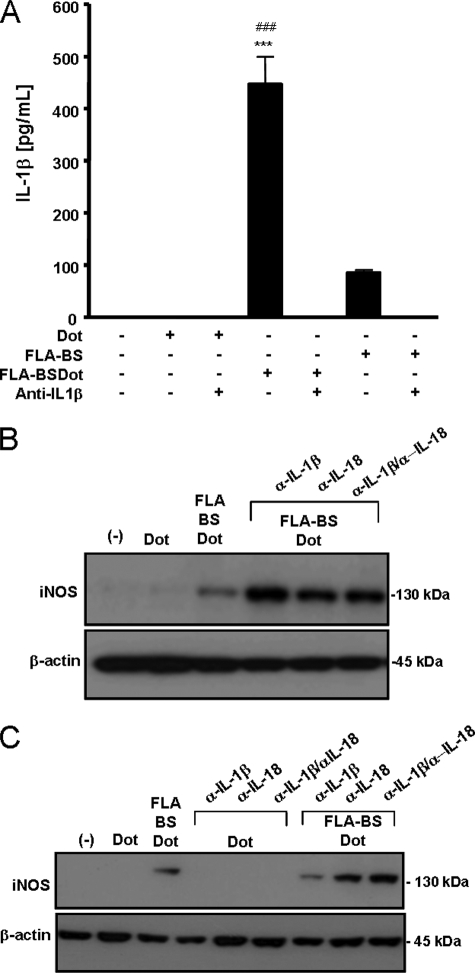
FLA-BSDot-induced iNOS Expression Does Not Depend on IL-1β and IL-18
Because caspase-1 activation induced by cytosolic flagellin was shown to result in the secretion of IL-1β and IL-18 (6), we next determined whether these cytokines participate in iNOS up-regulation. Macrophages were cultured with FLA-BSDot in the presence or absence of neutralizing anti-IL-1β and anti-IL-18 antibodies. Anti-IL-1β antibodies sequestered IL-1β present in FLA-BSDot-stimulated PM culture supernatants (Fig. 5A) but did not inhibit FLA-BSDot-induced iNOS expression (Fig. 5B). On the contrary, if anything, anti-IL-1β antibodies seem to slightly increase iNOS expression. The independence of IL-1β for iNOS activation in response to FLA-BSDot was supported by data obtained with BMDM, which up-regulated iNOS expression (Fig. 5C) without any detectable IL-1β production (data not shown). Similar results were obtained with neutralizing anti-IL-18 or the combination of both anti-IL-1β and anti-IL-18 antibodies (Fig. 5, B and C). It is important to emphasize that IL-18 was not found in the supernatants from FLA-BSDot-stimulated PMs or BMDMs. Together, our data indicate that cytosolic flagellin induces iNOS activation by a mechanism dependent on Nlrc4/Naip5-caspase-1 axis but independent of IL-1β and IL-18.
FIGURE 5.
Induction of iNOS expression by cytosolic flagellin is independent of IL-1β and IL-18. PMs (A, B) and BMDM (C) from C57BL/6 were stimulated with 3 μg/ml purified free (FLA-BS) or cytosolic (FLA-BSDot) flagellin from B. subtilis in the presence or absence of neutralizing anti-IL-1β (1 μg/ml) or anti-IL-18 (5 μg/ml). After 24 h, iNOS expression was evaluated in cell extracts by Western blot, and IL-1β secretion was determined in culture supernatants by ELISA. Numbers represent the mean ± S.D. of triplicate samples. ***, p < 0.001 when compared with control group. ###, p < 0.001 when compared with FLA-BSDot in the presence of neutralizing anti-IL-1β. Data are representative of three independent experiments.
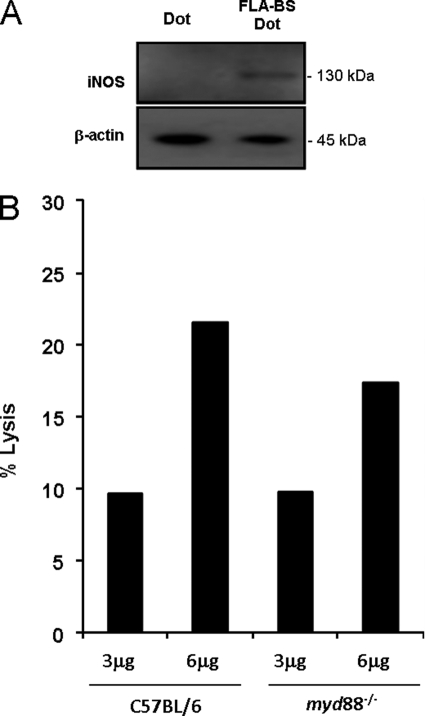
FLA-BSDot-induced iNOS Expression Is Independent of MYD88
Because IL-1 and IL-18 receptors as well as TLR5 signaling depend on MYD88 adaptor molecule (3, 8, 25), PMs from myd88−/− mice were stimulated with FLA-BSDot to evaluate the influence of this pathway on cytosolic flagellin-induced iNOS activation. As shown in Fig. 6, FLA-BSDot-mediated iNOS expression and LDH release are also observed in the absence of MYD88, supporting our claim that these responses are independent of IL-1 and IL-18 receptors and TLR5 signaling. Moreover, the flagellin-induced TLR-independent iNOS-activation seems to participate in the restriction of L. pneumophila infection. Selective inhibition of iNOS by aminoguanidine increased at the same extent the growth of wild-type L. pneumophila in both WT and myd88−/− C57BL/6 macrophages (supplemental Fig. 4A). However, aminoguanidine had no effect on the growth of the flagellin-deficient mutant FlaA L. pneumophila in either macrophage (supplemental Fig. 4B). Together, these results indicate that NO induced by flagellin through a TLR-independent pathway participate in the control of L. pneumophila infection by macrophages.
FIGURE 6.
MYD88 is not required for cytosolic flagellin-induced macrophage responses. PMs from Myd88−/− mice were stimulated with 3 μg/ml purified cytosolic flagellin from B. subtilis (FLA-BSDot). After 6 and 24 h macrophage lysis was monitored by LDH release in culture supernatants (B), and iNOS expression was evaluated in cell extracts by Western blot (A), respectively. Data are representative of three independent experiments.
DISCUSSION
Flagellin is one of the rare protein structures sensed by the innate immune system and can activate both TLR and NLR receptors. Extracellular flagellin is recognized by transmembrane TLR5 (8), whereas cytosolic flagellin is sensed by Naip5 and Nlrc4 (9–13, 26, 27). Interestingly, TLR5 seems to broadly recognize flagellated bacteria, whereas sensing of Nlrc4 and Naip5 depends on specialized transport systems present in virulent bacteria, such as the type III secretion systems of Salmonella (10, 27) and the type IV (T4SS) of Legionella (9, 13). These secretion systems allow the delivery of bacterial products directly into the host cell cytoplasm. In experimental models, Nlrc4- and Naip5-induced caspase-1 activation can be achieved by delivering purified flagellin into the host cell cytosol by means of cationic liposomes or by transducing cells with appropriated gene-expression systems (10, 27, 28). Consistent with this view, TLR5 stimulation primarily leads to the activation of microbicidal effector mechanisms that are supposedly effective in controlling less aggressive, non-pathogenic bacterial infection. In contrast, a more dramatic effector mechanism consisting of a Nlrc4/Naip5-caspase-1-induce inflammatory cell death is observed only in infections with virulent bacteria (7).
Therefore, the existence of different PRRs capable of sensing the same agonist or different agonists present on the same pathogen is obviously beneficial to the host. The combined action of distinct, non-redundant, specialized responses should lead to a synergistic, more robust immunity against potential pathogens. Extracellular and intracellular recognition of flagellin induces such distinct macrophage responses. High levels of IL-6 were observed only with FLA-BS, whereas IL-1β secretion and pyroptosis were detected only with FLA-BSDot (Fig. 1 and supplemental Fig. 2). However, as the expression of PRRs is not ubiquitous and, therefore, not all host cells are equally prepared to properly recognize and handle potential pathogens, it should be interesting for the host immunity to have different PRR family sharing at least some microbicidal mechanisms.
iNOS activation by TLRs can be considered one of the major effector mechanism for controlling intracellular pathogens (29). TLRs are able to recruit NF-κB and IRF transcription factors, both necessary to induce iNOS expression and activation (30, 31). In contrast, Nod1 and Nod2 are the only NLR members known to activate NF-κB (32), resulting in the activation of a large repertoire of genes, including iNOS (33). Nlrps, Naip5, and Nlrc4 inflammasomes, on the other hand, mediate caspase-1 activation and the consequent IL-1β and IL-18 secretion and/or a programmed cell death called pyroptosis (1, 7, 34). As macrophage activation by cytosolic flagellin occurs independently of TLR5 (10, 27), our data demonstrating iNOS up-regulation by FLA-BSDot (Fig. 2) highlights the plasticity of innate immunity recognition strategies, adding a role for NO on inflammasome-mediated microbicidal pathways.
Despite the fact that both Nlrc4 and Naip5 can induce caspase-1 activation, IL-1β secretion, and pyroptosis in response to L. pneumophila and S. typhimurium flagellin, it is still obscure how exactly Nlrc4 and Naip5 individually participate in the recognition of flagellin and in the assembly of the inflammasomes. Nlrc4 can bind directly to pro-caspase-1 via a CARD-CARD homotypic interaction, but the molecular partner(s) of Naip5 remains to be solved. Interesting, Naip5 do not participate in every function of the Nlrc4-inflammasome. Nlrc4 has been shown to be crucial in host defense for a number of pathogens such as Salmonella, Legionella, Pseudomonas, and Shigella, and its genetic defect cannot be substituted by a functional Naip5 (9–12, 17, 35–37). On the other hand, Naip5 is indispensable for the resistance to L. pneumophila infection (13). Interestingly, FLA-BSDot-induced iNOS expression was significantly reduced in Nlrc4-deficient macrophages and completely abolished in the absence of a functional Naip5 (Fig. 3). By using highly sensitive techniques such as real-time RT-PCR, we were able to detect a slight transcriptional induction of iNOS in the mice harboring a partially mutant A/J Naip5 allele and still significantly less when compared with Nlrc4-deficient macrophages. The partial functionality of Naip5 from A/J mice (11) could account for the observed flagellin-dependent iNOS mRNA induction, an effect that possibly would not be observed in Naip5−/− mice. Another possible explanation for these results is that Naip5 plays an additional Nlrc4-independent role in the post-transcriptional regulation of iNOS.
Importantly, we show here for the first time that, distinct from free flagellin, the activation of iNOS by cytosolic flagellin via Nlrc4/Naip5 is dependent on caspase-1 (Fig. 4). The genetic or pharmacological inactivation of caspase-1 resulted in the abrogation of iNOS protein and mRNA expression in response to FLA-BSDot. Caspase-1 promotes the release of IL-1β and IL-18, which in turn can activate iNOS (25). In fact, it was recently shown that IFN-γ- and TNF-α-induced iNOS activation in astrocytes and cerebral endothelial cells is dependent on caspase-1-mediated IL-1β secretion (38). However, our data show that neutralization of IL-1β and/or IL-18 with specific antibodies resulted in a rather increased iNOS expression by FLA-BSDot, ruling out the requirement for these cytokines in flagellin-mediated iNOS up-regulation (Fig. 5). These observations are reinforced by the fact that myd88−/− macrophages are able to express iNOS in response to cytosolic flagellin (Fig. 6) besides their deficiency in both IL-1 and IL-18 receptor, signaling pathways. Also, the observed up-regulation of iNOS cannot be attributed to unspecific macrophage activation as anti-IL-1β- and anti-IL-18-neutralizing antibodies were not able to activate iNOS in the absence of flagellin (Fig. 5C). Moreover, anti-IL-1β and anti-IL-18 have no effect on IL-6 produced in response to FLA-BSDot, showing their particular involvement on iNOS modulation (supplemental Fig. 3). These data indicate that IL-1β and IL-18 are not required for iNOS activation in response to FLA-BSDot but do not discard their participation in some aspects of iNOS regulation.
Because the iNOS promoter contains binding sites for NF-κB and IRF-1 (30, 31), additional investigation is required to elucidate whether the up-regulation of iNOS by caspase-1 involves the activation of either or both transcription factors. Recently, it was demonstrated that IRF-1 and IRF-8 transcription factors seem to be regulated during Naip5 and Nlrc4-mediated sensing of L. pneumophila (39). Moreover, these authors observed that, in the absence of flagellin, there is a defective activation of IL-12p40 and iNOS, the IRF-1 and IRF-8 transcriptional targets. Despite the clear evidences of Nlrc4 and Naip5-mediated caspase-1 and IRF-1/IRF-8 activation, it is not clear whether these events are correlated.
Additionally, although Nlrc4 can directly interact and activate caspase-1 via its N-terminal CARD domain (40, 41), the activation of caspase-1 by Naip5 possibly requires interaction with other proteins since Naip5 possesses a BIR domains, which for other inhibitor of apoptosis proteins family members are known to interact with different proteins (42). Regardless of the possible interaction between Naip5 and Nlrc4 (13), these NLR do not share all caspase-1-mediated functions (11), opening the possibility of interactions among Naip5 and other unknown proteins. Because multiple complexes containing more than one NLR member can be found within the same cell (34, 43), the sensing of cytosolic flagellin could lead to multiprotein complexes formation including Nlrc4, Naip5, caspase-1, IRF-1, IRF-8, or alternatively, to other NLR members and proteins involved in the activation of iNOS.
Although there are still missing pieces in the Nlrc4 and Naip5 inflammasome complexes, their role in macrophage clearance of bacterial pathogens is clear. Defects on Nlrc4 or caspase-1 render macrophages susceptible to infections such as S. typhimurium (10, 27), Pseudomonas aeruginosa (35, 36) and Shigella flexneri (37), whereas Naip5 deficiency lead to susceptibility against L. pneumophila infection (13). Because the same defects also lead to resistance to caspase-1-mediated cell death, pyroptosis seems to be the major effector mechanism to control these pathogens. However, the requirement of macrophage death during an infection and the relevance of pyroptosis in vivo are still matters of debate (7). In this regard, a Naip5/Nlrc4-caspase-1-dependent caspase-7 activation and phagosome maturation responsible for the inhibition of bacteria replication (44) was recently described.
Here, we are describing a novel pathway for iNOS activation orchestrated by the Naip5 and Nlrc4 and operated through caspase-1, adding a new effector mechanism mediated by inflammasomes. Importantly, this pathway cooperate with TLR-induced NO production for the control of L. pneumophila since pharmacological inhibition of iNOS renders higher susceptibility of both WT C57BL/6 and myd88−/− macrophages to flagellated L. pneumophila but not to flagellin-mutant bacteria (supplemental Fig. 4). The role of iNOS to the resistance against L. pneumophila was previously demonstrated (45, 46). However, iNOS seem not to be involved in the control of L. pneumophila by macrophages from Naip5-deficient A/J mice (47), corroborating our data showing that the TLR-independent iNOS activation is observed only in the presence of functional flagellin. It remains to be determined how caspase-1-mediated cellular events, such as pyroptosis, pro-inflammatory cytokines secretion, caspase-7, and iNOS activation cooperate to the control of infections.
Supplementary Material
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP-Brazil) and the Brazilian Research Council (CNPq).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- PRR
- pattern recognition receptor
- NLR
- NOD-like receptor
- TLR
- Toll-like receptor
- IRF
- IFN-responsive factors
- Naip5
- neuronal apoptosis inhibitory protein 5
- CARD
- caspase activation recruitment domain
- iNOS
- inducible nitric-oxide synthase
- BMDM
- bone marrow-derived macrophage
- PM
- peritoneal macrophage
- z-YVAD-fmk
- benzyloxycarbonyl-YVAD-fluoromethyl ketone
- PAMP
- pathogen-associated molecular pattern
- DOT
- DOTAP (lipidic vesicles).
REFERENCES
- 1.Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. (2009) Nat. Immunol. 10, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medzhitov R. (2007) Nature 449, 819–826 [DOI] [PubMed] [Google Scholar]
- 3.Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki A., Medzhitov R. (2004) Nat. Immunol. 5, 987–995 [DOI] [PubMed] [Google Scholar]
- 5.Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S. J., Moran A. P., Fernandez-Luna J. L., Nuñez G. (2003) J. Biol. Chem. 278, 5509–5512 [DOI] [PubMed] [Google Scholar]
- 6.Inohara, Chamaillard, McDonald C., Nuñez G. (2005) Annu. Rev. Biochem. 74, 355–383 [DOI] [PubMed] [Google Scholar]
- 7.Bortoluci K. R., Medzhitov R. (2010) Cell. Mol. Life Sci. 67, 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., Eng J. K., Akira S., Underhill D. M., Aderem A. (2001) Nature 410, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 9.Amer A., Franchi L., Kanneganti T. D., Body-Malapel M., Ozören N., Brady G., Meshinchi S., Jagirdar R., Gewirtz A., Akira S., Núñez G. (2006) J. Biol. Chem. 281, 35217–35223 [DOI] [PubMed] [Google Scholar]
- 10.Franchi L., Amer A., Body-Malapel M., Kanneganti T. D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., Grant E. P., Núñez G. (2006) Nat. Immunol. 7, 576–582 [DOI] [PubMed] [Google Scholar]
- 11.Lightfield K. L., Persson J., Brubaker S. W., Witte C. E., von Moltke J., Dunipace E. A., Henry T., Sun Y. H., Cado D., Dietrich W. F., Monack D. M., Tsolis R. M., Vance R. E. (2008) Nat. Immunol. 9, 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., Dixit V. M. (2004) Nature 430, 213–218 [DOI] [PubMed] [Google Scholar]
- 13.Zamboni D. S., Kobayashi K. S., Kohlsdorf T., Ogura Y., Long E. M., Vance R. E., Kuida K., Mariathasan S., Dixit V. M., Flavell R. A., Dietrich W. F., Roy C. R. (2006) Nat. Immunol. 7, 318–325 [DOI] [PubMed] [Google Scholar]
- 14.Brightbill H. D., Libraty D. H., Krutzik S. R., Yang R. B., Belisle J. T., Bleharski J. R., Maitland M., Norgard M. V., Plevy S. E., Smale S. T., Brennan P. J., Bloom B. R., Godowski P. J., Modlin R. L. (1999) Science 285, 732–736 [DOI] [PubMed] [Google Scholar]
- 15.Thoma-Uszynski S., Stenger S., Takeuchi O., Ochoa M. T., Engele M., Sieling P. A., Barnes P. F., Rollinghoff M., Bolcskei P. L., Wagner M., Akira S., Norgard M. V., Belisle J. T., Godowski P. J., Bloom B. R., Modlin R. L. (2001) Science 291, 1544–1547 [DOI] [PubMed] [Google Scholar]
- 16.Bogdan C. (2001) Trends Cell Biol. 11, 66–75 [DOI] [PubMed] [Google Scholar]
- 17.Lara-Tejero M., Sutterwala F. S., Ogura Y., Grant E. P., Bertin J., Coyle A. J., Flavell R. A., Galán J. E. (2006) J. Exp. Med. 203, 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright E. K., Goodart S. A., Growney J. D., Hadinoto V., Endrizzi M. G., Long E. M., Sadigh K., Abney A. L., Bernstein-Hanley I., Dietrich W. F. (2003) Curr. Biol. 13, 27–36 [DOI] [PubMed] [Google Scholar]
- 19.Zamboni D. S., Rabinovitch M. (2003) Infect. Immun. 71, 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bastos K. R., Alvarez J. M., Marinho C. R., Rizzo L. V., Lima M. R. (2002) J. Leukoc. Biol. 71, 271–278 [PubMed] [Google Scholar]
- 21.Simberg D., Weisman S., Talmon Y., Barenholz Y. (2004) Crit. Rev. Ther. Drug Carrier Syst. 21, 257–317 [DOI] [PubMed] [Google Scholar]
- 22.Weinlich R., Bortoluci K. R., Chehab C. F., Serezani C. H., Ulbrich A. G., Peters-Golden M., Russo M., Amarante-Mendes G. P. (2008) Cell Death Differ. 15, 1901–1909 [DOI] [PubMed] [Google Scholar]
- 23.Misko T. P., Moore W. M., Kasten T. P., Nickols G. A., Corbett J. A., Tilton R. G., McDaniel M. L., Williamson J. R., Currie M. G. (1993) Eur. J. Pharmacol. 233, 119–125 [DOI] [PubMed] [Google Scholar]
- 24.Diez E., Lee S. H., Gauthier S., Yaraghi Z., Tremblay M., Vidal S., Gros P. (2003) Nat. Genet. 33, 55–60 [DOI] [PubMed] [Google Scholar]
- 25.Dinarello C. A. (2009) Annu. Rev. Immunol. 27, 519–550 [DOI] [PubMed] [Google Scholar]
- 26.Ren T., Zamboni D. S., Roy C. R., Dietrich W. F., Vance R. E. (2006) PLoS Pathog. 2, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miao E. A., Alpuche-Aranda C. M., Dors M., Clark A. E., Bader M. W., Miller S. I., Aderem A. (2006) Nat. Immunol. 7, 569–575 [DOI] [PubMed] [Google Scholar]
- 28.Silveira T. N., Zamboni D. S. (2010) Infect. Immun. 78, 1403–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogdan C. (2001) Nat. Immunol. 2, 907–916 [DOI] [PubMed] [Google Scholar]
- 30.Kamijo R., Harada H., Matsuyama T., Bosland M., Gerecitano J., Shapiro D., Le J., Koh S. I., Kimura T., Green S. J. (1994) Science 263, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 31.Lowenstein C. J., Alley E. W., Raval P., Snowman A. M., Snyder S. H., Russell S. W., Murphy W. J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9730–9734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tattoli I., Travassos L. H., Carneiro L. A., Magalhaes J. G., Girardin S. E. (2007) Semin. Immunopathol. 29, 289–301 [DOI] [PubMed] [Google Scholar]
- 33.Tötemeyer S., Sheppard M., Lloyd A., Roper D., Dowson C., Underhill D., Murray P., Maskell D., Bryant C. (2006) J. Immunol. 176, 4804–4810 [DOI] [PubMed] [Google Scholar]
- 34.Ting J. P., Willingham S. B., Bergstralh D. T. (2008) Nat. Rev. Immunol. 8, 372–379 [DOI] [PubMed] [Google Scholar]
- 35.Miao E. A., Ernst R. K., Dors M., Mao D. P., Aderem A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2562–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutterwala F. S., Mijares L. A., Li L., Ogura Y., Kazmierczak B. I., Flavell R. A. (2007) J. Exp. Med. 204, 3235–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T., Franchi L., Toma C., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Inohara N., Sasakawa C., Nuñez G. (2007) PLoS Pathog. 3, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jüttler E., Bonmann E., Spranger M., Kolb-Bachofen V., Suschek C. V. (2007) Mol. Cell. Neurosci. 34, 612–620 [DOI] [PubMed] [Google Scholar]
- 39.Fortier A., Doiron K., Saleh M., Grinstein S., Gros P. (2009) Infect. Immun. 77, 4794–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poyet J. L., Srinivasula S. M., Tnani M., Razmara M., Fernandes-Alnemri T., Alnemri E. S. (2001) J. Biol. Chem. 276, 28309–28313 [DOI] [PubMed] [Google Scholar]
- 41.Martinon F., Tschopp J. (2007) Cell Death Differ. 14, 10–22 [DOI] [PubMed] [Google Scholar]
- 42.Mace P. D., Shirley S., Day C. L. (2010) Cell Death Differ. 17, 46–53 [DOI] [PubMed] [Google Scholar]
- 43.Bergsbaken T., Fink S. L., Cookson B. T. (2009) Nature Reviews 7, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akhter A., Gavrilin M. A., Frantz L., Washington S., Ditty C., Limoli D., Day C., Sarkar A., Newland C., Butchar J., Marsh C. B., Wewers M. D., Tridandapani S., Kanneganti T. D., Amer A. O. (2009) PLoS Pathog. 5, e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summersgill J. T., Powell L. A., Buster B. L., Miller R. D., Ramirez J. A. (1992) J. Leukoc. Biol. 52, 625–629 [DOI] [PubMed] [Google Scholar]
- 46.Skerrett S. J., Martin T. R. (1996) Infect. Immun. 64, 3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gebran S. J., Yamamoto Y., Newton C., Klein T. W., Friedman H. (1994) Infect. Immun. 62, 3197–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.