Abstract
Kallikrein-related peptidases (KLKs) play a central role in skin desquamation. They are tightly controlled by specific inhibitors, including the lymphoepithelial Kazal-type inhibitor (LEKTI) encoded by SPINK5 and LEKTI-2 encoded by SPINK9. Herein, we identify SPINK6 as a selective inhibitor of KLKs in the skin. Unlike LEKTI but similar to LEKTI-2, SPINK6 possesses only one typical Kazal domain. Its mRNA was detected to be expressed at low levels in several tissues and was induced during keratinocyte differentiation. Natural SPINK6 was purified from human plantar stratum corneum extracts. Immunohistochemical analyses revealed SPINK6 expression in the stratum granulosum of human skin at various anatomical localizations and in the skin appendages, including sebaceous glands and sweat glands. SPINK6 expression was decreased in lesions of atopic dermatitis. Using KLK5, KLK7, KLK8, KLK14, thrombin, trypsin, plasmin, matriptase, prostasin, mast cell chymase, cathepsin G, neutrophil elastase, and chymotrypsin, inhibition with recombinant SPINK6 was detected only for KLK5, KLK7, and KLK14, with apparent Ki values of 1.33, 1070, and 0.5 nm, respectively. SPINK6 inhibited desquamation of human plantar callus in an ex vivo model. Our findings suggest that SPINK6 plays a role in modulating the activity of KLKs in human skin. A selective inhibition of KLKs by SPINK6 might have therapeutic potential when KLK activity is elevated.
Keywords: Gene Expression, Kallikrein, Protease Inhibitor, Serine Protease, Skin, LEKTI, Netherton Syndrome, Atopic Dermatitis, Desquamation, Epidermal Barrier
Introduction
The skin protects us from water loss and mechanical damage. The surface-exposed epidermis, a self-renewing stratified squamous epithelium composed of several layers of keratinocytes, is most important for the barrier defense against these challenges. Recent discoveries have highlighted the balance of proteases and protease inhibitors as key players in both desquamation processes and epidermal barrier functions (1).
Human tissue kallikreins, or kallikrein-related peptidases (KLKs),3 are the largest family of trypsin- or chymotrypsin-like secreted serine proteases, which are encoded by 15 genes on chromosome region 19q13.4 (2). At least eight KLKs are expressed in normal skin, among which KLK5, KLK7, KLK8, and KLK14 have been reported to be most important (3–6). KLKs are capable of cleaving corneodesmosomes (7–10) and are thought to be key regulators of the desquamation process. The activity of KLKs is regulated by pH and specific protease inhibitors. The importance of epithelial protease inhibitors has been revealed impressively in Netherton syndrome (OMIM 256500), an autosomal recessive disorder caused by mutations in the gene SPINK5 (serine protease inhibitor Kazal-type 5) (11). Netherton syndrome presents as an ichthyosiform dermatosis with variable erythroderma, hair shaft defects (bamboo hair), atopic features, and growth retardation (12). Lymphoepithelial Kazal-type inhibitor (LEKTI) (13), the product of SPINK5, includes in its primary structure 15 different serine protease inhibitory domains (13). Domains 15 and 2 each comprise a typical Kazal-type structure, whereas the other domains lack a disulfide bridge. Recently, LEKTI-2, encoded by SPINK9, was reported as a selective KLK5 inhibitor expressed at palmoplantar sites (14, 15). LEKTI-2 consists of a single typical Kazal-type domain, which exhibits the highest homology to LEKTI/SPINK5 domain 15. This suggests that a complex balance exists between the KLK cascades and SPINK family members in human skin, maintaining normal epithelial barrier functions. Taking the multiple skin-expressed KLKs members into account, we hypothesized that more SPINK members are present in human skin. Herein, we identified SPINK6 as a selective inhibitor of KLKs in human skin.
EXPERIMENTAL PROCEDURES
Materials
Normal skin specimens were taken from routine clinical work at the Department of Dermatology, University Hospital Schleswig-Holstein, and represent tumor-free margins of benign melanocytic tumors surgically removed from patients. Restriction endonucleases were from New England Biolabs (Frankfurt, Germany). KLKs were purchased from R&D Systems (Minneapolis, MN). All other proteases, primers, substrates, and chemicals were purchased from Sigma (Taufkirchen, Germany) if not indicated otherwise.
Bioinformatics Homology
A search was done using the tBLASTn algorithm as provided by the Ensembl BlastView server. Determination of gene structure was done using the BLAT algorithm (16) as provided by the Ensembl UCSC Genome Browser. Subsequent sequence manipulations utilized the online BLAST 2 sequences (17). Protein domains were discovered on the SMART server (18). Multiple sequence alignments were performed using the ClustalW2 program and edited with GeneDoc.
Rapid Amplification of cDNA Ends (RACE)
Total RNA was obtained from cultured human foreskin-derived keratinocytes using TRIzol reagent (Invitrogen, Hamburg, Germany). After treatment with RNase-free DNase I (Roche Diagnostics, Mannheim, Germany) to exclude contamination with genomic DNA, 2 μg of DNA-free total RNA was used for the first-strand cDNA synthesis for RACE using a SMART RACE cDNA amplification kit (Clontech, Heidelberg, Germany) according to the manufacturer's protocol. 5′-RACE was performed with a gene-specific antisense primer (5′-AGG CAC ATT TAT TGC CAT ATG TCT GGC CAT C-3′), whereas 3′-RACE was done with a gene-specific sense primer (5′-GTG AGT TCC AGG ACC CCA AGG TCT ACT G-3′) essentially according to the manufacturer's protocol. PCR cycles were performed under the following conditions: 1 min at 95 °C, five cycles of 20 s at 95 °C and 3 min at 72 °C, 5 cycles of 20 s at 95 °C and 3 min at 70 °C, 25 cycles of 20 s at 95 °C and 3 min at 68 °C, and a final extension of 10 min at 72 °C. Subsequently, the PCR product was diluted 50-fold into Milli-Q water and used as a template for a nested PCR with a nested primer (for 5′-nest, 5′-GCC ACA GTG TGG GTT AGA TTC CCG AGT G-3′; and for 3′-nest, 5′-CCA CAC TGT GGC TCT GAT GGC CAG A-3′) under the following conditions: 1 min at 95 °C, 30 cycles of 20 s at 95 °C and 3 min at 70 °C, and a final extension of 10 min at 70 °C. The amplified fragment was gel-purified and subcloned into the pGEM-T vector (Promega, Mannheim), followed by full sequencing in both directions.
RT-PCR
Total RNA was isolated from cultured human foreskin-derived keratinocytes (HaCaT cell line) and skin using TRIzol reagent (Invitrogen). Other total RNAs from different tissues were obtained from Clontech. A total of 2 μg of total RNA from human tissues or culture was reverse-transcribed with an oligo(dT)18 primer and Superscript II RNase H− reverse transcriptase (Invitrogen). One pair of gene-specific PCR primers (forward primer, 5′-ACC TCA GCT GGA CAA AGC AG-3′; and reverse primer, 5′-TGG CAA GTC ACC AAG AAA CA-3′) was designed to amplify a 322-bp product and to span all three exon-intron boundaries. PCR was carried out with the Advantage 2 PCR polymerase (Clontech), and amplicons were analyzed by 2.0% agarose gel electrophoresis. As an internal control of cDNA templates, the glyceraldehyde-3-phosphate dehydrogenase housekeeping gene was assessed with each cDNA in a separate PCR as described (14). For quantitative real-time RT-PCR, an assay was carried out with the same primer pair as described above and the SYBR® Premix Ex TaqTM kit (Takara Bio, Heidelberg) in a fluorescence thermocycler (LightCycler, Roche Molecular Biochemicals, Hamburg) following the instructions of the manufacturer. Cycling conditions were as follows: 45 cycles at 95 °C for 5 s, 69 °C for 20 s (“touchdown” of −1 °C/cycle to 63 °C), and 72 °C for 20 s. To calculate the relative transcript amplification, the glyceraldehyde-3-phosphate dehydrogenase housekeeping gene was measured with each cDNA in a separate PCR. The data from triplicate samples were analyzed with GraphPad Prism 4. For absolute DNA quantification, reactions were carried out with different concentrations of linearized plasmid DNA containing the insert in parallel to the samples that should be quantified. The data from three independent observations were analyzed with the software and expressed as means ± S.D. of mRNA copies relative to 10 ng of total RNA.
Electrospray Ionization Mass Spectrometry (ESI-MS) Analysis
Protein and peptide mass determinations were performed by ESI-MS analyses using a quadrupole TOF hybrid mass spectrometer (Q-TofTM II, Waters Micromass, Milford, MA) equipped with an orthogonal electrospray source (Z-spray) operated in positive ionization mode. For MS analysis, aliquots (2–10 μl) of HPLC fractions containing the sample were diluted with 100 μl of carrier solvent (50:50 acetonitrile/water containing 0.2% formic acid or 60:40 methanol and 10 mm ammonium formate, pH 6.5) and infused into the electrospray source at a rate of 10–20 μl/min. Sodium iodide was used for mass calibration for a calibration range of m/z 100–2000. The capillary potential was set to 3.5 or 4 kV, and cone voltages between 25 and 75 V were chosen. The cone temperature was set to 80 °C; the desolvation temperature was 150 °C; and the ESI gas was nitrogen. The charge-to-mass ratio of ions was scanned in the range of 280 to 2000. Acquisition and data analysis were performed using the MassLynx 4 software package supplied by Waters Micromass. Mass spectra were averaged typically over 2–10 scans (2–20 s/scan). The multiply charged raw data of intact proteins were background-subtracted and deconvoluted using Maximum Entropy 1 (MaxEnt1) to obtain singly charged ion mass spectra to determine average molecular masses of intact proteins. The raw combined spectral data from small peptides or obtained after tandem mass spectrometry (MS/MS) fragmentation of selected precursor ions were background-subtracted and subjected to Maximum Entropy 3 (MaxEnt3) deconvolution to determine monoisotopic molecular masses. Sample identity was determined by data base search analysis of peptide fragment mass patterns and/or after de novo sequencing of tryptic peptide fragments. All mass fingerprint and MS/MS data were searched against the human protein data base using the Mascot program (Matrix Science, Boston, Massachusetts). Peptide sequences were directly determined from MS/MS data using the software program PepSeq from the MassLynx 4 software package. PepSeq-derived peptide sequences were analyzed with the NCBI BLAST Protein Database search program.
Recombinant Protein Production
The recombinant expression of SPINK6 cDNA (rSPINK6; residues 25–80) in Escherichia coli was performed by molecular subcloning of SPINK6 cDNA into the prokaryotic expression vectors pET-32a (Novagen, North Ryde, Australia) and pET-SUMO (Invitrogen) as described previously (19). The SUMO-His-tagged fusion protein was digested with SUMO protease 1 (LifeSensors Inc.) according to the manufacturer's suggestions. To subclone into the expression vector pET-32a, PCR was performed with Pfu DNA polymerase (Promega). The inserts were gel-purified and inserted into the pET-SUMO vector. Specific primer pairs used in this study were as follows: p32-SPINK6-forward, ACT GAG ATC TGG GTA CCG ACG ACG ACG ACA AGC AGG TTG ACT GTG GTG AGT TC; p32-SPINK6-reverse, ATT TGC GGC CGC TCA GCA TTT TCC AGG ATG CTT; pSUMO-SPINK6-forward, GGA GGA CAG GTT GAC TGT G; and pSUMO-SPINK6-reverse, TCA GCA TTT TCC AGG ATG CTT. All positive clones were identified and verified by sequencing. The plasmids were introduced into the E. coli host strain BL21(DE3)pLysS (Novagen). Subsequently, these were grown at 37 °C in tryptic soy broth medium containing appropriate antibiotics. Expression of the recombinant protein was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. Bacteria were harvested by centrifugation at 5000 × g for 5 min at 4 °C, lysed by sonication, and then centrifuged at 18,000 × g for 30 min at 4 °C (Beckman Coulter, Krefeld, Germany). Recombinant proteins were trapped with Ni2+ prepared columns (Macherey-Nagel, Düren, Germany), and Ni2+ affinity column-bound proteins were subjected to preparative reversed-phase HPLC with a column (SP250/10 Nucleosil 300-7 C8, Macherey-Nagel) that was previously equilibrated with 0.1% (v/v) TFA in HPLC-grade water containing 10% acetonitrile. The polyhistidine-tagged fusion proteins were eluted with a gradient of increasing concentrations of acetonitrile containing 0.1% (v/v) TFA (flow rate, 3 ml/min). Fractions containing UV light (215 nm)-absorbing material were collected, lyophilized, and analyzed by ESI-quadrupole TOF-MS (Micromass, Manchester, United Kingdom). Purified histidine-tagged SUMO fusion proteins were then digested with SUMO protease 1 according to the manufacturer's suggestion. The target peptide was purified by reversed-phase HPLC with a Jupiter 5μ-C4–300A HPLC column (Phenomenex, Aschaffenburg, Germany) equilibrated with 0.1% TFA in 10% acetonitrile. Peptides were eluted with a gradient of increasing concentrations of acetonitrile containing 0.1% (v/v) TFA (flow rate, 0.5 ml/min). Fractions of each peak were collected. The purity of recombinant fusion proteins was determined by SDS-PAGE. Masses of fusion proteins and rSPINK6 peptides were analyzed by ESI-quadrupole TOF-MS. Concentrations of proteins present in HPLC fractions were estimated by ultraviolet absorbance integration at 215 nm using ubiquitin for calibration.
Production of Antibody
Polyclonal antiserum against human rSPINK6 (residues 25–80) was generated in a goat as described previously (14) with minor modifications. Briefly, a total of 1.0 mg of fusion protein (pET-32a-SPINK6) was conjugated to maleimide-activated keyhole limpet hemocyanin (1:1, w/w) and subsequently mixed with 500 μg of pET-32a-SPINK6 for use as immunogens. Antisera were affinity-purified by absorption first against protein G. Unspecific antibodies were eliminated by collecting the flow-through after applying the antibody to a pET-tag-coupled column. Finally, the antibody was affinity-purified against rSPINK6 that was covalently bound to a HiTrap NHS-activated HP 1-ml column (Amersham Biosciences, Freiburg, Germany). Specificity was tested by Western blot analyses using rSPINK6, naturally isolated LEKTI-2 (data not shown), and SPINK6 as well as stratum corneum extracts.
Isolation of SPINK6 from Stratum Corneum Extracts
Pooled heel stratum corneum was extracted similarly as described (14). After diafiltration (Amicon filters, 3-kDa cut-off) against 10 mm Tris citrate buffer, pH 8.0, extracts were applied to an anti-SPINK6 affinity column. Affinity-purified anti-human SPINK6 polyclonal antibodies were covalently bound to HiTrap NHS-activated HP 1-ml columns. Unspecific material that was weakly bound to the affinity column was first eluted with 2 m NaCl. Strongly bound material was eluted by pH shift using 0.1 m glycine HCl, pH 3. Material that was strongly bound to the affinity column was further purified by reversed-phase HPLC (Jupiter C18 column, Phenomenex). Each HPLC fraction was analyzed by ESI-MS. Lyophilized fractions were subjected to tributylphosphine/vinylpyridine gas-phase reduction and alkylation (20). After dissolving samples in 20 μl of NH4HCO3 with 2 μl of acetonitrile, peptides were digested with 4.5 ng/μl trypsin at 37 °C for 2 h and then further analyzed by MS/MS.
Western Blot Analyses
For Western blot analysis, stratum corneum extracts from healthy volunteers, rSPINK6, and HPLC fractions containing natural SPINK6 were loaded onto a 12% SDS/Tricine-polyacrylamide gel. Proteins were transferred to a Protran-nitrocellulose membrane (Schleicher & Schuell BioScience GmbH, Dassel, Germany), blocked for 1 h in blocking buffer (5% (w/v) BSA in PBS containing 0.05% Tween 20 (PBST)), and then incubated for 18 h at 4 °C in 3% (w/v) BSA in PBST containing affinity-purified anti-SPINK6 antibody (1 μg/ml). The membrane was washed with PBST six times for 5 min each and then incubated for 45 min in PBST containing a 1:10,000 dilution of goat anti-mouse IgG-HRP conjugate (Dianova, Hamburg). After further washing, the membrane was incubated for 5 min with chemiluminescent peroxidase substrate (Roche Diagnostics, Mannheim, Germany) and visualized using a Diana III cooled CCD camera imaging system (Raytest, Straubenhardt, Germany).
Immunohistochemistry
Immunohistochemical staining of paraffin-embedded tissue was performed as described previously (21) using affinity-purified goat anti-SPINK6 polyclonal antibody (final concentration of 10 ng/μl) followed by a biotinylated rabbit anti-goat IgG secondary antibody (1:100 dilution; DakoCytomation) and subsequently incubated with Vector Universal DAB kit. Counterstaining was done with hematoxylin. A specificity test of the anti-SPINK6 antibody was performed using rSPINK6 peptide to block the primary antibody. Negative controls were established using preimmune goat sera to stain sections.
Protease Inhibition Assays
All protease assays were performed by measuring chromogenic substrate release by proteases in the buffer recommended by the manufacturer as described previously (14). Specific concentrations of proteases, substrates, and inhibitors are indicated in Table 1. Recombinant human kallikreins were purchased from R&D Systems and activated according to the manufacturer's protocol. All enzymes were incubated with increasing concentrations of rSPINK6 for 5 min in 0.1 m Tris, 150 mm NaCl, 5 mm EDTA, and 0.05% Tween 20, pH 8.0. After incubation, a solution of the substrate in the same buffer was added, resulting in a final concentration of 2 nm KLK, 1 mm tosyl-Gly-Pro-Arg p-nitroanilide (KLK5, KLK8, and KLK14) or S-2586 (KLK7), and SPINK6 in a total volume of 200 μl. The reaction progress was monitored by measuring the absorbance at 405 nm using a Tecan Sunrise microplate reader in kinetic mode. Initial reaction velocities for each tested inhibitor concentration were determined by linear regression and were subsequently plotted against inhibitor concentration. Data were further analyzed according to the Morrison tight-binding inhibition model. For determination of Km, enzymes were mixed with varying substrate concentrations. Mixtures contained 2 nm KLK and the respective substrate in a concentration range of 0.2–6 mm. Initial velocities were determined by linear regression and plotted against respective substrate concentrations. Collected data were analyzed by nonlinear regression to the Michaelis-Menten equation using the GraphPad Prism 5 built-in model.
TABLE 1.
Protease inhibition by recombinant SPINK6
pNA, p-nitroanilide.
| Proteinase | SPINK6 | Inhibition | Substrate (0.33 mm) |
|---|---|---|---|
| nm | % | ||
| Bovine trypsin (2 nm final concentration) | 400 | 0 | N-(p-Tosyl)-Arg-Gly-Val 5-NA |
| Cathepsin G (1 nm final concentration) | 666 | 0 | N-Succinyl-Ala-Ala-Pro-Phe pNA |
| Chymase (2 nm final concentration) | 666 | 0 | N-Succinyl-Ala-Ala-Pro-Phe pNA |
| Human chymotrypsin (2 nm final concentration) | 400 | 0 | 3-MeO-Arg-Pro-Tyr pNA |
| KLK14 (2 nm final concentration) | 400 | 99.9 | N-(p-Tosyl)-Arg-Gly-Val 5-NA |
| KLK5 (5.3 nm final concentration) | 400 | 99.9 | N-(p-Tosyl)-Arg-Gly-Val 5-NA |
| KLK7 (15.8 nm final concentration) | 400 | 88.3 | MeO-Suc-Arg-Pro-Tyr pNA |
| Human leukocyte elastase (2 nm final concentration) | 400 | 0 | N-MeO-Ala-Ala-Pro-Val pNA |
| Human plasmin (2 nm final concentration) | 400 | 0 | N-(p-Tosyl)-Gly-Pro-Lys 4-NA |
| Human thrombin (1 nm final concentration) | 400 | 0 | N-(p-Tosyl)-Gly-Pro-Arg pNA |
| Matriptase (0.5 nm final concentration) | 400 | 0 | H-d-Ile-Pro-Arg pNA |
| Prostasin/Prss8 (3.5 nm final concentration) | 1750 | 3.3 | Tosyl-Gly-Pro-Arg 7-amino-4-methylcoumarin |
Cell Shedding from Human Plantar Stratum Corneum
The procedure of Lundström and Egelrud (22) was applied. Flakes (∼0.5 mm thick) of stratum corneum were cut parallel to the skin surface from the heels of healthy volunteers. After the tissue had been soaked in 0.01 m sodium phosphate, 0.14 m NaCl, 0.1% NaN3, and 1% Triton X-100, pH 7.2, for 5 h at room temperature, loosely attached cells were scraped off with a scalpel. Stratum corneum cylinders with a defined surface area were prepared with a 3-mm biopsy punch. Cylinders were incubated in 1.5-ml Eppendorf tubes with 1 ml of 0.1 m Tris-HCl, 5 mm EDTA, and 0.1% NaN3, pH 8.0, for 18 h at 37 °C. After Vortex agitation (30 s), the tissue pieces were removed from the incubation solution. The released cells were collected by centrifugation (10,000 × g, 10 min), washed in phosphate-buffered saline, and extracted with 1 m NaOH. Solubilized protein was quantified using NanoDrop with bovine serum albumin treated with 1 m NaOH as a standard.
RESULTS
SPINK6 Is Expressed in Human Skin
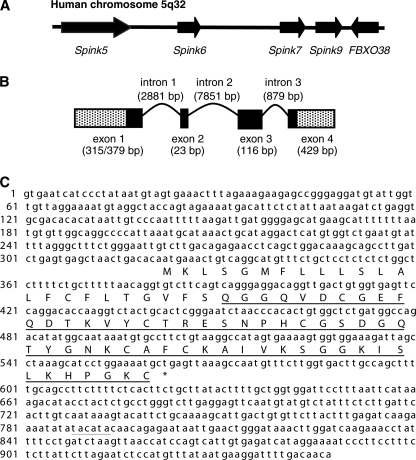
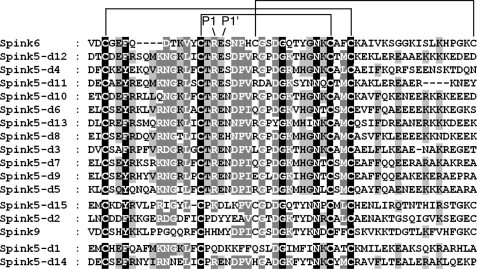
To search for genes encoding putative functional domains homologous to atypical Kazal-like domains of LEKTI, the amino acid sequence of LEKTI domain 6 was used as a query to blast against the human genome (NCBI builder 34, 2003). A gene locus was identified and localized between the two known genes, SPINK5 (GenBankTM accession number NM_001127698) and SPINK7 (accession number NM_032566) (Fig. 1A). To identify the gene, RACE and RT-PCR were performed. Two full-length cDNA sequences were obtained from cultured foreskin-derived keratinocytes and were registered under GenBankTM accession numbers GQ504704 and GQ504705, respectively. Although this gene had been predicted and designated as SPINK6 (serine protease inhibitor Kazal-type 6; GenBankTM accession number NM_205841), to the best of our knowledge, the identification of this gene has not been reported. Comparison between the cDNA and genomic sequences revealed that the SPINK6 gene contains four conserved exons, whereas it has two different transcription start sites at bp 1 and 64 but only one termination site (Fig. 1B). All of the splice donor and acceptor sites conform to the GT/AG rule. These two transcripts, SPINK6-v1 (947 bp) and SPINK6-v2 (883 bp), contain a common ORF of 243 bp and a polyadenylation signal (AAUAAA) situated 214 nucleotides 5′ of the polyadenylation tail (Fig. 1C). The deduced protein chain consists of 80 amino acids with a leader sequence containing a putative signal peptide (residues 1–23) and a typical Kazal domain with six cysteine residues (residues 28–80). Comparison of SPINK6 with LEKTI/SPINK5 domains and SPINK9 revealed that SPINK6 shows high homology to the atypical Kazal domains of LEKTI/SPINK5 especially in the putative reactive center residues P2 (Thr), P1 (Arg), and P1′ (Glu) (Fig. 2), whereas LEKTI-2/SPINK9 exhibits the highest degree of sequence homology to SPINK5 domain 2.
FIGURE 1.
Nucleotide and amino acid sequences of human SPINK6. A, schematic physical map of human SPINK gene loci (5q33.1). Genes are ordered from the centromere (left) to the telomere (right). B, schematic diagram of the SPINK6 gene based on its cDNA isolated from foreskin-derived keratinocytes. It consists of four exons and three introns. The positions of the exons (boxes) and introns (curve lines) of SPINK6 are deduced by comparing its full-length cDNA sequence with the corresponding genomic DNA. C, the two full-length SPINK6 cDNA sequences and the deduced protein sequence.
FIGURE 2.
Amino acid sequence alignment of SPINK6 in comparison with LEKTI/SPINK5 domains and SPINK6. The alignment of the Kazal domains of SPINK6, LEKTI-2/SPINK9, and LEKTI/SPINK5 domains (d) was generated by ClustalW and displayed using GeneDoc. Identical residues are in black boxes, whereas gray boxes indicate partially conserved residues. The darker the shading, the more the amino acids are conserved among the family members. The different LEKTI/LEKTI-2 domains are ordered by their homology to SPINK6. The P1 and P1′ sites are indicated.
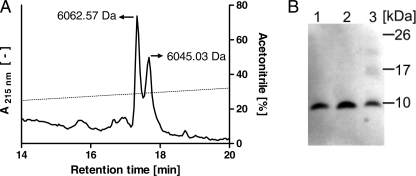
Given the fact that SPINK6 cDNA was cloned from cultured human keratinocytes, we tried to identify SPINK6 protein in the skin using an anti-SPINK6 polyclonal antibody affinity column to capture natural SPINK6 from human callus extracts. After affinity chromatography, the eluting fraction was further purified by C18 reversed-phase HPLC, resulting in two peaks (Fig. 3A) with masses of 6045.03 and 6062.57 Da, which fit well with a SPINK6 protein containing three disulfide bonds starting with Gln24 and its derivative starting with pyroglutamine 24 (Fig. 1C, underlined AA sequence). The identity of these peptides as SPINK6 was confirmed by MS/MS analyses (data not shown) and Western blot analyses (Fig. 3B). Immunoblot analyses exhibited immunoreactivity of purified pyroglutamine 24 (lane 1), purified Gln24 SPINK6 (lane 2), and rSPINK6 (lane 3).
FIGURE 3.
Purification of SPINK6 from human stratum corneum extracts. A, natural SPINK6 was isolated from human plantar stratum corneum extracts using first an anti-SPINK6 affinity column (not shown), followed by a C18 reversed-phase HPLC column. ESI-MS analyses of UV light-absorbing peak fractions revealed masses of 6045.03 and 6062.57 Da, which fit well with a truncated SPINK6 protein containing three disulfide bonds starting with Gln24 (calculated mass of 6062.8 Da) and its pyroglutamine 24 form (calculated mass of 6045.8 Da). For both peptides, sequences were confirmed by MS/MS. The dotted line marks the eluting acetonitrile concentration. B, immunoblot analyses exhibited immunoreactivity of purified pyroglutamine 24 (lane 1), purified Gln24 SPINK6 (lane 2), and rSPINK6 (lane 3).
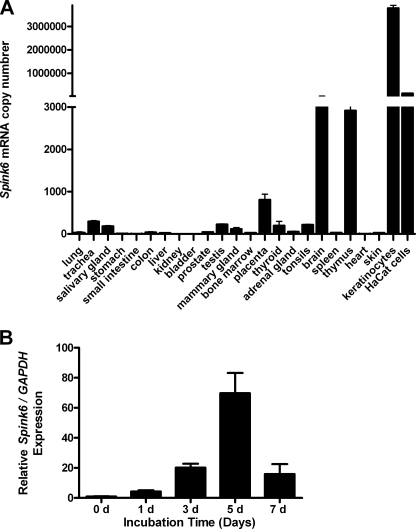
To further investigate whether SPINK6 is expressed in other tissues and organs, RT-PCR analyses were performed. Low levels of SPINK6 mRNA were detected in all tissues and cells examined using RT-PCR (data not shown). Real-time PCR analyses revealed that SPINK6 expression was much higher in cultured primary keratinocytes and HaCaT keratinocytes compared with tissue samples (Fig. 4A). SPINK6 expression was induced during keratinocyte differentiation (Fig. 4B).
FIGURE 4.
SPINK6 mRNA is ubiquitously expressed in human tissues and cultured keratinocytes. Quantitative PCR analyses revealed expression of SPINK6 mRNA in all cDNA samples investigated (A). SPINK6 mRNA expression was induced in cultured primary keratinocytes during differentiation (B). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. d, days.
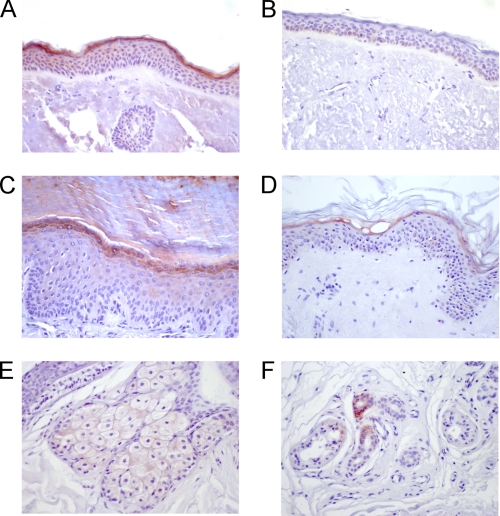
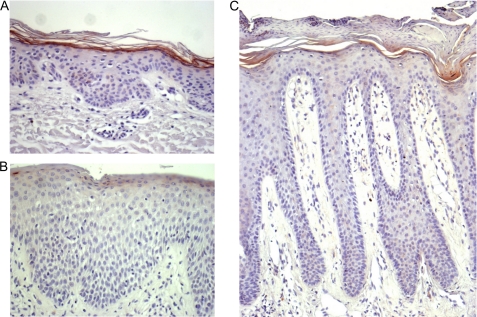
To localize the SPINK6 protein expression in human skin, immunohistochemical analyses of paraffin-embedded sections were performed. SPINK6 immunoreactivity was detected in the upper epidermal layer of the skin (Fig. 5). SPINK6 was detected in the skin at different body sites, including the face, arms, trunk, and legs, whereas it exhibited the most prominent expression level at palmoplantar sites (Fig. 5D). Furthermore, SPINK6 immunoreactivity was also detected in sebaceous glands and in some sweat glands (Fig. 5, E and F). The specificity of the antibody was demonstrated by blocking the antibody with recombinant antigen (Fig. 5B). SPINK6 expression was markedly decreased in atopic dermatitis lesions and slightly in psoriasis lesions (Fig. 6).
FIGURE 5.
SPINK6 is expressed in the skin and its appendages. Immunohistochemical analyses using an affinity-purified goat anti-SPINK6 polyclonal antibody revealed localization of SPINK6 within the stratum granulosum of the epidermis and stratum corneum (A, C, and D). Exemplary anatomical localizations of the samples were the face (A), plantar site (C), and upper arm (D). In B, the control is shown when rSPINK6 (0.5 μg/ml) was used to block the antibody (same paraffin sample as in A). Sebaceous glands showed positive staining (E), and some sweat glands exhibited immunoreactive SPINK6 (F).
FIGURE 6.
SPINK6 expression in atopic dermatitis and psoriasis. Shown are the results from immunohistochemical analyses of atopic dermatitis (B) and psoriasis (C) lesions compared with healthy skin (A; same patient as in B). Exemplary results of three independent investigations are shown.
SPINK6 Is a Selective Inhibitor of KLKs
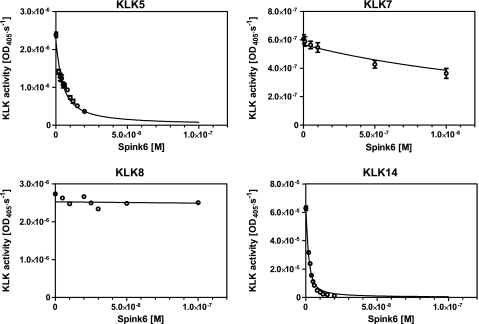
As SPINK6 possesses a typical Kazal-type domain, inhibition against different serine proteases was tested. SPINK6 exhibited inhibition against KLK5, KLK7, and KLK14, whereas it had no obvious activity against all other serine proteases tested, including trypsin, cathepsin G, mast cell chymase, leukocyte elastase, plasmin, matriptase, prostasin, chymotrypsin, and thrombin (Table 1). Next, inhibition against the relevant members of the skin KLK family, KLK5, KLK7, KLK8, and KLK14, was analyzed in more detail. The strongest inhibitory activity was observed against KLK5 and KLK14, with Ki values of 1.33 and 0.5 nm, respectively (Fig. 7 and Table 2), whereas KLK7 was just slightly inhibited, and KLK8 was not inhibited by SPINK6. rSPINK6 inhibited spontaneous desquamation of human plantar skin using an ex vivo model in a concentration-dependent manner (Table 3).
FIGURE 7.
Inhibition of human kallikreins by rSPINK6. Inhibition of KLKs (KLK5, KLK7, KLK8, and KLK14) was performed using increasing amounts of rSPINK6. The reaction progress was monitored by measuring the absorbance at 405 nm. Initial reaction velocities for each tested inhibitor concentration were determined by linear regression and were subsequently plotted against inhibitor concentration. Data were further analyzed according to the Morrison tight-binding inhibition model. The graphs include the means ± S.E. of at least three replicates for each enzyme.
TABLE 2.
KLK inhibition by SPINK6
Values are the kinetic parameters of KLK interaction with recombinant SPINK6. Data include means ± S.E. of at least three replicates for each enzyme. NI, no inhibition.
| Enzyme | Substrate | Km | Ki |
|---|---|---|---|
| mm | nm | ||
| KLK5 | Tosyl-Gly-Pro-Arg pNA | 0.6 ± 0.06 | 1.33 ± 0.04 |
| KLK7 | MeO-Suc-Arg-Pro-Tyr pNA | 2.0 ± 0.4 | 1070 ± 80 |
| KLK8 | Tosyl-Gly-Pro-Arg pNA | 5.8 ± 0.8 | NI |
| KLK14 | Tosyl-Gly-Pro-Arg pNA | 1.675 ± 0.27 | 0.5 ± 0.03 |
TABLE 3.
Effects of SPINK6 on cell shedding from human plantar callus
Single 3-mm callus cylinders (∼0.5 mm thick) were incubated for 18 h at 37 °C in 0.1 m Tris-HCl and 5 mm EDTA (pH 8.0) with protease inhibitors as indicated.
| Inhibitor | Inhibition ± S.E. | n |
|---|---|---|
| % | ||
| Buffer | 0 ± 12.9 | 5 |
| SPINK6 (10 μm) | 98.7 ± 0.5 | 5 |
| SPINK6 (5 μm) | 90.7 ± 2 | 2 |
| SPINK6 (2.5 μm) | 78.1 ± 8.8 | 2 |
| Aprotinin (10 μm) | 96.4 ± 2.2 | 3 |
| α-Antitrypsin (10 μm) | 95 ± 3.7 | 3 |
DISCUSSION
In this study, we have identified and purified the protease inhibitor SPINK6 from human stratum corneum extracts. We have cloned its full-length cDNA from cultured human keratinocytes and shown its expression in human epidermis. The most relevant result of this study is that SPINK6 is a selective inhibitor of several KLKs, which are known to be important in epidermal homeostasis. Our finding suggests that SPINK6 plays a role in modulating the activities of KLKs in human skin. As we could detect SPINK6 mRNA expression in various tissues, we suggest that the biological function of SPINK6 might not be limited to the epidermis.
Expression of SPINK6 mRNA was detectable at low levels in all tissue samples investigated. SPINK6 mRNA expression was magnitudes higher in cultured keratinocytes compared with skin extracts. SPINK6 protein was identified in human callus. Whether SPINK6 is expressed in other tissues remains to be investigated in more detail. Immunohistochemical analyses of terminal ileum as well as colon tissue samples revealed no SPINK6 expression (data not shown), although mRNA levels were comparable with those of SPINK6 in the skin. The expression pattern of SPINK6 in the skin is similar to that of LEKTI/SPINK5. Ultrastructural analyses revealed that LEKTI/SPINK5 and KLK7 are transported separately in the lamellar granule system and are co-localized in the extracellular spaces (23), which were demonstrated to be expressed in lamellar bodies and secreted into the intercellular space in the uppermost stratum granulosum (23–25). However, the expression of SPINK6 differs from that of LEKTI/SPINK5 in skin appendages. The latter is present in the inner root sheath of the hair follicle, where we could not detect SPINK6 (data not shown). Instead, we could find SPINK6 immunoreactivity in sebaceous glands and in some sweat glands. Its expression in sweat glands was not always present in different skin samples and showed no homogeneous level in specific sweat glands (Fig. 5), suggesting a potential regulatory mechanism of SPINK6 expression in sweat glands, although further experiments are required. The other skin-expressed SPINK member, SPINK9, was described only at palmoplantar sites, where we could also detect SPINK6 expression. Slightly varying expression patterns of LEKTI/SPINK5, SPINK6, and SPINK9 in human skin suggest localized functional differences. Their function is thought to be as protease inhibitors. SPINK6 shows structural similarities to the atypical domains of LEKTI/SPINK5 (Fig. 2). Especially the P1 site exhibits high similarity to these domains, suggesting a similar inhibition profile. Like LEKTI/SPINK5, SPINK6 inhibits KLK5, KLK7, and KLK14, but it does not inhibit other serine proteases tested so far. LEKTI/SPINK5 has been reported to have a much broader activity profile. The LEKTI/SPINK5 domains inhibit trypsin, plasmin, subtilisin A, cathepsin G, human neutrophil elastase, KLK5, KLK7, and KLK14 (26). The determined Ki values of LEKTI domains to inhibit KLK5 were in the range of 3 nm (domains 8–11) to 120 nm (domains 9–15) (9, 27, 28). SPINK9 exhibited inhibition activity only against KLK5, with a determined Ki of 65–230 nm (14, 15). Interestingly, KLK8 is not inhibited by any of these SPINK family members. The inhibition of KLK5 and KLK14 by SPINK6 on the nanomolar scale might have pathophysiological relevance. KLK5 and KLK14, but not KLK8, have been reported to activate protease-activated receptor 2 (29), a signaling receptor in epidermal inflammation (30) and regulator of epidermal barrier function (31). Recently, KLK5 was identified to induce atopic dermatitis-like lesions through activation of protease-activated receptor 2 by inducing thymic stromal lymphopoietin expression in Netherton syndrome patients (32). Thymic stromal lymphopoietin is an epithelial cell-derived cytokine and a potential key player in the induction of allergic inflammation. Thymic stromal lymphopoietin-activated human dendritic cells produce Th2-attracting chemokines, but no IL-12, and induce naïve CD4+ and CD8+ T cell differentiation into effector cells with a typical pro-allergic phenotype (33). Increased trypsin-like proteolytic activity was reported in atopic dermatitis lesions (34), which might be the result of decreased SPINK6 expression as demonstrated herein. As SPINK6 is a very potent and selective inhibitor of KLKs, it could have therapeutic potential to modulate increased KLK activity.
We propose that LEKTI/SPINK5, SPINK6, and SPINK9, members of the SPINK family, are natural inhibitors of KLKs in human skin, showing a specific expression and inhibition profile. As SPINK6 mRNA is detectable in other tissues, the biological function of SPINK6 might not be limited to the skin. Indeed, KLK5 and KLK14 expression has also been reported in other tissues. KLK5 was reported to be expressed at high levels (100–1000 ng/g of tissue) in breast, testis, salivary gland, and thyroid tissue samples, whereas KLK14 showed high concentrations in bladder, breast, and vagina tissue samples (35). Further studies are needed to clarify whether SPINK6 plays a functional role in tissues other than the skin. In conclusion, SPINK6 is present in human skin and inhibits a selective repertoire of skin-expressed KLKs.
Acknowledgments
We thank Christel Martensen-Kerl and Jutta Quitzau for excellent technical assistance.
- KLK
- kallikrein-related peptidase
- LEKTI
- lymphoepithelial Kazal-type inhibitor
- RACE
- rapid amplification of cDNA ends
- ESI-MS
- electrospray ionization mass spectrometry
- MS/MS
- tandem mass spectrometry
- rSPINK6
- recombinant SPINK6
- SUMO
- small ubiquitin-like modifier
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Meyer-Hoffert U. (2009) Arch. Immunol. Ther. Exp. 57, 345–354 [DOI] [PubMed] [Google Scholar]
- 2.Yousef G. M., Diamandis E. P. (2001) Endocr. Rev. 22, 184–204 [DOI] [PubMed] [Google Scholar]
- 3.Komatsu N., Suga Y., Saijoh K., Liu A. C., Khan S., Mizuno Y., Ikeda S., Wu H. K., Jayakumar A., Clayman G. L., Shirasaki F., Takehara K., Diamandis E. P. (2006) J. Invest. Dermatol. 126, 2338–2342 [DOI] [PubMed] [Google Scholar]
- 4.Komatsu N., Takata M., Otsuki N., Toyama T., Ohka R., Takehara K., Saijoh K. (2003) J. Invest. Dermatol. 121, 542–549 [DOI] [PubMed] [Google Scholar]
- 5.Komatsu N., Saijoh K., Toyama T., Ohka R., Otsuki N., Hussack G., Takehara K., Diamandis E. P. (2005) Br. J. Dermatol. 153, 274–281 [DOI] [PubMed] [Google Scholar]
- 6.Hansson L., Bäckman A., Ny A., Edlund M., Ekholm E., Ekstrand Hammarström B., Törnell J., Wallbrandt P., Wennbo H., Egelrud T. (2002) J. Invest. Dermatol. 118, 444–449 [DOI] [PubMed] [Google Scholar]
- 7.Simon M., Jonca N., Guerrin M., Haftek M., Bernard D., Caubet C., Egelrud T., Schmidt R., Serre G. (2001) J. Biol. Chem. 276, 20292–20299 [DOI] [PubMed] [Google Scholar]
- 8.Caubet C., Jonca N., Brattsand M., Guerrin M., Bernard D., Schmidt R., Egelrud T., Simon M., Serre G. (2004) J. Invest. Dermatol. 122, 1235–1244 [DOI] [PubMed] [Google Scholar]
- 9.Egelrud T., Brattsand M., Kreutzmann P., Walden M., Vitzithum K., Marx U. C., Forssmann W. G., Mägert H. J. (2005) Br. J. Dermatol. 153, 1200–1203 [DOI] [PubMed] [Google Scholar]
- 10.Borgoño C. A., Michael I. P., Komatsu N., Jayakumar A., Kapadia R., Clayman G. L., Sotiropoulou G., Diamandis E. P. (2007) J. Biol. Chem. 282, 3640–3652 [DOI] [PubMed] [Google Scholar]
- 11.Chavanas S., Bodemer C., Rochat A., Hamel-Teillac D., Ali M., Irvine A. D., Bonafé J. L., Wilkinson J., Taïeb A., Barrandon Y., Harper J. I., de Prost Y., Hovnanian A. (2000) Nat. Genet. 25, 141–142 [DOI] [PubMed] [Google Scholar]
- 12.Griffiths W. A. D., Leigh I. M., Judge M. R. (1998) in Textbook of Dermatology (Champion R. H., Breathnach S. M., Burns D. A., Burton J. L. eds) Blackwell Science, Oxford [Google Scholar]
- 13.Mägert H. J., Ständker L., Kreutzmann P., Zucht H. D., Reinecke M., Sommerhoff C. P., Fritz H., Forssmann W. G. (1999) J. Biol. Chem. 274, 21499–21502 [DOI] [PubMed] [Google Scholar]
- 14.Meyer-Hoffert U., Wu Z., Schröder J. M. (2009) PLoS ONE 4, e4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brattsand M., Stefansson K., Hubiche T., Nilsson S. K., Egelrud T. (2009) J. Invest. Dermatol. 129, 1656–1665 [DOI] [PubMed] [Google Scholar]
- 16.Kent W. J. (2002) Genome Res. 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatusova T. A., Madden T. L. (1999) FEMS Microbiol. Lett. 174, 247–250 [DOI] [PubMed] [Google Scholar]
- 18.Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z., Hansmann B., Meyer-Hoffert U., Glaser R., Schröder J. M. (2009) PLoS ONE 4, e5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale J. E., Butler J. P., Gelfanova V., You J. S., Knierman M. D. (2004) Anal. Biochem. 333, 174–181 [DOI] [PubMed] [Google Scholar]
- 21.Wu Z., Meyer-Hoffert U., Reithmayer K., Paus R., Hansmann B., He Y., Bartels J., Gläser R., Harder J., Schröder J. M. (2009) J. Invest. Dermatol. 129, 1446–1458 [DOI] [PubMed] [Google Scholar]
- 22.Lundström A., Egelrud T. (1988) J. Invest. Dermatol. 91, 340–343 [DOI] [PubMed] [Google Scholar]
- 23.Ishida-Yamamoto A., Deraison C., Bonnart C., Bitoun E., Robinson R., O'Brien T. J., Wakamatsu K., Ohtsubo S., Takahashi H., Hashimoto Y., Dopping-Hepenstal P. J., McGrath J. A., Iizuka H., Richard G., Hovnanian A. (2005) J. Invest. Dermatol. 124, 360–366 [DOI] [PubMed] [Google Scholar]
- 24.Sondell B., Thornell L. E., Egelrud T. (1995) J. Invest. Dermatol. 104, 819–823 [DOI] [PubMed] [Google Scholar]
- 25.Ishida-Yamamoto A., Simon M., Kishibe M., Miyauchi Y., Takahashi H., Yoshida S., O'Brien T. J., Serre G., Iizuka H. (2004) J. Invest. Dermatol. 122, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 26.Mitsudo K., Jayakumar A., Henderson Y., Frederick M. J., Kang Y., Wang M., El-Naggar A. K., Clayman G. L. (2003) Biochemistry 42, 3874–3881 [DOI] [PubMed] [Google Scholar]
- 27.Deraison C., Bonnart C., Lopez F., Besson C., Robinson R., Jayakumar A., Wagberg F., Brattsand M., Hachem J. P., Leonardsson G., Hovnanian A. (2007) Mol. Biol. Cell 18, 3607–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schechter N. M., Choi E. J., Wang Z. M., Hanakawa Y., Stanley J. R., Kang Y., Clayman G. L., Jayakumar A. (2005) Biol. Chem. 386, 1173–1184 [DOI] [PubMed] [Google Scholar]
- 29.Stefansson K., Brattsand M., Roosterman D., Kempkes C., Bocheva G., Steinhoff M., Egelrud T. (2008) J. Invest. Dermatol. 128, 18–25 [DOI] [PubMed] [Google Scholar]
- 30.Steinhoff M., Buddenkotte J., Shpacovitch V., Rattenholl A., Moormann C., Vergnolle N., Luger T. A., Hollenberg M. D. (2005) Endocr. Rev. 26, 1–43 [DOI] [PubMed] [Google Scholar]
- 31.Hachem J. P., Houben E., Crumrine D., Man M. Q., Schurer N., Roelandt T., Choi E. H., Uchida Y., Brown B. E., Feingold K. R., Elias P. M. (2006) J. Invest. Dermatol. 126, 2074–2086 [DOI] [PubMed] [Google Scholar]
- 32.Briot A., Deraison C., Lacroix M., Bonnart C., Robin A., Besson C., Dubus P., Hovnanian A. (2009) J. Exp. Med. 206, 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziegler S. F., Liu Y. J. (2006) Nat. Immunol. 7, 709–714 [DOI] [PubMed] [Google Scholar]
- 34.Voegeli R., Rawlings A. V., Breternitz M., Doppler S., Schreier T., Fluhr J. W. (2009) Br. J. Dermatol. 161, 70–77 [DOI] [PubMed] [Google Scholar]
- 35.Shaw J. L., Diamandis E. P. (2007) Clin. Chem. 53, 1423–1432 [DOI] [PubMed] [Google Scholar]