Abstract
Cystic fibrosis (CF) is caused by mutations in the CF transmembrane conductance regulator (CFTR) that prevent its proper folding and trafficking to the apical membrane of epithelial cells. Absence of cAMP-mediated Cl− secretion in CF airways causes poorly hydrated airway surfaces in CF patients, and this condition is exacerbated by excessive Na+ absorption. The mechanistic link between missing CFTR and increased Na+ absorption in airway epithelia has remained elusive, although substantial evidence implicates hyperactivity of the epithelial Na+ channel (ENaC). ENaC is known to be activated by selective endoproteolysis of the extracellular domains of its α- and γ-subunits, and it was recently reported that ENaC and CFTR physically associate in mammalian cells. We confirmed this interaction in oocytes by co-immunoprecipitation and found that ENaC associated with wild-type CFTR was protected from proteolytic cleavage and stimulation of open probability. In contrast, ΔF508 CFTR, the most common mutant protein in CF patients, failed to protect ENaC from proteolytic cleavage and stimulation. In normal airway epithelial cells, ENaC was contained in the anti-CFTR immunoprecipitate. In CF airway epithelial cultures, the proportion of full-length to total α-ENaC protein signal was consistently reduced compared with normal cultures. Our results identify limiting proteolytic cleavage of ENaC as a mechanism by which CFTR down-regulates Na+ absorption.
Keywords: ABC Transporter, Cystic Fibrosis, Lung, Protease, Sodium Channels, Sodium Transport, CFTR, ENaC
Introduction
Elevated epithelial Na+ absorption was first detected by in vivo assays of nasal and bronchial epithelial potential difference in cystic fibrosis (CF)2 patients (1), and it has been shown to contribute to depletion of airway surface liquid (ASL) in well differentiated cultures of CF airway epithelial cells (2). Most CF mutations causing severe disease practically eliminate the functional CF transmembrane conductance regulator (CFTR) at the apical membrane (3). Thus, Na+ hyperabsorption has been attributed to loss of negative regulation of the epithelial Na+ channel (ENaC) that is normally exerted by CFTR in airway epithelia (4). The negative influence of CFTR on ENaC has been reconstituted in multiple in vitro cell models (2, 5–13). However, such work has been controversial (14) and has yet to reveal a clear and compelling mechanistic basis for the down-regulation of ENaC by CFTR. Furthermore, the coordinated stimulation of CFTR and ENaC in sweat ductal epithelium indicates that the functional relationship of CFTR and ENaC varies with the tissue-specific physiologic roles of these ion channels (15).
In recent years, ENaC regulation has become much better understood. In particular, selective endoproteolysis of small segments of the large extracellular domains of α- and γ-ENaCs has been shown to increase ENaC open probability (16–19). Berdiev et al. (20, 21) recently demonstrated a physical association of CFTR and ENaC in HEK293T cells overexpressing these two channels. In this work, we assessed this association in primary airway epithelial cells and in Xenopus oocytes and evaluated its impact on ENaC regulation by partial endoproteolysis. We confirm that CFTR physically associates with ENaC and report for the first time that CFTR markedly impedes ENaC stimulation by suppressing proteolysis of its extracellular domains.
EXPERIMENTAL PROCEDURES
Culture Conditions and Protein Expression
Primary human airway epithelial (HAE) cultures were derived from human bronchial tissue as described previously (22) following a protocol approved by the University of North Carolina Medical School Institutional Review Board. Fully differentiated HAE cultures were obtained by maintaining cells at an air-liquid interface for 21 days. Xenopus laevis oocytes were harvested and maintained as described previously (23). Animals were maintained and studied under protocols approved by the University of North Carolina Institutional Animal Care and Use Committee. For protein expression in oocytes, 0.3 ng of cRNAs encoding rat ENaC α-, β-, and γ-subunits were utilized. Either the α- or γ-ENaC construct was double-tagged with HA and V5 epitopes at the N and C termini, respectively. cRNAs of matriptase (1 ng) and CFTR (1 or 2 ng) or MRP1 (multidrug resistance protein 1; 1 or 2 ng) were co-injected. Oocytes were maintained for 24 h before they were used for functional studies or for preparation of lysates for Western blotting.
Western Blot Analysis and Immunoprecipitation
Cell lysates from HAE cultures were obtained, and Western blotting was performed as described (24). Lysates from oocytes were prepared similarly as described by Garcia-Caballero et al. (23). Proteins were separated by 7, 10, or 4–20% SDS-PAGE and then transferred to nitrocellulose for immunoblot analysis. N-terminally HA-tagged and C-terminally V5-tagged α- and γ-ENaCs were visualized with anti-HA mAb (Covance) and anti-V5 mAb (Invitrogen), respectively. CFTR was detected with mAb 596 recognizing an epitope in NBD2 of CFTR (25), MRP1 was detected with mAb 42.4 (26), and actin was visualized as the loading control with rabbit anti-actin antibody (Stressgen) or anti-actin mAb (Millipore). Immunoprecipitations of proteins from oocyte lysates were performed as described previously (23), with the exception that all immunoprecipitations were performed in Nonidet P-40 buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris, 1 mm EGTA, 4 μg of leupeptin, 8 μg/ml aprotinin, 200 μg/ml Pefabloc, 484 μg/ml benzamidine, and 14 μg/ml E64, pH 7.4). For immunoprecipitation of CFTR, rabbit anti-CFTR polyclonal antibody 155 was utilized (27), and for immunoprecipitation of N-terminally HA-tagged γ-ENaC, we applied rabbit anti-HA antibody (Abcam ab9110), both followed by pulldown with protein A-Agarose (Invitrogen). Human α-ENaC was detected with the specific mAb UNC1 19.2.1 recognizing an N-terminal epitope of this subunit (supplemental Fig. 1). For detection of primary antibodies, we applied IRDye 680-conjugated goat anti-mouse IgG (Invitrogen) or IRDye 800-conjugate goat anti-rabbit IgG (Rockland Immunochemicals) using an Odyssey infrared imaging system (LI-COR Biosciences).
Functional Studies in Xenopus Oocytes
24 h after injection, two-electrode voltage clamping was performed using a GeneClamp amplifier (MDS Analytical Technologies) as described previously (23). Currents were measured in the presence and absence of 10 μm amiloride, with membrane voltage clamped to −100 mV. Currents were digitized and recorded using a Digidata 1200 A/D converter (MDS Analytical Technologies) and AxoScope software. Trypsin (2 μg/ml) was perfused for 5 min after first measuring the basal amiloride-sensitive current (INa). To assess whole cell ENaC open probability (Po), oocytes were injected with cRNA for wild-type α-ENaC, S518C β-ENaC, and wild-type γ-ENaC. Basal INa was measured, methanethiosulfonate ethyltrimethylammonium (MTSET; 1 mm) was applied for 5 min, and INa was determined again. Whole cell Po was estimated from the ratio of basal INa to MTSET-stimulated INa (28).
RESULTS
CFTR and ENaC Associate in Oocytes
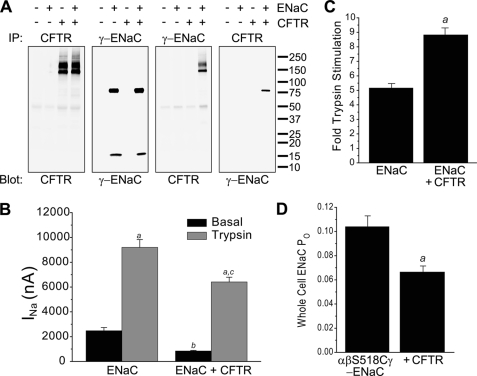
To test for association of CFTR and ENaC in Xenopus oocytes, we performed CFTR and ENaC immunoprecipitations from four experimental groups (uninjected, ENaC only, CFTR only, and CFTR + ENaC) (Fig. 1A). The resulting immunoprecipitated proteins were then probed for the presence of ENaC and CFTR. CFTR was immunoprecipitated from oocytes injected with CFTR cRNA as the typical B and C bands (Fig. 1A, first panel), indicating that oocytes produce core-glycosylated and mature CFTR protein, respectively (29). We immunoprecipitated and detected γ-ENaC using an antibody against its N-terminal HA tag. γ-ENaC proteins immunoprecipitated with anti-HA antibody contained an ∼80–85-kDa band representing full-length γ-ENaC, as well as an ∼15-kDa band known to be produced by cleavage mediated by endogenous proteases at or near the furin site (Fig. 1A, second panel) (19, 30).
FIGURE 1.
CFTR interacts with ENaC and decreases ENaC-mediated INa. A, CFTR and ENaC associate with each other when expressed in Xenopus oocytes. Oocytes were injected with rat α-, β-, and HA/V5-tagged γ-ENaC cRNAs in the absence or presence of CFTR cRNA. CFTR was immunoprecipitated (IP) with rabbit anti-CFTR antibody 155, and HA/V5-tagged γ-ENaC was immunoprecipitated with rabbit anti-HA antibody (Abcam ab9110). Immunoprecipitated proteins were detected by Western blotting using anti-CFTR mAb 596 and anti-HA mAb (Covance HA.11). B, CFTR coexpression decreases ENaC-mediated INa. Current was recorded from oocytes injected with ENaC cRNAs (0.3 ng/each) alone or with CFTR cRNA (1–2 ng). Amiloride was removed for 30 s to measure basal INa. Trypsin (2 μg/ml) was applied for 5 min, and INa was determined again (n = 84 oocytes in each group, from 14 separate batches of oocytes). a, trypsin-stimulated versus basal INa (p < 0.001); b, ENaC + CFTR basal versus ENaC basal INa (p < 0.01); c, CFTR + ENaC trypsin-stimulated versus ENaC trypsin-stimulated INa. Analysis of variance with Tukey's test was performed. C, CFTR coexpression increases stimulation of ENaC by trypsin. ENaC INa reported in B was analyzed as -fold trypsin stimulation. In oocytes expressing ENaC, INa was increased by 5.16 ± 0.35-fold. In oocytes coexpressing ENaC and CFTR, trypsin increased INa by 8.69 ± 0.46-fold (n = 84 oocytes in each group, from 14 separate batches of oocytes). a, p < 0.001 by unpaired t test. D, CFTR coexpression decreases ENaC whole cell open probability (Po). Oocytes were injected with cRNA for wild-type α- and γ-ENaCs and S518C β-ENaC (0.3 ng each) alone or the same ENaC subunit combination with CFTR (1 ng). Amiloride was removed for 30 s to measure basal INa. MTSET (1 mm) was applied for 5 min, and INa was determined again. Whole cell Po was calculated from the ratio of basal INa to MTSET-stimulated INa (n = 44 oocytes in each group, from 14 separate batches of oocytes). a, p < 0.001 by unpaired t test.
We next probed γ-ENaC immunoprecipitates with anti-CFTR antibody (Fig. 1A, third panel). The CFTR B and C bands were detected in proteins immunoprecipitated by anti-HA antibody, confirming that CFTR associates with γ-ENaC. In the anti-CFTR immunoprecipitate, anti-HA antibody detected the ∼80–85-kDa full-length γ-ENaC band, demonstrating association of ENaC with CFTR (Fig. 1A, fourth panel). Compared with the same experimental group (CFTR + ENaC) immunoprecipitated with anti-ENaC antibody (Fig. 1A, second panel), just a fraction of the total amount of the ∼80–85-kDa γ-ENaC band was detected in the anti-CFTR immunoprecipitate. Surprisingly, the ∼15-kDa band representing an N-terminal cleavage fragment of γ-ENaC was not present in the anti-CFTR immunoprecipitate, even when overexposed. It is well established that this N-terminal ∼15-kDa band represents the product of cleavage near the furin site in γ-ENaC and that it remains associated with the larger C-terminal fragment during protein isolation (31). Furthermore, detection of the 15-kDa band in the γ-ENaC immunoprecipitate (Fig. 1A, second panel) indicates that it was generated by endogenous proteases in the oocyte groups expressing ENaC, either alone or with CFTR. Therefore, the absence of the N-terminal 15-kDa fragment of γ-ENaC in the CFTR immunoprecipitate suggests that the pool of ENaC closely associated with CFTR was shielded from endogenous proteases.
CFTR Decreases ENaC-mediated INa
Endoproteolysis of α- and γ-ENaCs has been shown to increase ENaC open probability (16, 32), and if CFTR shielded γ-ENaC from proteolytic activation, INa should be decreased in CFTR + ENaC oocytes compared with ENaC-only oocytes. Indeed, INa was reduced in oocytes coexpressing ENaC and CFTR compared with ENaC alone (Fig. 1B). Furthermore, exogenous trypsin caused a proportionately larger stimulation of INa in oocytes coexpressing CFTR (Fig. 1, B and C). This reduction in basal INa and increased -fold trypsin stimulation indicate reduced proteolytic activation of ENaC on the surface of oocytes expressing CFTR. Previous work in oocytes showing decreased ENaC current in the presence of CFTR has been criticized as a recording artifact potentially introduced by the large conductance mediated by CFTR (14). However, the increased -fold stimulation of INa by trypsin in CFTR-expressing oocytes cannot be explained by such a technical artifact. In addition, we used an independent method to estimate the effects of CFTR on ENaC open probability (Fig. 1D). ENaC constituted with the β-ENaC mutant S518C is converted to a locked open state when the sulfhydryl-reactive reagent MTSET enters the pore and covalently reacts with the introduced cysteine (33). We used the ratio of basal INa to MTSET-stimulated INa to estimate ENaC open probability in oocytes coexpressing ENaC and CFTR compared with oocytes expressing ENaC alone (28). CFTR coexpression reduced ENaC open probability, consistent with biochemical evidence that CFTR reduced cleavage of a pool of ENaC.
CFTR Diminishes Proteolysis of ENaC
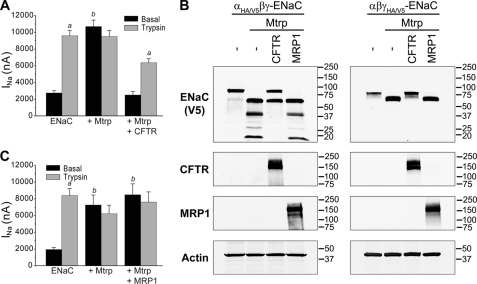
As the extent of ENaC cleavage by endogenous proteases is variable, we chose to test the apparent ability of CFTR to limit ENaC proteolysis under more defined conditions. Therefore, we coexpressed exogenous matriptase, which is known to fully activate ENaC by a proteolytic mechanism (34). In oocytes expressing ENaC only, we observed a low basal INa that was markedly stimulated by the addition of trypsin (Fig. 2A). As predicted, ENaC coexpressed with matriptase produced a large basal INa that was not increased further by exogenous trypsin (Fig. 2A). When CFTR was also coexpressed, basal ENaC currents were not increased in matriptase-expressing oocytes, but these currents could be robustly stimulated by trypsin. These results were paralleled by changes in the fragmentation patterns of α- and γ-ENaCs in lysates from oocytes coexpressing matriptase alone or matriptase with CFTR (Fig. 2B). Matriptase alone strongly converted full-length α- and γ-ENaCs to smaller fragments that have been shown to arise from cleavages at or near the two furin cleavage sites in α-ENaC and the single furin site and a polybasic RKRK tract in γ-ENaC (17, 23). Coexpressed CFTR clearly limited these cleavages (Fig. 2B), and we showed in other experiments that this inhibition of ENaC proteolysis was dependent on the amount of CFTR expressed (supplemental Fig. 2). As a control, we coexpressed MRP1, an ABC protein similar in size and membrane topography to CFTR (35), and observed no effect on stimulation of INa or α-ENaC cleavage by matriptase (Fig. 2, B and C).
FIGURE 2.
Specific inhibition of ENaC proteolysis and INa by CFTR. A, ENaC was expressed in Xenopus oocytes (as described for Fig. 1B) alone, with matriptase (Mtrp), and with both matriptase and CFTR. Basal INa was increased by matriptase and not further stimulated by exogenous trypsin (2 μg/ml). CFTR prevented stimulation of basal INa and restored sensitivity to exogenous trypsin. B, CFTR prevents proteolytic cleavage of α- and γ-ENaCs by matriptase. Oocytes were injected with all three ENaC subunits, whereby either the α- or γ-subunit was tagged at the N and C termini with HA and V5 epitopes, respectively. Similar experiments were performed with coexpression of matriptase and CFTR or MRP1. Oocyte lysates were analyzed by Western blotting using anti-V5 mAb to detect ENaC, mAb 596 to detect CFTR, and mAb 42.4 to visualize expression of MRP1. As a control for loading of equal amounts of proteins, actin was detected with anti-actin mAb. C, experiments were similar to those described for A except that CFTR was replaced by MRP1. Matriptase coexpression was associated with large basal INa that was unaffected by trypsin, and coexpression of MRP1 did not alter this result. a, trypsin-stimulated INa versus basal INa (p < 0.001); b, basal INa of ENaC + Mtrp or ENaC + Mtrp + MRP1 versus basal INa of ENaC alone (p < 0.01). Analysis of variance with Tukey's test was performed (A and C, n = 21–22 oocytes combined from five batches).
ΔF508 CFTR Does Not Affect Proteolytic Cleavage of ENaC
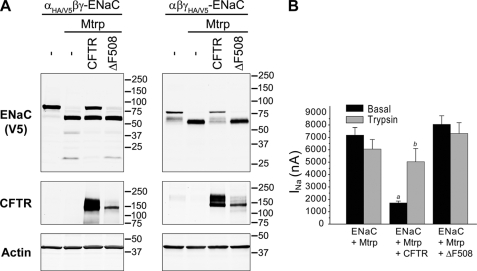
The CF mutation ΔF508 CFTR causes misfolding and retention of the protein at the ER in its core-glycosylated immature form and subsequent ER-associated degradation (3, 36). A small number of ΔF508 CFTR channels may escape to the cell surface but show reduced chloride channel function. When coexpressed with ENaC subunits and matriptase, ΔF508 CFTR, which appeared as an immature protein lacking complex oligosaccharide chains, did not diminish proteolytic cleavage of either the ENaC α- or γ-subunit (Fig. 3A). Unlike the block of matriptase-induced stimulation of INa and restoration of trypsin sensitivity by wild-type CFTR, coexpressed ΔF508 had no effect on matriptase-stimulated INa (Fig. 3B).
FIGURE 3.
ΔF508 CFTR does not diminish proteolysis of ENaC. A, Xenopus oocytes were injected with all three ENaC subunits, whereby either the α- or γ-subunit was tagged at the N and C termini with HA and V5 epitopes, respectively. CFTR or ΔF508 CFTR was coexpressed with ENaC and matriptase (Mtrp). Oocyte lysates were analyzed by Western blotting as described for Fig. 3. B, wild-type CFTR but not ΔF508 CFTR interferes with stimulation of ENaC by matriptase. INa of (ENaC + Mtrp)-expressing oocytes was not further stimulated by a 5-min exposure to trypsin (2 μg/ml). Coexpression of wild-type CFTR but not ΔF508 CFTR significantly decreased basal INa, and the reduced current was partially recovered by trypsin. Data were combined from three batches of oocytes (n = 16–17). a, basal INa of ENaC + Mtrp + CFTR versus basal INa of ENaC + Mtrp or ENaC + Mtrp + ΔF508 CFTR (p < 0.01); b, trypsin-stimulated INa versus basal INa (p < 0.05). Analysis of variance with Tukey's test was applied.
CFTR and ENaC Associate in Primary HAE Cultures
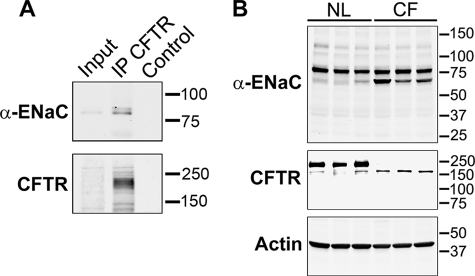
We followed up on these heterologous expression experiments by asking whether the extent of ENaC proteolysis differed in normal and CF airway epithelia. To analyze endogenous ENaC in primary HAE cultures, we developed mAbs detecting epitopes in the N- or C-terminal tails of individual ENaC subunits. The mouse anti-α-ENaC monoclonal antibody (mAb UNC1 19.2.1; supplemental Fig. 1) allowed for particularly robust detection of ENaC endogenously expressed in primary HAE cultures, and we used this antibody to test for association of ENaC with CFTR by co-immunoprecipitation. As shown in Fig. 4A, the presence of α-ENaC in proteins immunoprecipitated by anti-CFTR antibody was readily detected.
FIGURE 4.
Endogenous CFTR and ENaC co-immunoprecipitate, and CFTR impedes proteolysis of ENaC in primary HAE cultures. A, α-ENaC co-immunoprecipitates with CFTR in primary airway cells (HAE). CFTR was immunoprecipitated (IP) with rabbit anti-CFTR Ab and pulled down with protein A-agarose beads, and associated α-ENaC was detected with mAb UNC1 19.2.1. CFTR was visualized with mAb 596. Control lysates were treated identically but did not contain anti-CFTR Ab. 2% of immunoprecipitation inputs were loaded. B, ENaC is less cleaved in normal (NL) primary HAE cultures than in CF cultures (ΔF508/ΔF508). Primary human HAE cells were grown on an air-liquid interface until well polarized, and lysates were analyzed by Western blotting with anti-α-ENaC mAb UNC1 19.2.1. Three different cultures with cells derived from three different normal or CF individuals were analyzed. CFTR was visualized with mAb 596 after immunoprecipitation with rabbit polyclonal Ab.
CFTR Impedes Proteolysis of ENaC in Primary HAE Cultures
We employed mAb UNC1 19.2.1 (supplemental Fig. 1) to examine the effect of CFTR on proteolytic processing of α-ENaC natively expressed in HAE cultures. First, we prepared lysates of well differentiated airway epithelial cells cultured from three normal individuals and visualized the banding pattern of α-ENaC. mAb UNC1 19.2.1 mainly stained an ∼83-kDa band consistent with full-length α-ENaC. A less intense, more rapidly migrating band of ∼65 kDa was also detected. Next, we generated lysates from CF HAE cultures of three patients that were homozygous for ΔF508 CFTR. Western blots with anti-CFTR mAb 596 confirmed the absence of mature CFTR and the expression of core-glycosylated immature mutant protein. In the absence of functional CFTR, α-ENaC appeared more prominently in the ∼65-kDa band. Similar fragmentation patterns were seen when α-ENaC was detected in mouse lung tissue homogenate (37) and when α-ENaC was coexpressed with TMPRSS4 in Xenopus oocytes (23). Although we were not successful in developing antibodies to assess endogenous γ-ENaC proteolysis in airways, both α- and γ-ENaCs were protected similarly by CFTR in oocytes (Figs. 2 and 3 and supplemental Fig. 2). Thus, our results suggest that CFTR in airway epithelia impedes the proteolytic processing of ENaC, which has been strongly associated with increased ENaC open probability.
DISCUSSION
We found CFTR and ENaC coexpressed in oocytes to co-immunoprecipitate, similar to earlier reports (7, 20, 21). Importantly, the pool of γ-ENaC found in the proteins immunoprecipitated by anti-CFTR antibody showed fewer signs of cleavage by endogenous oocyte proteases, and functional assays clearly indicated that ENaC coexpressed with CFTR was protected from proteolytic activation. This finding was extended by studies revealing that CFTR coexpression prevented full cleavage and activation of ENaC by the coexpressed surface-acting protease matriptase. This protection against cleavage and stimulation by matriptase was not shared by coexpression of ΔF508 CFTR or MRP1, an ABC transport protein related to CFTR (Figs. 2 and 3). The concordance of these functional and biochemical data in oocytes allays concerns that CFTR-mediated inhibition of ENaC in these experiments is an artifact of the two-electrode voltage clamp technique (14).
We were able to bridge our observations in the oocyte expression system to the regulation of ENaC in human tissue by demonstrating that CFTR and ENaC co-immunoprecipitate in normal human primary airway epithelia (Fig. 4). The expression of mature CFTR in normal airway epithelia has long been inferred to check the rate of Na+ absorption mediated by ENaC (38). Importantly, we found ENaC to be more fragmented in airway epithelial cells cultured from CF patients lacking appreciable CFTR in their apical membranes. These data provide mechanistic insights into published reports of protease-mediated regulation of ENaC in airway epithelial cells. Selective knockdown of prostasin has firmly implicated net proteolytic activity on the respiratory surface in the stimulation of ENaC (39, 40). In addition, Myerburg et al. (41) recently showed that the content of soluble cognate prostasin inhibitors in ASL is important in determining ENaC activity in airways. Tarran and co-workers (42) identified one such soluble mediator as SPLUNC1 (short palate, lung, and nasal epithelial clone 1), which impairs proteolytic stimulation undergone by ENaC. Collectively, these studies established that signals reporting ASL volume, including protease inhibitor concentration, can slow Na+ absorption by preventing proteolytic stimulation of ENaC and thereby achieve homeostatic control of ASL depth.
Tarran et al. (12) reported several years ago that Na+ absorption of CF airway epithelial cultures is elevated and not further stimulated by exogenous trypsin, whereas in normal cells under the same conditions, Na+ absorption is lower but can be stimulated by trypsin. Thus, in normal cells, ENaC is partially protected from endogenous proteolytic stimulation (12). In contrast, in CF cells, the same soluble signals that limit ENaC proteolysis and stimulation in normal cells, such as protease inhibitors and SPLUNC1, are present but insufficient to prevent full proteolytic stimulation of ENaC (12). Tarran et al. restored regulation of ENaC and ASL depth to CF cultures by experimentally increasing protease inhibition with exogenous aprotinin. Our finding that ENaC is more extensively cleaved in CF airway epithelial cells is entirely consistent with these functional studies and with observations by Chinet et al. (43) of greater open probability of Na+-permeable channels in CF airway cells. We propose that the presence of CFTR in normal cells, through protection against ENaC proteolysis, is required in addition to soluble protease inhibitors for normal regulation of ENaC in airway epithelia.
Previous studies demonstrating CFTR-ENaC association did not definitively explain how proximity of the two ion channels led to restraint of ENaC function (7, 20, 21). Our work identifies, for the first time, CFTR-mediated inhibition of ENaC proteolysis as a mechanism for negative regulation of ENaC by CFTR that is seen in airways. Moreover, bidirectional CFTR-ENaC co-immunoprecipitation and the ability of CFTR to compromise bimolecular interactions between proteases and the large extracellular domains of ENaC imply an extent of CFTR-ENaC interaction that could affect ENaC activity in additional ways. For example, CFTR physically situated near ENaC could impact signaling pathways that control the trafficking and surface expression of ENaC (13). Other processes or conditions important for ENaC activity could be influenced by the effects of CFTR on the pH near the apical surface (44, 45) or intracellular ion content (46, 47). Importantly, our results provide a concrete mechanism of CFTR negative regulation of ENaC in airway epithelia while leaving open the possibility that other modes of ENaC regulation are utilized in epithelia with different salt and water physiology.
Acknowledgments
We acknowledge the skilled technical contribution of Hong He to this work and the diligent efforts of the University of North Carolina Cystic Fibrosis/Pulmonary Research and Treatment Center Cell Culture Core in providing airway epithelial cells. We are grateful to the University of North Carolina Immunology Core Facility for production of anti-α-ENaC mAb, the Cystic Fibrosis/Pulmonary Research and Treatment Center Molecular Core Facility for knockdown of α-ENaC expression in HAE cultures, and Rob Wonsetler for analyzing protein expression. We thank Lisa Brown for assistance with editing and submission of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants 5P01HL034322 and 5R01HL080561. Work performed by the Cell Culture Core and the Molecular Biology Core was supported by Cystic Fibrosis Foundation Grant R026-CR07 and National Institutes of Health Grant 5P30DK065988.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- CF
- cystic fibrosis
- ASL
- airway surface liquid
- CFTR
- CF transmembrane conductance regulator
- ENaC
- epithelial Na+ channel
- HAE
- human airway epithelial
- MTSET
- methanethiosulfonate ethyltrimethylammonium.
REFERENCES
- 1.Knowles M., Gatzy J., Boucher R. (1983) J. Clin. Invest. 71, 1410–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsui H., Grubb B. R., Tarran R., Randell S. H., Gatzy J. T., Davis C. W., Boucher R. C. (1998) Cell 95, 1005–1015 [DOI] [PubMed] [Google Scholar]
- 3.Riordan J. R. (1999) Am. J. Hum. Genet. 64, 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher R. C. (2003) Pflugers Arch. 445, 495–498 [DOI] [PubMed] [Google Scholar]
- 5.Stutts M. J., Canessa C. M., Olsen J. C., Hamrick M., Cohn J. A., Rossier B. C., Boucher R. C. (1995) Science 269, 847–850 [DOI] [PubMed] [Google Scholar]
- 6.Mall M., Hipper A., Greger R., Kunzelmann K. (1996) FEBS Lett. 381, 47–52 [DOI] [PubMed] [Google Scholar]
- 7.Kunzelmann K., Kiser G. L., Schreiber R., Riordan J. R. (1997) FEBS Lett. 400, 341–344 [DOI] [PubMed] [Google Scholar]
- 8.Mall M., Bleich M., Kuehr J., Brandis M., Greger R., Kunzelmann K. (1999) Am. J. Physiol. Gastroentest. Liver Physiol. 277, G709–G716 [DOI] [PubMed] [Google Scholar]
- 9.Ji H. L., Chalfant M. L., Jovov B., Lockhart J. P., Parker S. B., Fuller C. M., Stanton B. A., Benos D. J. (2000) J. Biol. Chem. 275, 27947–27956 [DOI] [PubMed] [Google Scholar]
- 10.Jiang Q., Li J., Dubroff R., Ahn Y. J., Foskett J. K., Engelhardt J., Kleyman T. R. (2000) J. Biol. Chem. 275, 13266–13274 [DOI] [PubMed] [Google Scholar]
- 11.Konstas A. A., Koch J. P., Korbmacher C. (2003) Pflugers Arch. 445, 513–521 [DOI] [PubMed] [Google Scholar]
- 12.Tarran R., Trout L., Donaldson S. H., Boucher R. C. (2006) J. Gen. Physiol. 127, 591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu C., Jiang C., Pribanic S., Rotin D. (2007) J. Cyst. Fibros. 6, 419–422 [DOI] [PubMed] [Google Scholar]
- 14.Nagel G., Barbry P., Chabot H., Brochiero E., Hartung K., Grygorczyk R. (2005) J. Physiol. 564, 671–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy M. M., Light M. J., Quinton P. M. (1999) Nature 402, 301–304 [DOI] [PubMed] [Google Scholar]
- 16.Caldwell R. A., Boucher R. C., Stutts M. J. (2004) Am. J. Physiol. Cell Physiol. 286, C190–C194 [DOI] [PubMed] [Google Scholar]
- 17.Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 18.Kleyman T. R., Carattino M. D., Hughey R. P. (2009) J. Biol. Chem. 284, 20447–20451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossier B. C., Stutts M. J. (2009) Annu. Rev. Physiol. 71, 361–379 [DOI] [PubMed] [Google Scholar]
- 20.Berdiev B. K., Cormet-Boyaka E., Tousson A., Qadri Y. J., Oosterveld-Hut H. M., Hong J. S., Gonzales P. A., Fuller C. M., Sorscher E. J., Lukacs G. L., Benos D. J. (2007) J. Biol. Chem. 282, 36481–36488 [DOI] [PubMed] [Google Scholar]
- 21.Berdiev B. K., Qadri Y. J., Benos D. J. (2009) Mol. BioSyst. 5, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulcher M. L., Gabriel S., Burns K. A., Yankaskas J. R., Randell S. H. (2005) Methods Mol. Med. 107, 183–206 [DOI] [PubMed] [Google Scholar]
- 23.Garciá-Caballero A., Dang Y., He H., Stutts M. J. (2008) J. Gen. Physiol. 132, 521–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cholon D. M., O'Neal W. K., Randell S. H., Riordan J. R., Gentzsch M. (2010) Am. J. Physiol. Lung Cell. Mol. Physiol. 298, L304–L314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui L., Aleksandrov L., Chang X. B., Hou Y. X., He L., Hegedus T., Gentzsch M., Aleksandrov A., Balch W. E., Riordan J. R. (2007) J. Mol. Biol. 365, 981–994 [DOI] [PubMed] [Google Scholar]
- 26.Hou Y., Cui L., Riordan J. R., Chang X. (2000) J. Biol. Chem. 275, 20280–20287 [DOI] [PubMed] [Google Scholar]
- 27.Kartner N., Hanrahan J. W., Jensen T. J., Naismith A. L., Sun S. Z., Ackerley C. A., Reyes E. F., Tsui L. C., Rommens J. M., Bear C. E., Riordan J. R. (1991) Cell 64, 681–691 [DOI] [PubMed] [Google Scholar]
- 28.Yang L. M., Rinke R., Korbmacher C. (2006) J. Biol. Chem. 281, 9859–9868 [DOI] [PubMed] [Google Scholar]
- 29.Benharouga M., Sharma M., Lukacs G. L. (2002) Methods Mol. Med. 70, 229–243 [DOI] [PubMed] [Google Scholar]
- 30.Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 31.Hughey R. P., Mueller G. M., Bruns J. B., Kinlough C. L., Poland P. A., Harkleroad K. L., Carattino M. D., Kleyman T. R. (2003) J. Biol. Chem. 278, 37073–37082 [DOI] [PubMed] [Google Scholar]
- 32.Carattino M. D., Hughey R. P., Kleyman T. R. (2008) J. Biol. Chem. 283, 25290–25295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheng S., Li J., McNulty K. A., Kieber-Emmons T., Kleyman T. R. (2001) J. Biol. Chem. 276, 1326–1334 [DOI] [PubMed] [Google Scholar]
- 34.Vuagniaux G., Vallet V., Jaeger N. F., Hummler E., Rossier B. C. (2002) J. Gen. Physiol. 120, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein I., Sarkadi B., Váradi A. (1999) Biochim. Biophys. Acta 1461, 237–262 [DOI] [PubMed] [Google Scholar]
- 36.Kopito R. R. (1999) Physiol. Rev. 79, S167–S173 [DOI] [PubMed] [Google Scholar]
- 37.Planès C., Randrianarison N. H., Charles R. P., Frateschi S., Cluzeaud F., Vuagniaux G., Soler P., Clerici C., Rossier B. C., Hummler E. (2010) EMBO Mol. Med. 2, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boucher R. C., Stutts M. J., Knowles M. R., Cantley L., Gatzy J. T. (1986) J. Clin. Invest. 78, 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Planès C., Leyvraz C., Uchida T., Angelova M. A., Vuagniaux G., Hummler E., Matthay M., Clerici C., Rossier B. (2005) Am. J. Physiol. Lung Cell. Mol. Physiol 288, L1099–L1109 [DOI] [PubMed] [Google Scholar]
- 40.Coote K., Atherton-Watson H. C., Sugar R., Young A., MacKenzie-Beevor A., Gosling M., Bhalay G., Bloomfield G., Dunstan A., Bridges R. J., Sabater J. R., Abraham W. M., Tully D., Pacoma R., Schumacher A., Harris J., Danahay H. (2009) J. Pharmacol. Exp. Ther. 329, 764–774 [DOI] [PubMed] [Google Scholar]
- 41.Myerburg M. M., McKenna E. E., Luke C. J., Frizzell R. A., Kleyman T. R., Pilewski J. M. (2008) Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L932–L941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Caballero A., Rasmussen J. E., Gaillard E., Watson M. J., Olsen J. C., Donaldson S. H., Stutts M. J., Tarran R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11412–11417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chinet T. C., Fullton J. M., Yankaskas J. R., Boucher R. C., Stutts M. J. (1994) Am. J. Physiol. Cell Physiol. 266, C1061–C1068 [DOI] [PubMed] [Google Scholar]
- 44.Coakley R. D., Grubb B. R., Paradiso A. M., Gatzy J. T., Johnson L. G., Kreda S. M., O'Neal W. K., Boucher R. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 16083–16088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verkman A. S. (2001) Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L306–L308 [DOI] [PubMed] [Google Scholar]
- 46.Bachhuber T., König J., Voelcker T., Mürle B., Schreiber R., Kunzelmann K. (2005) J. Biol. Chem. 280, 31587–31594 [DOI] [PubMed] [Google Scholar]
- 47.König J., Schreiber R., Voelcker T., Mall M., Kunzelmann K. (2001) EMBO Rep. 2, 1047–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]