Abstract
C1, the complex that triggers the classic pathway of complement, is a 790-kDa assembly resulting from association of a recognition protein C1q with a Ca2+-dependent tetramer comprising two copies of the proteases C1r and C1s. Early structural investigations have shown that the extended C1s-C1r-C1r-C1s tetramer folds into a compact conformation in C1. Recent site-directed mutagenesis studies have identified the C1q-binding sites in C1r and C1s and led to a three-dimensional model of the C1 complex (Bally, I., Rossi, V., Lunardi, T., Thielens, N. M., Gaboriaud, C., and Arlaud, G. J. (2009) J. Biol. Chem. 284, 19340–19348). In this study, we have used a mass spectrometry-based strategy involving a label-free semi-quantitative analysis of protein samples to gain new structural insights into C1 assembly. Using a stable chemical modification, we have compared the accessibility of the lysine residues in the isolated tetramer and in C1. The labeling data account for 51 of the 73 lysine residues of C1r and C1s. They strongly support the hypothesis that both C1s CUB1-EGF-CUB2 interaction domains, which are distant in the free tetramer, associate with each other in the C1 complex. This analysis also provides the first experimental evidence that, in the proenzyme form of C1, the C1s serine protease domain is partly positioned inside the C1q cone and yields precise information about its orientation in the complex. These results provide further structural insights into the architecture of the C1 complex, allowing significant improvement of our current C1 model.
Keywords: Complement, Innate immunity, Mass Spectrometry (MS), Protein Assembly, Protein Chemical Modification, C1q, C1s/C1r Tetramer
Introduction
Complement is an essential component of innate immunity due to its ability to recognize pathogens and to limit infection in the vertebrate host. In addition, activation of the complement system enhances the migration of phagocytic cells to infected areas and stimulates the adaptive immune response (1, 2). The initial steps of the complement cascade involve modular proteases that are activated in a sequential manner via one of three pathways: the classic, lectin, and alternative pathways. The classic pathway is triggered by C1, a 790-kDa Ca2+-dependent complex resulting from the association of a recognition protein C1q and a tetramer comprising two copies of the serine proteases C1r and C1s (3–6). Recognition of targets such as pathogens or immune complexes by the C1q moiety of C1 elicits self-activation of C1r, which in turn converts C1s into its active form. Once activated, C1s specifically cleaves C4 and C2, thereby initiating a series of sequential and highly specific proteolytic reactions leading to the formation of the membrane-attack complex and the elimination of the target. The classic pathway of complement is also involved in immune tolerance due to the ability of C1 to recognize and induce clearance of apoptotic cells and plays a major role in xenograft rejection (7). The uncontrolled activation of the complement system, however, can result in self-tissue damages and pathologic inflammation.
During the last years, the three-dimensional structure of several fragments of C1r, C1s, and C1q has been solved by x-ray crystallography and other biophysical methods (6, 8–11). C1r and C1s, and the mannan-binding lectin (MBL)3-associated serine proteases of the lectin complement pathway, share the same type of modular organization (12) with, starting from the N-terminal end, a C1r, C1s, Uegf, and bone morphogenetic protein (CUB) module, an epidermal growth factor (EGF)-like module, a second CUB module, two successive complement control protein (CCP) modules, and a chymotrypsin-like serine protease (SP) domain. Whereas the CCP1-CCP2-SP regions of C1r and C1s mediate their enzymatic properties, their N-terminal CUB1-EGF segments are involved in the Ca2+-dependent C1r-C1s interactions required for assembly of the C1s-C1r-C1r-C1s tetramer. Available structural data have led to low resolution models of the C1 complex in which C1s-C1r-C1r-C1s (hereafter named the tetramer) adopts a compact conformation when bound to C1q (13–15). The main ionic interactions between the C1q collagen stems and the tetramer were initially supposed to be mediated by an acidic cluster located in the C1r EGF module (16). Mutagenesis experiments have recently ruled out this hypothesis (17) and led to a refined three-dimensional model of the C1 complex in which acidic residues involved in the Ca2+-binding sites of the C1r CUB1 and CUB2 and C1s CUB1 modules interact with the C1q stems. Given the location of these sites, the CUB1-EGF-CUB2 interaction domains of C1r and C1s are now proposed to be located entirely inside the cone delimited by the six C1q stems, in sharp contrast with the original model.
To gain further information about the assembly and structure of human C1, we have used stable chemical modifications associated with a mass spectrometry-based strategy and a label-free semi-quantitative approach to investigate the changes of surface accessibility taking place in the tetramer upon C1 assembly. Lysine acetylation is one of the most common chemical modifications used to analyze protein complexes (18–23). Because these residues are charged, they are likely to occupy solvent-exposed regions of proteins, which makes them excellent candidates to identify protein-protein interactions. In addition, the relatively large number of lysines (146 in total) present in the tetramer and their distribution provide the opportunity to investigate the effects of C1q binding on the whole tetramer structure. Our data are consistent with the hypothesis that the C1s interaction domains interact with each other in C1 and provide experimental evidence that the C1s catalytic domains are partly located inside the C1q cone, yielding further insights into C1 architecture.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were of analytical grade. Slide-A-Lyzer 10,000 molecular weight cut-off dialysis cassettes and sulfo-N-hydroxysuccinimide (sulfo-NHS) acetate were purchased from Pierce. Fibrinogen-binding inhibitor peptide (FBIP, fragment 400–411), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP-HCl), α-cyano-4-hydroxycinnaminic acid, sinapinic acid, trifluoroacetic acid, formic acid, and porcine pepsin were all obtained from Sigma. Acetonitrile was purchased from VWR. Coated silica PicoTip emitters for the nano-ESI2 source were obtained from New Objective. ZipTip C4 and C18 tips were purchased from Millipore. Ultrapure water was obtained from a Milli-Q-System (Millipore).
Proteins
C1q and the proenzyme form of the tetramer were isolated from human plasma as described previously (24, 25). Prior to use, C1q was dialyzed at 4 °C against 250 mm NaCl, 4 mm CaCl2, 250 mm Hepes, pH 7.3, using a membrane of 10,000 molecular weight cut-off (final C1q concentration: 1.6 μm).
Chemical Modification of the Lysine Residues of C1s-C1r-C1r-C1s
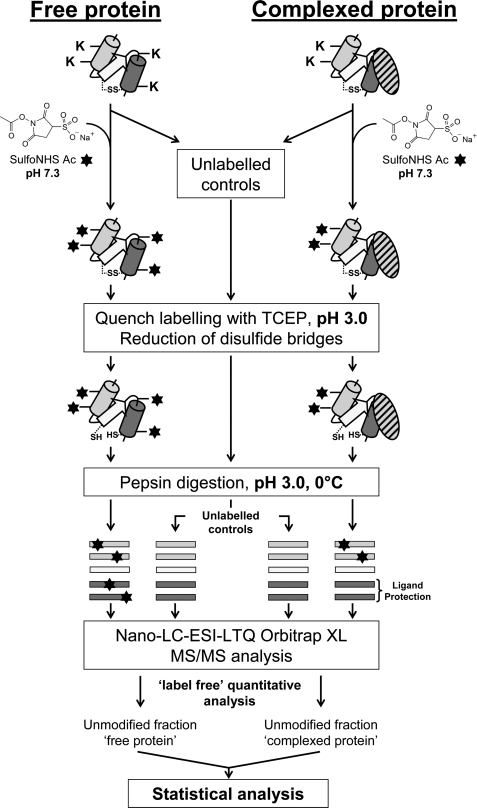
The overall procedure is outlined in Fig. 1. The free tetramer and the C1 complex were labeled in parallel as follows. Solutions containing 15 pmol of the tetramer were prepared in 16 μl of 250 mm NaCl, 4 mm CaCl2, 250 mm Hepes, pH 7.3, in the presence or absence of 16 pmol of C1q, and preincubated for 15 min at 21 °C to allow C1 complex formation. Then, 1 μl of a freshly prepared sulfo-NHS acetate aqueous solution was added to each sample, and labeling was allowed to proceed for 5 min at 21 °C. Sulfo-NHS acetate solutions at 547 and 986 mm were used to reach a ratio of 250 mol of reagent/mol of lysine residues for the tetramer and the C1 complex, respectively. The number of free lysine residues taken into account was 146 for the tetramer (36 in C1r and 37 in C1s) and 271 for C1, assuming 126 unmodified lysines per C1q molecule (26). The reaction was quenched by decreasing the pH to 3.0 using 4 μl of 1 m TCEP-HCl, pH 2.5, and samples were incubated for a further 10 min at room temperature to reduce disulfide bonds. Prior to protein digestion, each sample was placed on ice for 2 min. Then, 2 μl of a pre-cooled porcine pepsin solution prepared in 100 mm H3PO4, pH 1.8, was added to achieve a protease/protein ratio of 1:1 (w/w). Proteolysis was performed at pH 3.0 for 5 min at 0 °C and stopped by adding 1.5 μl of an 8 m NaOH solution (final pH 8.0). After 1 min, samples were acidified to pH 2.5 with 3 μl of a 50% trifluoroacetic solution, flash-frozen in liquid nitrogen, and stored at −80 °C. Unlabeled controls were performed in parallel by replacing the sulfo-NHS acetate reagent by water. All experiments were repeated 15 times to ensure reproducibility and reliability and to generate sufficient data for statistical analysis.
FIGURE 1.
Overview of the experimental approach used in this study. Experimental details are described under “Experimental Procedures.”
For nano-LC-MS/MS analysis, samples were rapidly defrosted on ice and diluted 4-fold in H2O/acetonitrile (98/2, v/v) containing 0.1% formic acid to achieve final tetramer and C1q concentrations of 136 and 143 nm, respectively. Samples were injected randomly to ensure reliability of semi-quantitative measurements.
Validation of the Lysine Accessibility Mapping Procedure
Two critical steps of our experimental approach needed to be validated, namely the quenching of lysine labeling and the quenching of pepsin digestion. This was performed using FBIP (fragment 400–411) as a test sample; its mass increase due to labeling with sulfo-NHS acetate was monitored by MALDI-TOF MS analysis (see the MALDI-TOF-MS section below).
FBIP (12.6 pmol) was prepared in 20 μl of 250 mm NaCl, 4 mm CaCl2, 250 mm Hepes, pH 7.3. One microliter of a freshly prepared 3.1 mm sulfo-NHS acetate solution was then added to reach a ratio of 250 mol of reagent/mol of lysine residues, and the mixture was incubated for 5 min at 21 °C. The reaction was stopped by decreasing the pH to 3.0 using 1 μl of a 50% trifluoroacetic solution. Upon modification at neutral pH, ∼50% of FBIP was acetylated, resulting in an increase in molecular mass of 42 Da (supplemental Figs. S1A and S1B). To evaluate the efficiency of the quenching by TCEP-HCl, the FBIP solution was acidified to pH 3.0 prior to the addition of sulfo-NHS acetate. In addition to decreasing the pH value, TCEP-HCl reduces protein disulfide bonds within a few minutes at room temperature. Labeling was allowed to proceed for 5 or 30 min. In contrast to the positive control (supplemental Fig. S1A), the labeling reaction was completely inhibited when the pH was decreased to 3.0. The acetylated form of FBIP was no longer observed on the MALDI mass spectra, indicating that the quenching procedure was highly effective (supplemental Fig. S1C).
In the procedure used (Fig. 1), following sample reduction, labeled and unlabeled samples are subjected to pepsin digestion on ice. After protein hydrolysis, pepsin is irreversibly denatured by increasing the pH from 3.0 to 8.0 for 1 min. Because the acetylation reaction is pH-sensitive, this increase may reactivate the labeling. To minimize as far as possible unwanted lysine modifications on the generated peptic fragments, pepsin inactivation was performed at 0 °C. To check that lysine acetylation was inhibited at this temperature, FBIP was used as a control sample. Briefly, the FBIP solution was prepared and acidified as described above, then 1 μl of a 3.1 mm sulfo-NHS acetate solution was added, and the mixture was left on ice for 7 min. The pH was then raised to 8.0 by adding 1.5 μl of an 8 m NaOH solution, and the mixture was left at 0 °C for 1 more min prior to re-acidification to pH 2.5 with a 50% trifluoroacetic acid solution. The extent of acetylation observed by MALDI-TOF MS analysis was very low (supplemental Fig. S1D), confirming that the structural information obtained on the samples would be retained upon pepsin inactivation.
Control of the Tetramer Activation State in C1
The activation state of the tetramer upon interaction with C1q was checked by MALDI-TOF-MS analysis. For this purpose, after incubation for 30 min at 21 °C, the protein solution was acidified to pH 3.0 using 110 mm TCEP-HCl and incubated for a further 10 min to reduce disulfide bonds. Acidified samples were then prepared for MALDI-TOF-MS analysis in the linear mode.
MALDI-TOF-MS
All MALDI experiments were performed on a Voyager DE-STR mass spectrometer (Applied Biosystems, Boston, MA). For analysis of intact proteins, each acidified sample (10 μl) was desalted using a ZipTip C4 tip and eluted with 5 μl of H2O/acetonitile (40/60, v/v) containing 1% formic acid. The same procedure was used for peptides, using a ZipTip C18 tip. All desalted samples were mixed in a 1:1 (v/v) ratio with a matrix solution. For analysis of proteins in the linear mode, the matrix consisted of a saturated sinapinic acid solution prepared in H2O/acetonitrile (70/30, v/v), 1% formic acid, whereas saturated α-cyano-4-hydroxycinnaminic acid in H2O/acetonitrile (1/1, v/v), 1% formic acid was used for peptide analysis in the reflectron mode. One microliter of each sample-matrix mixture was spotted on the target plate and air dried.
In the linear mode, data were acquired with an accelerating voltage of 25 kV, a 75% grid voltage, and a 445-ns extraction delay time. In the reflectron mode, these values were set to 20 kV, 70%, and 225 ns. Spectra were produced by signal accumulation obtained from 100 consecutive laser shots. All data were reprocessed using the Applied Biosystems Data Explorer 4.0 software. The mass scale was calibrated externally using either the TIS test mixture (SequazymeTM Peptide Mass Standards kit, Applied Biosystems) for the reflectron mode (m/z 500–2,500) or a bovine serum albumin solution (m/z 4,500–120,000) for the linear mode.
Nano-LC-ESI-MS/MS
Nano-LC-ESI-MS/MS experiments were performed with an Ultimate 3000 nano-LC system (Dionex) connected to a LTQ OrbiTrap XL mass spectrometer (ThermoFisher Scientific) equipped with a nanospray ion source. Unlabeled and labeled samples (205 fmol of tetramer and 214 fmol of C1q), were loaded onto a C18 μ-precolumn cartridge (Acclaim PepMap100, 5 mm × 300 μm inner diameter, 5 μm, 100 Å, Dionex) and desalted at 30 °C for 5 min at a flow rate of 20 μl/min with 100% buffer A (H2O/acetonitrile (98/2, v/v) containing 0.1% formic acid). Peptides were then separated on an analytical capillary C18 column (Acclaim PepMap100, 15 cm × 75 μm inner diameter, 3 μm, 100 Å, LC-Packings) by means of a 2–80% acetonitrile gradient over 50 min at 30 °C and a flow rate of 0.3 μl/min. The nano-LC eluate was directly interfaced to the nanospray ion source using a coated silica tip (10.5-cm length, 20-μm inner diameter with 10-μm inner diameter at the tip extremity). The ion spray voltage was 1.6 kV with a transfer capillary temperature of 200 °C.
For MS/MS experiments, the LTQ Orbitrap XL was operated in the data-dependent mode. Each scan cycle comprised one MS scan in the Orbitrap analyzer (m/z range 400–2,000, scan resolution of 30,000, and MS target value of 3 × 105 with a maximum injection time of 500 ms) followed by five sequential data-dependent MS2 scans performed in the linear ion trap to fragment the five most intense precursors found in the preceding MS spectrum (MSn target value of 1 × 104 with a maximum injection time of 100 ms). For collision-induced dissociation, the normalized collision energy was 35%, the activation time was 40 ms, the isolation width was 3.0 units, the activation q was 0.25, and the minimal signal required was 500. To ensure mass accuracy, the Orbitrap analyzer was calibrated in the positive mode according to the manufacturer's instructions. For all MS/MS experiments, the monoisotopic precursor selection mode was selected, and the dynamic exclusion was used with one repeat count, a 30-s repeat duration, and an exclusion duration of 90 s. Mass spectra were processed using Bioworks 3.3.1 (ThermoFisher Scientific) to generate Mascot-compatible MGF file.
MS/MS Data Processing: Procedure of Label-free Relative Quantification
Peptide identification was carried out using either the SEQUEST algorithm (ThermoFisher Scientific) or the locally installed Mascot search engine (version 2.2.1., Matrix Science) using an in-house data base containing the amino acid sequences of C1r and C1s. The mass tolerance was set to 10 ppm and 0.8 Da for MS and MS/MS analyses, respectively. Given the nonspecificity of pepsin, no enzyme was selected for peptide assignment. Acetylations on lysine, serine, and tyrosine residues were set as variable modifications. Possible oxidation of methionine residues was also taken into account.
To assess the effects of C1q binding on the solvent-accessible surface of the tetramer, the extracted ion currents of the precursor ions were utilized for relative quantification. Quantification was performed either manually or by using the SuperHirn software (available on-line) (27). SuperHirn was used to rapidly identify peptides exhibiting a modified acetylation pattern in the presence of C1q. To construct the MasterMaps, raw data were first converted to mzXML files with ReAdW (available on-line), and pepXML files were generated with the Mascot search engine. Manual quantification was performed as follows: for each identified peptide containing lysine residues, the extracted ion current of the precursor ion was extracted from the total ion current, and the area under the peak (AUP) was calculated using QualBrowser 2.0.7 (ThermoFisher Scientific). The unmodified fraction (UF) of each peptide was then estimated using the following formula (28),
 |
where UF corresponds to the fraction of an unlabeled tetramer peptide remaining after acetylation in the presence or absence of C1q.
Statistical Analysis
For each peptide, the UFs calculated from the 15 replicate samples were pooled and organized into two independent groups containing all the values obtained in the absence (group A) or in the presence (group B) of C1q. A non-parametric Mann-Whitney U test was then applied to test the null hypothesis (H0). H0 is defined as the hypothesis of “no difference,” meaning no solvent accessibility difference between groups A and B. A two-sided p value was used to perform the U test. The p value corresponds to the estimated probability of rejecting the null hypothesis and was set to 1% (<1 in 100 chances of being wrong). An excel sheet containing the Mann-Whitney U test was directly downloaded from Formations et études en statistiques (available on-line).
UF values obtained for each peptide were graphically depicted using a box-and-whisker plot to compare the statistical dispersion between groups A and B. Box-plots were constructed as follows. Data were first ordered from the smallest to the highest UF values. Then, the lower (Q1), median (Q2), and upper (Q3) quartiles were calculated. Quartiles divide the data set into four equal parts, so that each represents one fourth of the ordered set of UF values. Q2 corresponds to the exact middle of the ordered set of all UF values (central tendency), whereas Q1 and Q3 represent the exact middle numbers of the lower and upper half of the UF data set, respectively. Then, the inter-quartile range was calculated by subtracting the lower quartile from the upper quartile. The inter-quartile range (IQR) corresponds to a measurement of the statistical dispersion of the population of each group. The minimum (Min) and maximum (Max) non-outlier values were calculated using the following equations:
Any data lying below the Min or above the Max values was considered to be an outlier.
RESULTS
Most of our current knowledge about the overall architecture of C1 arises from low resolution studies by electron microscopy and neutron scattering as well as from the crystal structure of C1r and C1s fragments and of the C1q globular heads. The main barriers to solve the whole structure of C1 and decipher the mechanism of its assembly lie in the size of the complex and the fact that it involves non-covalent interactions. To gain access to new structural information about this complex, we used mass spectrometry in association with a stable chemical modification of lysine residues. We chose to investigate the solvent accessibility of lysines in the free and complexed forms of the tetramer for several reasons: (i) these residues are abundant and evenly distributed in the tetramer; (ii) lysine residues are mostly located on the surface of proteins and are therefore ideal candidates for probing protein-protein interfaces; and (iii) unlike C1q, the tetramer does not have an oligomeric organization, which is expected to facilitate identification of the areas of C1r and C1s involved in conformational changes and/or in binding to C1q. In addition, previous studies have shown that chemical modifications of the lysine residues of the tetramer do not prevent assembly of C1 (29).
The solvent accessibility of lysine residues was probed by stable chemical modification with the primary amine-specific reagent sulfo-NHS acetate. The free tetramer and the reconstituted C1 complex were both exposed to an excess of reagent, and unlabeled controls were prepared in parallel to calculate the unmodified fraction of each lysine-containing peptide (see below). The labeling was then quenched by decreasing the pH to 3.0 using a TCEP-HCl solution. Following reduction of the disulfide bridges, labeled and unlabeled protein samples were both subjected to proteolysis by pepsin, an acid protease that retains its enzymatic activity under the quenching conditions, i.e. at low pH, low temperature, and high concentration of the reducing agent (30, 31). After proteolysis, pepsin was irreversibly denatured by increasing the pH to 8.0.
Effects of C1q Binding on the Lysine Acetylation Pattern of C1r
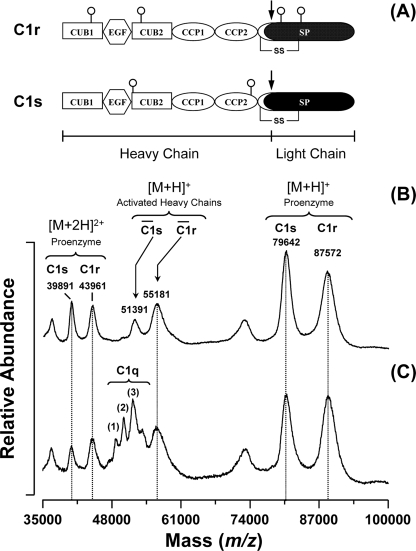
To validate our approach, it was essential to ensure that the acetylation reaction was carried out under conditions where the integrity of the C1 complex was fully preserved. First, labeling was performed at neutral pH so that the native conformation of C1 was retained. In addition, the activation state of C1 was checked throughout the labeling procedure, considering that C1 is known to undergo spontaneous activation in vitro even without binding to a target (32). Activation of the complex induces structural changes within the tetramer (9, 33) and leads to the hydrolysis of the Arg–Ile bond located in the SP domain of each protease. As a result, both activated C1r and C1s proteins comprise two chains linked by a disulfide bridge (Fig. 2A). To evaluate the extent of C1 activation under the experimental conditions used, the complex was incubated at room temperature for 30 min at pH 7.3, this incubation time corresponding to the duration of the whole procedure, except pepsin digestion. As shown in Fig. 2B, a minor fraction of the tetramer was found to be activated in the absence of C1q. This activation mainly occurred during the multiple purification steps required to isolate the tetramer from human plasma. However, the activated fraction of the tetramer did not increase in the C1 complex after incubation for 30 min at room temperature (Fig. 2C), indicating that, under the experimental conditions used, the structural integrity of C1 was preserved throughout the labeling procedure.
FIGURE 2.
Effect of C1q binding on the activation state of the C1s-C1r-C1r-C1s tetramer. A, modular structure of C1r and C1s. Both proteases are activated through cleavage of an Arg–Ile bond (represented by a black arrow) located in their SP domain. The only disulfide bridge shown is the one connecting the activation peptide to the SP domain. N-Linked oligosaccharides are represented by open circles. B, MALDI-TOF mass spectra of the isolated tetramer under reducing conditions. In the absence of C1q, a small fraction of the tetramer appears activated, as indicated by the peaks corresponding to the heavy chains of C1r and C1s. C, MALDI-TOF mass spectra of the reconstituted C1 complex under reducing conditions. The peaks at m/z 48114.6 (1), 49695.5 (2), and 51317.8 (3) correspond to random associations of the C1q chains (43).
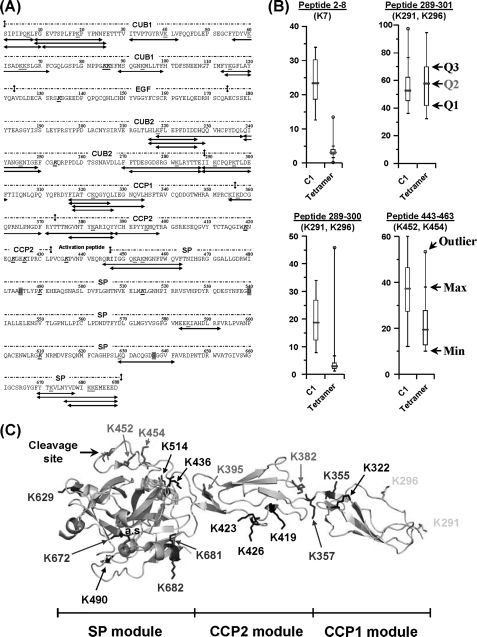
Thirty-six peptides containing lysine residues were assigned to C1r by LC-MS/MS analysis of the pepsin digests, thus accounting for 25 of the 36 lysines of this protein (Fig. 3A). Residues that were not recovered were mainly located at the C-terminal end of the CCP2 module (Lys419, Lys423, Lys426, and Lys436) and in the SP domain (Lys490, Lys514, and Lys610). To assess the effects of C1q binding on the accessibility of each lysine residue, a statistical Mann and Whitney U test with a two-sided p value of 1% was applied on the 15 independent MS data sets collected. As listed in Table 1, the solvent accessibility of most of the residues located in modules CUB1 and CUB2 remained unchanged upon C1q binding. Interestingly, a single lysine located in the N-terminal part of CUB1 (Fig. 3A) behaved differently. The box-and-whisker diagram of fragment 2–8 revealed that Lys7 is highly exposed in the free tetramer, as only 3% remained unmodified after acetylation (Fig. 3B). This value shifted to ∼23% in the presence of C1q, indicating that Lys7 is partially protected from chemical modification within C1.
FIGURE 3.
Modification of the C1r solvent accessibility upon association of the C1s-C1r-C1r-C1s tetramer with C1q. A, amino acid sequence of C1r showing the 36 lysine-containing peptic fragments (in red, bold type, and underlined) selected for quantitative analysis. Lysines that could not be recovered are shown in black, bold type, and underlined. The catalytic residues His485, Asp540, and Ser637 and the Arg-Ile cleavage site are highlighted in magenta and shown in blue, respectively. C1r residues interacting with C1q (17) are highlighted in yellow. B, effect of C1q binding on the solvent accessibility of residues Lys7 (CUB1), Lys291/Lys296 (CCP1), and Lys452/Lys454 (SP domain). Each box-and-whisker plot compares the statistical distribution of the unmodified fraction of a given C1r peptide in the presence (C1) or absence (tetramer) of C1q. Q1, Q2, and Q3 correspond to the lower, median (red bar), and third quartiles, respectively. The largest (Max) and smallest (Min) non-outlier observations are marked with a small black vertical line (whiskers). Data points lying above the upper whisker or below the lower whisker are considered as outliers and indicated by an open circle. C, structure of the zymogen CCP1-CCP2-SP C1r catalytic domain (10) showing the position of lysine residues. The catalytic triad (His485, Asp540, and Ser637) is represented by three magenta spheres. Lysine residues are color-coded as follows: blue, no modification of surface accessibility upon C1 assembly; red, decreased surface accessibility; yellow, decreased and/or unmodified surface accessibility; and black, no data available.
TABLE 1.
Effect of C1 assembly on the solvent accessibility of the lysine residues of C1r
Residues showing significant changes in their accessibility are shown in bold type.
| Domain | C1r protein |
|||
|---|---|---|---|---|
| Lysines | SASAa | Mann-Whitney U test, H0b, p < 0.01c | Accessibility within C1 | |
| Å2 | ||||
| CUB1 | K7 | –d | False | Decreased |
| K19 | –d | True | Unchanged | |
| K40 | –d | True | Unchanged | |
| K60 | –d | True | Unchanged | |
| K65 | –d | True | Unchanged | |
| K66 | –d | True | Unchanged | |
| K85 | –d | NDe | ND | |
| K86 | –d | ND | ND | |
| K94 | –d | True | Unchanged | |
| K115 | –d | True | Unchanged | |
| EGF | K134 | 59.0 | ND | ND |
| CUB2 | K218 | –d | True | Unchanged |
| K245 | –d | True | Unchanged | |
| K253 | –d | True | Unchanged | |
| K282 | –d | ND | ND | |
| CCP1 | K291f | 81.2 | True & false | Unchanged & decreasedf |
| K296f | 159.0 | |||
| K322 | 99.8 | ND | ND | |
| K355 | 101.1 | True | Unchanged | |
| CCP2 | K357 | 46.7 | True | Unchanged |
| K382 | 114.3 | False | Decreased | |
| K395 | 145.6 | False | Decreased | |
| K419 | 64.0 | ND | ND | |
| K423 | 50.6 | ND | ND | |
| K426 | 165.6 | ND | ND | |
| a.p.g | K436 | 89.3 | ND | ND |
| SP | K452 | 124.4 | False | Decreased |
| K454 | 136.1 | False | Decreased | |
| K490 | 8.8 | ND | ND | |
| K514 | 42.4 | ND | ND | |
| K585 | 101.0 | True | Unchanged | |
| K610 | 85.3 | ND | ND | |
| K629 | 115.2 | True | Unchanged | |
| K672 | 30.0 | True | Unchanged | |
| K681 | 98.9 | True | Unchanged | |
| K682 | 146.4 | True | Unchanged | |
a Solvent accessibility surface area of lysine side chains. Accessible surface areas of C1r are based on available X-ray data (pdb accession numbers: 1APQ (EGF), 1GPZ (CCP1-CCP2-SP), and 1MD8 (SP)) and calculated by using the software program VADAR (44).
b H0, hypothesis of “no difference” of solvent accessibility between the free tetramer and C1.
c Two-sided p value; the significant level at which H0 is rejected is set to 1%.
d No structure available.
e ND, not determined.
f Residues covered by two distinct peptic peptides with different solvent accessibility modifications upon C1q binding.
g Activation peptide.
h Residue was undefined in the crystal structure (10).
C1r Lys291 and Lys296 are also fully exposed to the solvent in the free tetramer, with solvent accessibility surface area (SASA) values of 81.2 and 159.0 Å2, respectively. These two residues are located in the N-terminal part of the CCP1 module and are covered by two distinct overlapping peptides 289–300 and 289–301 (Fig. 3, A and C). Surprisingly, the overall acetylation extent of Lys291 and Lys296 in the free tetramer was strikingly different in these peptides: ∼3% of peptide 289–300 remained unacetylated, whereas this value increased to 58% for peptide 289–301 (Fig. 3B). This difference was observed consistently in all experiments, suggesting the presence of two distinct conformations in solution. In addition, the chemical reactivity of both peptides toward the acetylating agent was also significantly different in the presence of C1q. Upon C1 formation, the unmodified fraction of peptide 289–301 remained unchanged, whereas the accessibility of peptide 289–300 slightly decreased (Fig. 3B). Taken together, these observations suggested that the N-terminal end of the C1r CCP1 module exhibits two different conformations, both in the isolated tetramer and in C1.
Four other lysines exhibiting reduced solvent accessibility upon C1q binding are also identified in the C1r CCP2 and SP domains. Residues Lys382 and Lys395 are both located in the CCP2 region (Fig. 3C) and were fully exposed to the solvent, as judged by their calculated SASA values of 114.3 Å2 and 145.6 Å2. Upon C1q binding, both lysines became protected from modification, with a 30% increase of their respective unmodified fraction (data not shown). Similar results were observed with Lys452 and Lys454 from the SP domain. As seen in the crystal structure of the proenzyme SP domain, these latter residues are very close to the activation segment of C1r (Fig. 3, A and C) and display similar SASA values (Table 1). When the C1 complex is formed, both lysines become less accessible to the acetylating reagent, with an increase of 20% of their unmodified fraction (Fig. 3B). This result was surprising, because the activation segment contains the susceptible Arg446–Ile447 bond cleaved upon autolytic activation of C1r. Therefore, no major change in the solvent accessibility of this region was expected, considering that this site should remain fully accessible. A possibility is that the activation segment becomes less exposed to the solvent in the resting C1 complex to adopt a conformation inappropriate for C1r self-activation. However, this hypothesis does not appear consistent with previous data indicating that the C1r activation potential is prevented in the free tetramer and restored in C1 (34).
Effects of C1q Binding on the Lysine Acetylation Pattern of C1s
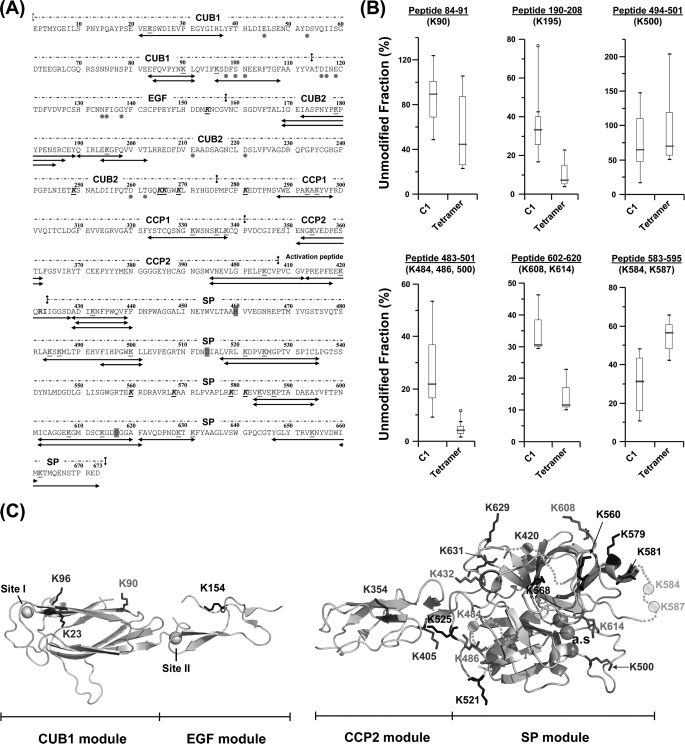
We next investigated the effects of C1 assembly on the acetylation pattern of C1s. As summarized in Fig. 4A, 30 peptic peptides were assigned to C1s by MS/MS, thus accounting for 26 of the 37 lysine residues of the protein. Most of the lysines that were not recovered are clustered in the C-terminal part of the CUB2 module (Lys249, Lys265, Lys266, and Lys269) and in the SP domain (Lys560, Lys568, Lys579, and Lys581). Two lysines exhibiting reduced solvent accessibility in C1 were identified in C1s CUB1 and CUB2. Residues Lys90 (CUB1) and Lys195 (CUB2) appearred fully exposed in the isolated tetramer and became less accessible in the presence of C1q. This effect was particularly striking in the case of Lys90, which showed a 40% increase of its unmodified fraction, thereby becoming virtually inaccessible in C1 (Fig. 4, B and C). A similar overall tendency was observed for the lysine residues located in the CCP1 module, except for Lys281 and Lys338 (Table 2). In contrast to the above observation, the two lysine residues located in CCP2, and the single lysine Lys420 found in the activation segment showed no change in accessibility upon assembly of the C1 complex (Table 2 and supplemental Fig. S2). Lys420 is located in the activation segment of C1s that contains the Arg422–Ile423 bond cleaved upon activation by C1r. The activation segment of C1s thus appears to remain fully accessible in C1, in contrast to our observation in C1r.
FIGURE 4.
Modification of the C1s solvent accessibility upon interaction of the C1s-C1r-C1r-C1s tetramer with C1q. A, amino acid sequence of C1s showing the 30 lysine-containing peptic fragments used for quantitative analysis. The color coding used is the same as stated in the legend to Fig. 3. B, effect of C1q binding on the surface accessibility of residues Lys90 (CUB1), K195 (CUB2), and residues Lys484, Lys486, Lys500, Lys584, Lys587, Lys608, and Lys614 of the SP domain. Each box-and-whisker plot compares the statistical distribution of the unmodified fraction of a given C1s peptide in the presence (C1) or absence (tetramer) of C1q. C, structures of the C1s CUB1-EGF (16) and CCP2-SP regions (33) showing the position of lysine residues. The Ca2+ ions bound to CUB1 (site I) and EGF (site II) are represented by yellow spheres, and the catalytic triad is shown as three magenta spheres. Orange dots correspond to residues not defined in the C1s CCP2-SP x-ray structure. Lysine residues are color-coded as follows: blue, no modification of solvent accessibility inside the C1 complex; red, decreased accessibility; green, increased accessibility; and black, no data available.
TABLE 2.
Effect of C1 assembly on the solvent accessibility of the lysine residues of C1s
Residues showing significant changes in accessibility upon C1q binding are shown in bold type.
| Domain | C1s protein |
|||
|---|---|---|---|---|
| Lysines | SASAa | Mann-Whitney U test, H0b, p < 0.01c | Accessibility within C1 | |
| Å2 | ||||
| CUB1 | K23 | 93.0 | True | Unchanged |
| K90 | 117.3 | False | Decreased | |
| K96 | 81.2 | True | Unchanged | |
| EGF | K154 | 67.2 | NDe | ND |
| CUB2 | K179 | –d | True | Unchanged |
| K195 | –d | False | Decreased | |
| K249 | –d | ND | ND | |
| K265 | –d | ND | ND | |
| K266 | –d | ND | ND | |
| K269 | –d | ND | ND | |
| CCP1 | K281 | –d | ND | ND |
| K293 | –d | False | Decreased | |
| K295 | –d | False | Decreased | |
| K331 | –d | False | Decreased | |
| K336 | –d | False | Decreased | |
| K338 | –d | ND | ND | |
| CCP2 | K354 | 117.5 | True | Unchanged |
| K405 | 122.0 | True | Unchanged | |
| a.p.f | K420 | –g | True | Unchanged |
| SP | K432 | 66.2 | False | Decreased |
| K484 | –g | False | Decreased | |
| K486 | 87.0 | False | Decreased | |
| K500 | 112.8 | True | Unchanged | |
| K521 | 100.0 | ND | ND | |
| K525 | 96.8 | ND | ND | |
| K560 | 153.2 | ND | ND | |
| K568 | 37.2 | ND | ND | |
| K579 | 87.4 | ND | ND | |
| K581 | 53.0 | ND | ND | |
| K584 | –g | False | Increased | |
| K587 | –g | False | Increased | |
| K608 | 148.3 | False | Decreased | |
| K614 | 145.6 | False | Decreased | |
| K629 | 147.2 | True | Unchanged | |
| K631 | 54.5 | True | Unchanged | |
| K654 | 35.0 | True | Unchanged | |
| K662 | 116.3 | True | Unchanged | |
a SASA of lysine side chains. Accessible surface areas of C1s are based on available X-ray data (pdb accession numbers: 1NZI (CUB1-EGF) and 1ELV (CCP2-SP)) and calculated by using the software program VADAR (44).
b H0, hypothesis of “no difference” of solvent accessibility between the free tetramer and C1.
c Two-sided p value; the significant level at which H0 is rejected is set to 1%.
d No structure available.
e ND, not determined.
f Activation peptide.
g Residue was undefined in the crystal structure (33).
Among the 18 lysine residues located in the C1s SP domain, five (Lys432, Lys484, Lys486, Lys608, and Lys614) showed significantly reduced accessibility within C1 (Table 2 and Fig. 4C). As shown in Fig. 4A, Lys500 is covered by two overlapping peptides, 494–501 and 483–501. No significant modification of solvent accessibility was observed for peptide 494–501, whereas the unmodified fraction of peptide 483–501 increased by 16% (Fig. 4B), indicating that reduction of solvent accessibility was restricted to the segment containing Lys484 and Lys486. This example illustrates the advantage of using the nonspecific protease pepsin, which generates overlapping fragments, thereby allowing in some cases the accessibility of particular lysine residues to be assessed.
The accessibility of peptides 602–620 and 602–621 was also significantly reduced (∼20% in both cases) upon interaction with C1q, indicating decreased exposure of Lys608 and/or Lys614 (Fig. 4B). In contrast, two lysines showing increased solvent accessibility, Lys584 and Lys587, were identified in the C1s SP domain (Fig. 4B). Both residues lie in a segment not defined in the C1s catalytic domain x-ray structure and are possibly located in the vicinity of the active site entrance (Fig. 4C). Binding to C1q markedly increases their reactivity toward the acetylating reagent, indicating that conformational changes occur in this segment upon C1 assembly.
DISCUSSION
X-ray crystallography and NMR spectroscopy have been extensively used over the past decade to gain structural information about the constituent proteins of human C1, allowing resolution of 67 and 72% of the C1r and C1s structures, respectively (6). Although there are still missing links, namely the C1r CUB1 and CUB2 modules and the C1s CUB2-CCP1 segment, these data provide an overall view of the three-dimensional organization of the free C1s-C1r-C1r-C1s tetramer in solution. Earlier information arising from neutron scattering (35, 36) and electron microscopy analyses (37–39) had led to the concept that the extended tetramer folds into a more compact conformation upon interaction with C1q, providing the basis for most of the low resolution C1 models proposed originally (13–15).
The sites of C1r and C1s involved in the interactions between the tetramer and C1q have recently been delineated by site-directed mutagenesis (17), revealing that C1 assembly involves high-affinity C1q-binding sites contributed by the C1r CUB1 and CUB2 modules and lower affinity sites contributed by the C1s CUB1 modules. Based on the location of these sites and available structural information, a refined three-dimensional model of C1 assembly has been proposed, where the CUB1-EGF-CUB2 interaction domains of both C1r and C1s are entirely located inside C1q and interact via six binding sites with reactive lysines located approximately half-way along the C1q collagen-like stems (17, 26, 40). Based on the use of truncated protease segments, a similar interaction model was derived by another group (41).
The present study provides for the first time a detailed comparative analysis of the accessibility of the lysine residues of C1r and C1s in the free and complexed forms of the C1s-C1r-C1r-C1s tetramer, allowing us to test whether our current concept of C1 assembly (17) is consistent with this new information. As no experimental x-ray structure is available yet for C1r CUB1 and CUB2 and C1s CUB2 and CCP1, the data relating to these modules will not be discussed in detail, considering the relative imprecision of three-dimensional homology models. Nevertheless, it is interesting to note that, with the exception of Lys7 in C1r CUB1 and Lys195 in C1s CUB2, the accessibility of the lysine residues contained in these modules remains unchanged upon C1 assembly (Tables 1 and 2). This is consistent with the fact that C1r and C1s interact with C1q through acidic residues contributed by their CUB modules (17) and with earlier studies showing that chemical modification of the lysine residues of C1s-C1r-C1r-C1s has no effect on C1 assembly (29). The fact that little structural changes are detected in the N-terminal interaction regions of C1r and C1s also likely arises for a large part from the high stability of the head-to-tail C1r/C1s CUB1-EGF heterodimeric assemblies that connect C1r to C1s in the tetramer and are expected to retain a relatively rigid conformation in the C1 complex (16).
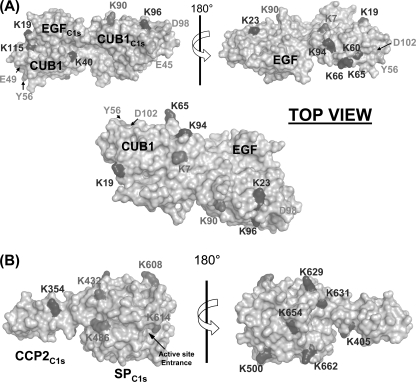
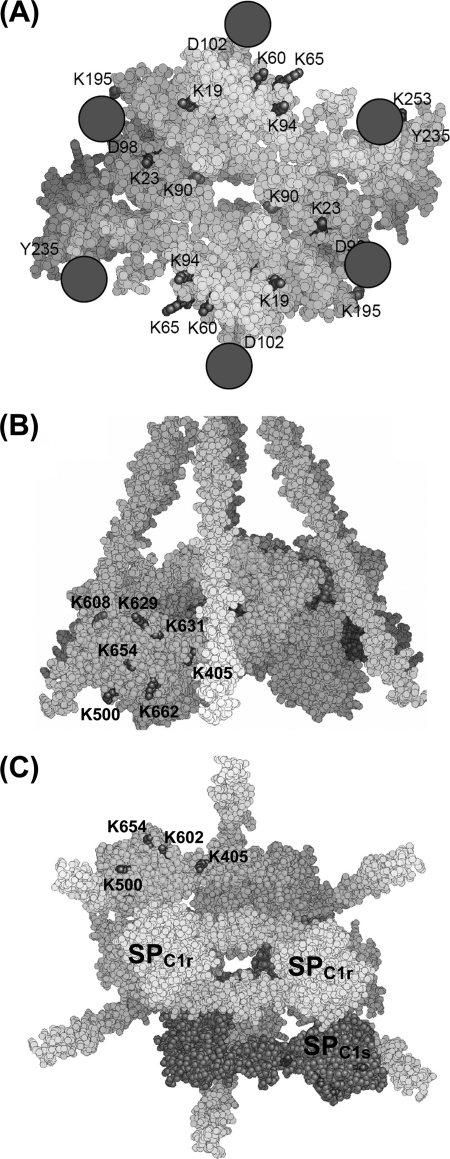
The labeling data relating to the C1s CUB1 module, in contrast, can be analyzed in light of the C1s CUB1-EGF x-ray structure (16). In this module, whereas the accessibility of Lys23 and Lys96 remained unchanged upon C1 assembly, that of Lys90 decreased very significantly (Table 2 and Fig. 4B). As judged from the C1s CUB1-EGF structure, all three lysine residues are expected to be accessible in the free tetramer. The new version of the C1 model is largely based on the assumption that both C1r/C1s CUB1-EGF-CUB2 heterodimers, which were distant in the free tetramer, became closely packed side by side through their C1s moieties to form a single compact assembly inside the C1q cone (17). That such a close packing actually occurs in C1 is fully supported by our labeling data, considering that, in the resulting assembly, Lys90 is positioned in the middle of the C1s/C1s interface, consistent with its decreased accessibility (Figs. 5A and 6A). Conversely, Lys23 is expected to remain accessible in C1, and the same applies to Lys96, despite the proximity of this latter residue with the low affinity C1q-binding site harbored by C1s CUB1 (17). Thus, that C1 assembly involves formation of a new interaction between both C1s CUB1-EGF-CUB2 domains, an essential requirement of our current C1 model, appears fully consistent with the above experimental data. Conversely, the fact that most of the lysine residues of C1r CUB1 and CUB2 are labeled to similar extents in the free tetramer and in C1 is also compatible with the model, considering that, except in the vicinity of the C1q-binding sites, these modules are expected to remain fully accessible in C1, given their location on the outer part of the C1r/C1s interaction domains assembly (Fig. 6A).
FIGURE 5.
A, space-filling representation of the head-to-tail C1r/C1s CUB1-EGF heterodimer. One C1s monomer (gray) was used as a template to position and visualize the lysine residues identified in the C1r CUB1 module, based on the sequence alignment of the CUB1 modules of C1r and C1s (supplemental Table S1). Lysines are color-coded as defined in Fig. 3. C1r and C1s residues interacting with C1q are colored green (17). B, space-filling representation of the C1s CCP2-SP region illustrating the position of the lysine residues (in red) showing reduced solvent accessibility upon C1q binding, except for Lys484, which lies in a region not defined in the crystal structure (33). Lysines undergoing no modification of solvent accessibility within C1 are all located on the same face, opposite to the one harboring Lys432, Lys486, Lys608, and Lys614.
FIGURE 6.
A, space-filling representation of the assembly of the C1r/C1s CUB1-EGF-CUB2 interaction domains as proposed to occur in the C1 complex (17) (top view). C1r and C1s are shown in yellow and gray, respectively. The color coding used is the same as stated in the legend to Fig. 3. Residues interacting with C1q are colored green. The six collagen triple helices of C1q are shown as magenta spheres. B and C, side and bottom views of the whole C1 complex highlighting the positioning of the C1s SP domains with respect to the remainder of the complex. Both C1r monomers are in yellow, whereas C1s molecules are shown in cyan and magenta. The color coding used for lysine residues is the same as stated in the legend to Fig. 3.
An intriguing finding from our study lies in the peculiar labeling data relating to the C1r segment Ile289–Phe301, suggesting that this area exhibits two different conformations in both the free and complexed forms of the tetramer (Fig. 3B). As this sequence stretch is covered by two overlapping peptides differing by a single residue (Fig. 3A), we initially suspected an artifact of the “label-free” quantification procedure. However, similar results were consistently obtained when performing 15 independent labeling experiments, hence validating the above hypothesis. Indeed, this segment, at the interface between the CUB2 and CCP1 modules of C1r, is known to be intrinsically very flexible, as shown by its high susceptibility to cleavage by a variety of proteolytic enzymes (42). Likewise, one of the two lysines contained in this segment (Lys296) exhibits opposite orientations in the two x-ray structures currently available (10, 11). Thus, that the two overlapping peptides are differentially labeled in the free tetramer likely reflects the fact that the corresponding sequence stretch adopts two alternative conformations. In the C1 complex, the CUB2 modules of C1r are thought to occupy the upper part of the C1q cone, the following CCP1-CCP2-SP catalytic domains being in contrast positioned in the lower part (17). Connection between these two compartments therefore likely requires significant conformational changes at the CUB2/CCP1 interface, which appears consistent with the decreased lysine accessibility observed for peptide 289–300 (Fig. 3B). Why in contrast the accessibility of the other peptide 289–301 remains unchanged in C1 is currently unknown, although it cannot be excluded that this reflects a structural asymmetry between the CUB2-CCP1 junctions of both C1r molecules, connected with the expected asymmetry of the C1r activation process itself (6). It is noteworthy that residues Lys293 and Lys295 of C1s, located in the equivalent region of module CCP1 at the interface with the preceding CUB2 module, also exhibit decreased accessibility in C1. This is consistent with the hypothesis that C1s also possesses significant flexibility at its CUB2/CCP1 interface, a feature expected to be necessary for the C1s activation process (6).
The C1r residues Lys355 and Lys357, on either side of the CCP1/CCP2 junction, are both exposed in the C1r CCP1-CCP2-SP structure (10) and remain fully accessible in the C1 model, in agreement with our labeling data (Table 1). In the same way, Lys395 (C1r CCP2), which is near the C1r SP domain in the CCP1-CCP2-SP structure, could move closer to this domain, by means of the limited flexibility of the CCP2/SP interface (9, 10), or could become engaged in a salt bridge with the neighboring residue Glu391, hence its decreased accessibility in the C1 complex. Residues Lys629, Lys672, Lys681, and Lys682 of the C1r SP domain are all exposed, to various extents, in the CCP1-CCP2-SP structure (Fig. 3C) and remain accessible in the C1 model, in agreement with the labeling data (Table 1). In contrast, the fact that residues Lys452 and Lys454 show a decreased labeling in C1 does not appear fully consistent with the model, according to which they should remain accessible despite their orientation toward a C1r CUB2 module. It should be emphasized, however, that Lys454 exhibits variable orientations in the four available x-ray structures (9, 10, 11) and lies in the vicinity of the C1r activation segment 444–450, which is very unstable in the zymogen form. Finally, according to our model, Lys382 (C1r CCP2) should remain accessible in the C1 complex, in contrast to the labeling data.
A major outcome of this study lies in the observed differential accessibility of the lysine residues contained in the C1s SP domain, a finding that for the first time indicates that this domain is at least partially inserted in the C1q cone, in the proenzyme C1 complex, and yields precise information about its orientation in the complex. Thus, the lysine residues showing unmodified accessibility upon interaction of the tetramer with C1q (Lys500, Lys629, Lys631, Lys654, and Lys662) are all located on the same side of the C1s SP domain (Fig. 5B), indicating that this part of the domain remains exposed to the solvent and therefore likely faces the outside of the complex. Consistent with this hypothesis, four residues showing decreased labeling in C1 (Lys432, Lys484, Lys486, Lys608, and Lys614) lie on the opposite side of the C1s SP domain (Fig. 5B), providing strong indication that, conversely, this region faces the inside of the complex. Thus, it is tempting to hypothesize that Lys432, Lys484, Lys486, and Lys608 may be involved in interactions with the C1r catalytic domains positioned inside the C1q cone or with C1q itself. In contrast, the implication of Lys614 in such interactions appears less likely considering its particular location above the active site entrance (33). This new information allows us to propose a complete version of our C1 model (17) integrating the C1s catalytic domain (Fig. 6, B and C). In the resulting assembly (Fig. 6), residues Lys584 and Lys587 of the C1s SP domain, which are disordered in the C1s CCP2-SP structure (33) (Fig. 4C) are located in the vicinity of a C1q collagen fiber. The C1s CCP2 module is positioned in such a way that its residues Lys354 and Lys405 remain accessible, in agreement with the labeling data (Table 2).
In summary, a strategy combining chemical modification of lysines and mass spectrometry analysis has been applied for the first time to the human C1 complex. The structural data generated are, for the most part, consistent with our recent three-dimensional C1 model (17). In addition, they yield the first experimental evidence that the C1s SP domain is partly positioned inside the C1q cone. The same type of analysis could be applied to probe other residues such as tyrosines and histidines and would provide further precise insights into the C1 architecture.
Supplementary Material
This work was supported by CNRS and by Genopole-France.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Table S1.
- MBL
- mannan-binding lectin
- ESI
- electrospray ionization
- nano-ESI
- nanoelectrospray ionization
- MSn
- general designation of mass spectrometry to the nth degree
- SASA
- solvent accessibility surface area
- CUB
- C1r, C1s, Uegf, and bone morphogenetic protein
- CCP
- complement control protein
- SP
- serine protease
- sulfo-NHS
- sulfo-N-hydroxysuccinimide
- FBIP
- Fibrinogen-binding inhibitor peptide
- TCEP-HCl
- Tris(2-carboxyethyl) phosphine hydrochloride
- UF
- unmodified fraction.
REFERENCES
- 1.Duncan R. C., Wijeyewickrema L. C., Pike R. N. (2008) Biochimie 90, 387–395 [DOI] [PubMed] [Google Scholar]
- 2.Frank M. M., Fries L. F. (1991) Immunol. Today 12, 322–326 [DOI] [PubMed] [Google Scholar]
- 3.Arlaud G. J., Gaboriaud C., Thielens N. M., Rossi V. (2002) Biochem. Soc. Trans. 30, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 4.Arlaud G. J., Thielens N. (1993) Methods Enzymol. 223, 61–82 [DOI] [PubMed] [Google Scholar]
- 5.Gaboriaud C., Teillet F., Gregory L. A., Thielens N. M., Arlaud G. J. (2007) Immunobiology 212, 279–288 [DOI] [PubMed] [Google Scholar]
- 6.Gaboriaud C., Thielens N. M., Gregory L. A., Rossi V., Fontecilla-Camps J. C., Arlaud G. J. (2004) Trends Immunol. 25, 368–373 [DOI] [PubMed] [Google Scholar]
- 7.Botto M., Walport M. J. (2002) Immunobiology 205, 395–406 [DOI] [PubMed] [Google Scholar]
- 8.Bersch B., Hernandez J. F., Marion D., Arlaud G. J. (1998) Biochemistry 37, 1204–1214 [DOI] [PubMed] [Google Scholar]
- 9.Budayova-Spano M., Grabarse W., Thielens N. M., Hillen H., Lacroix M., Schmidt M., Fontecilla-Camps J. C., Arlaud G. J., Gaboriaud C. (2002) Structure 10, 1509–1519 [DOI] [PubMed] [Google Scholar]
- 10.Budayova-Spano M., Lacroix M., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. (2002) EMBO J. 21, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kardos J., Harmat V., Palló A., Barabás O., Szilágyi K., Gráf L., Náray-Szabó G., Goto Y., Závodszky P., Gál P. (2008) Mol. Immunol. 45, 1752–1760 [DOI] [PubMed] [Google Scholar]
- 12.Gál P., Barna L., Kocsis A., Závodszky P. (2007) Immunobiology 212, 267–277 [DOI] [PubMed] [Google Scholar]
- 13.Arlaud G. J., Colomb M. G., Gagnon J. (1987) Immunol. Today 8, 106–111 [DOI] [PubMed] [Google Scholar]
- 14.Schumaker V. N., Hanson D. C., Kilchherr E., Phillips M. L., Poon P. H. (1986) Mol. Immunol. 23, 557–565 [DOI] [PubMed] [Google Scholar]
- 15.Weiss V., Fauser C., Engel J. (1986) J. Mol. Biol. 189, 573–581 [DOI] [PubMed] [Google Scholar]
- 16.Gregory L. A., Thielens N. M., Arlaud G. J., Fontecilla-Camps J. C., Gaboriaud C. (2003) J. Biol. Chem. 278, 32157–32164 [DOI] [PubMed] [Google Scholar]
- 17.Bally I., Rossi V., Lunardi T., Thielens N. M., Gaboriaud C., Arlaud G. (2009) J. Biol. Chem. 284, 19340–19348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H., Schuster M. C., Sfyroera G., Geisbrecht B. V., Lambris J. D. (2008) J. Am. Soc. Mass Spectrom. 19, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cutalo J. M., Darden T. A., Kunkel T. A., Tomer K. B. (2006) Biochemistry 45, 15458–15467 [DOI] [PubMed] [Google Scholar]
- 20.Gabant G., Augier J., Armengaud J. (2008) J. Mass Spectrom. 43, 360–370 [DOI] [PubMed] [Google Scholar]
- 21.Janecki D. J., Beardsley R. L., Reilly J. P. (2005) Anal. Chem. 77, 7274–7281 [DOI] [PubMed] [Google Scholar]
- 22.Scholten A., Visser N. F., van den Heuvel R. H., Heck A. J. (2006) J. Am. Soc. Mass Spectrom. 17, 983–994 [DOI] [PubMed] [Google Scholar]
- 23.Suckau D., Mak M., Przybylski M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5630–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arlaud G. J., Sim R. B., Duplaa A. M., Colomb M. G. (1979) Mol. Immunol. 16, 445–450 [DOI] [PubMed] [Google Scholar]
- 25.Tacnet-Delorme P., Chevallier S., Arlaud G. J. (2001) J. Immunol. 167, 6374–6381 [DOI] [PubMed] [Google Scholar]
- 26.Pflieger D., Przybylski C., Gonnet F., Le Caer J. P., Lunardi T., Arlaud G. J., Daniel R. (2010) Mol. Cell Proteomics 9, 593–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller L. N., Rinner O., Schmidt A., Letarte S., Bodenmiller B., Brusniak M. Y., Vitek O., Aebersold R., Müller M. (2007) Proteomics 7, 3470–3480 [DOI] [PubMed] [Google Scholar]
- 28.Pflieger D., Chabane S., Gaillard O., Bernard B. A., Ducoroy P., Rossier J., Vinh J. (2006) Proteomics 6, 5868–5879 [DOI] [PubMed] [Google Scholar]
- 29.Illy C., Thielens N. M., Arlaud G. J. (1993) J. Protein Chem. 12, 771–781 [DOI] [PubMed] [Google Scholar]
- 30.Hamuro Y., Coales S. J., Morrow J. A., Molnar K. S., Tuske S. J., Southern M. R., Griffin P. R. (2006) Protein Sci. 15, 1883–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan X., Zhang H., Watson J., Schimerlik M. I., Deinzer M. L. (2002) Protein Sci. 11, 2113–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziccardi R. J. (1982) J. Immunol. 128, 2500–2504 [PubMed] [Google Scholar]
- 33.Gaboriaud C., Rossi V., Bally I., Arlaud G. J., Fontecilla-Camps J. C. (2000) EMBO J. 19, 1755–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thielens N. M., Illy C., Bally I., Arlaud G. J. (1994) Biochem. J. 301, 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyd J., Burton D. R., Perkins S. J., Villiers C. L., Dwek R. A., Arlaud G. J. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 3769–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perkins S. J., Villiers C. L., Arlaud G. J., Boyd J., Burton D. R., Colomb M. G., Dwek R. A. (1984) J. Mol. Biol. 179, 547–557 [DOI] [PubMed] [Google Scholar]
- 37.Strang C. J., Siegel R. C., Phillips M. L., Poon P. H., Schumaker V. N. (1982) Proc. Natl. Acad. Sci. U.S.A. 79, 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tschopp J., Villiger W., Fuchs H., Kilchherr E., Engel J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 7014–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villiers C. L., Arlaud G. J., Colomb M. G. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4477–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bally I., Rossi V., Thielens N. M., Gaboriaud C., Arlaud G. J. (2008) Mol. Immunol. 45, 4097.(Abstract) [Google Scholar]
- 41.Phillips A. E., Toth J., Dodds A. W., Girija U. V., Furze C. M., Pala E., Sim R. B., Reid K. B., Schwaeble W. J., Schmid R., Keeble A. H., Wallis R. (2009) J. Immunol. 182, 7708–7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arlaud G. J., Gagnon J., Villiers C. L., Colomb M. G. (1986) Biochemistry 25, 5177–5182 [DOI] [PubMed] [Google Scholar]
- 43.Tissot B., Gonnet F., Iborra A., Berthou C., Thielens N., Arlaud G. J., Daniel R. (2005) Biochemistry 44, 2602–2609 [DOI] [PubMed] [Google Scholar]
- 44.Willard L., Ranjan A., Zhang H., Monzavi H., Boyko R. F., Sykes B. D., Wishart D. S. (2003) Nucleic Acids Res. 31, 3316–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.