Abstract
DNA polymerase δ (pol δ) is one of the two main replicative polymerases in eukaryotes; it synthesizes the lagging DNA strand and also functions in DNA repair. In previous work, we demonstrated that heterozygous expression of the pol δ L604G variant in mice results in normal life span and no apparent phenotype, whereas a different substitution at the same position, L604K, is associated with shortened life span and accelerated carcinogenesis. Here, we report in vitro analysis of the homologous mutations at position Leu-606 in human pol δ. Four-subunit human pol δ variants that harbor or lack 3′ → 5′-exonucleolytic proofreading activity were purified from Escherichia coli. The pol δ L606G and L606K holoenzymes retain catalytic activity and processivity similar to that of wild type pol δ. pol δ L606G is highly error prone, incorporating single noncomplementary nucleotides at a high frequency during DNA synthesis, whereas pol δ L606K is extremely accurate, with a higher fidelity of single nucleotide incorporation by the active site than that of wild type pol δ. However, pol δ L606K is impaired in the bypass of DNA adducts, and the homologous variant in mouse embryonic fibroblasts results in a decreased rate of replication fork progression in vivo. These results indicate that different substitutions at a single active site residue in a eukaryotic polymerase can either increase or decrease the accuracy of synthesis relative to wild type and suggest that enhanced fidelity of base selection by a polymerase active site can result in impaired lesion bypass and delayed replication fork progression.
Keywords: DNA Damage, DNA Polymerase, DNA Replication, DNA Synthesis, Mutagenesis Mechanisms, DNA Polymerase delta, Deletion Mutagenesis, Lesion Bypass, Mutator, Replication Fidelity
Introduction
Eukaryotic DNA replication is extraordinarily accurate, with an average of less than one error per 109–1010 bases copied (1). The high fidelity of DNA synthesis is principally determined by the properties of the replicative nuclear DNA polymerases, DNA polymerase (pol)2 δ and DNA pol ϵ (2, 3). These enzymes are highly accurate with respect to nucleotide selection at the polymerase active site, with additional accuracy conferred by an associated 3′ → 5′-exonuclease proofreading domain, which excises noncomplementary nucleotides immediately after incorporation (4). The overall fidelity of DNA synthesis is further enhanced by post-replication repair (5). Accurate DNA replication is central for the avoidance of cancer, a disease that arises in association with the accumulation of mutations in multiple genetic pathways controlling cellular growth and metastasis (6). Given that many mutations are necessary for the development of cancer, yet that normal DNA replication is extremely accurate, it has been postulated that error prone synthesis via the acquisition of a mutator phenotype may be required for carcinogenesis (7, 8). Because pol δ and pol ϵ are responsible for the majority of DNA syntheses during each replicative cycle, we infer that mutations in the genes encoding these enzymes that render DNA synthesis error prone could constitute a major mechanism for the expression of a mutator phenotype during carcinogenesis. This postulate has been investigated by creating mice expressing mutant variants of pol δ or pol ϵ that lack 3′ → 5′-proofreading exonuclease activity. In the case of both pol δ (9) and pol ϵ (10), exonuclease-deficient heterozygous mice have a near normal life span and no apparent phenotype, whereas homozygous animals exhibit a substantially elevated tumor incidence. These studies suggest that the exonuclease domains of the replicative polymerases have an important role in suppressing tumorigenesis. However, these exonucleases likely have functions in the cell other than proofreading of replication errors, such as maturation of Okazaki fragments, processing of mismatch repair intermediates (11), and removing nucleotide modifications that prevent the initiation of DNA synthesis by DNA polymerases (12). Defects in these processes might also contribute to the observed cancer phenotypes.
Our group recently developed a second model for analyzing the relationship between the fidelity of DNA synthesis by pol δ and carcinogenesis. We have previously demonstrated that substitutions for isoleucine at homologous positions 709 and 614 in Escherichia coli DNA polymerase I (13) and Thermus aquaticus DNA polymerase I (14), respectively, alter the fidelity of DNA synthesis. These studies were extended to Saccharomyces cerevisiae; we created all 19 amino acid substitutions for the homologous position in the catalytic site of S. cerevisiae pol δ (15). This work identified variants that enhance mutagenesis in yeast. We then selected two of these variants, L604G (pol δG) and L604K (pol δK), for further study in mice (16). Mice homozygous for either mutation are inviable. Heterozygous pol δ+/G mice lack a distinctive phenotype; they have a normal life span and no change in cancer incidence. In contrast, heterozygous pol δ+/K mice exhibit a shortened life span and accelerated tumorigenesis. The overall cancer incidence, as well as the spectrum of tumor types, was similar among pol δ+/+, pol δ+/G, and pol δ+/K mice. However, the rate of tumor progression was greater in the pol δ+/K animals; they died with tumors at an earlier age.
Available mutation data indicate that both pol δG and pol δK result in altered fidelity of DNA replication in vivo. In S. cerevisiae, the frequency of CAN1 resistance in cells expressing pol δG is 17-fold elevated relative to wild type, whereas pol δK yielded a somewhat lower 13-fold elevation (15). In mouse embryonic fibroblasts, fluctuation analysis revealed that the mutation rate at the Hprt locus is elevated 5-fold relative to wild type in cells expressing pol δG, whereas cells expressing pol δK exhibit a 4-fold elevation (16). Given that the pol δG mutation does not result in a cancer phenotype in heterozygous mice, whereas the pol δK mutation is associated with accelerated tumorigenesis, it is apparent that the overall magnitude of elevation in the mutation rate is not sufficient to explain the phenotype in mice. However, the spectrum of mutations synthesized by the variants differs. In S. cerevisiae, the identified replication errors in pol δG cells consist predominantly of point mutations (95% point mutations; 5% deletions). In contrast, cells expressing pol δK exhibit an increased frequency of deletions (65% point mutations; 35% deletions) (15). Likewise, in mouse embryonic fibroblasts, chromosomal spreads revealed that although cells from both pol δ+/G and pol δ+/K mice exhibit an elevated frequency of spontaneous chromosomal aberrations relative to wild type, the highest levels of aberrations are seen in pol δ+/K cells, which are elevated 2–3-fold relative to pol δ+/G cells (16). These differing patterns of mutation in vivo are likely relevant to the disparate phenotypes of the pol δG and pol δK mice, but their etiology is not clear as the biochemical properties of the pol δG and pol δK polymerases are unknown.
The study of mutated mammalian pol δ has previously been hampered by the lack of an efficient purification system. A recently described method (17) now allows for sufficient amounts of human pol δ to be obtained to allow for detailed biochemical analysis. Thus, to study the biochemical consequences of mutation in the active site of mammalian pol δ and by extension gain insight into the disparate phenotypes of the pol δ L604G and pol δ L604K mouse models, we have purified the homologous variants at position Leu-606 of human pol δ. We report that human pol δG and pol δK maintain polymerase and exonuclease activity, replicate DNA with comparable processivity, and are similar in the extent to which their activity is stimulated by PCNA. However, pol δG and pol δK differ dramatically in base replication accuracy. pol δG synthesizes DNA harboring an extremely high frequency of point mutations, which is an unexpected finding as mice expressing a single copy of the homologous mutation do not have a discernible phenotype. In contrast, the pol δK variant, which is associated with a heterozygous cancer phenotype in mice, results in highly accurate synthesis in vitro; surprisingly, the accuracy of base selection by the active site of pol δK is improved relative to that of wild type pol δ. However, pol δK is defective in bypassing sites of DNA damage in vitro, and expression of the homologous variant in mouse embryonic fibroblasts results in stalled replication in vivo. We conclude that substitution at a single residue in the active site of human pol δ can either increase or decrease the accuracy of base selection and that enhanced base selection accuracy is associated with impaired bypass of bulky DNA adducts. Extension of our results to the pol δG and pol δK mouse models suggests that reduced DNA synthesis by a major replicative polymerase can result in genetic instability and accelerated carcinogenesis.
EXPERIMENTAL PROCEDURES
Expression and purification of human pol δ and the polymerase and exonuclease assays were performed as described previously (18). pol δ catalytic mutants were constructed by site-directed mutagenesis of the pET303-hpolD1 plasmid with the Stratagene QuikChange kit. Human PCNA was a kind gift of Prof. Ulrich Hübscher (19).
Processivity Assay
Reactions (30 μl) contained 600 fmol of plus-strand m13 DNA annealed to 5′-32P end-labeled primer 5′-GCTGTTGGGAAGGGCGATCG-3′, 40 mm Tris-HCl (pH 7.5), 50 mm KCl, 2.5 mm MgCl2, 0.1% Triton X-100, 10% glycerol, 6 μg of BSA, 1 mm DTT, 250 μm of each dNTP, and 30 fmol of exonuclease-proficient DNA polymerase. Reactions were initiated by addition of polymerase and incubated for 15 min at 37 °C. Length of extension products was constant at 5, 10, and 15 min, indicating that single-hit conditions were satisfied (data not shown). Reactions were quenched by adding an equal volume of 95% formamide, 20 mm EDTA, and products were separated on a 14% denaturing sequencing gel.
Stimulation by PCNA
Stimulation of pol δ activity by PCNA was performed essentially as described previously (20). In brief, reactions (10 μl) contained 400 fmol of poly(dA)-(dT)30, 0.1 pmol of exonuclease-proficient DNA polymerase, 0.3 pmol of human PCNA, 40 mm Tris-HCl (pH 7.9), 8 mm MgAc2, 2 μg of BSA, 1 mm DTT, and 25 μm [α-32P]dTTP. Reactions were quenched by addition of EDTA to 25 mm final concentration. Products were then bound to a Microdyne B membrane (Nalge Nunc International, Rochester, NY) and quantified by PhosphorImager analysis.
Single Nucleotide Kinetics
Reactions utilized 5′ 32P end-labeled primer 5′-CAT GAA CTA CAA GGA C-3′ annealed to the template 5′-GCA TTC AGT GGT CCT TGT AGT TCA TG-3′. Reactions (10 μl) contained 40 mm Tris-HCl (pH 7.5), 50 mm KCl, 5 mm MgCl2, 0.1% Triton X-100, 10% glycerol, 2 μg of BSA, 1 mm DTT, 10 nm primer-template, and 20 nm of the indicated exonuclease-deficient DNA polymerase. Reactions were initiated by nucleotide addition, proceeded 1–10 min at 37 °C as needed to satisfy single completed hit conditions with less than 20% total extension of available primer (21), and were quenched with an equal volume of 95% formamide, 20 mm EDTA. Reaction products were heated to 95 °C for 5 min, separated on a 14% polyacrylamide-urea gel, and quantified by PhosphorImager analysis. Reaction velocity was plotted as a function of nucleotide concentration and fit to the Michaelis-Menten equation with the KaleidaGraph software package.
Forward Mutation Assay
Reactions (25 μl) contained 40 mm Tris-HCl (pH 7.5), 50 mm KCl, 2.5 mm MgCl2, 0.1% Triton X-100, 10% glycerol, 5 μg of BSA, 1 mm DTT, 250 μm of each dNTP, 1.5 nm M13mp2 gapped DNA substrate, and 300 nm polymerase. Reactions were initiated by incubation at 37 °C for 1 h and terminated by addition of EDTA to 10 mm. Reaction products were introduced into E. coli; mutant plaques were scored, and error rates were calculated as described previously (18).
Lesion Bypass Assay
The substrate was 5′ 32P end-labeled primer 5′-CGC GCC GAA TTC CCG CTA GCA ATA TTC T-3′ annealed to the template 5′-TTG GCN GCA GAA TAT TGC TAG CGG GAA TTC GGC GCG-3′, where N indicates the modified nucleotide as indicated in the figure legend. Reactions (10 μl) contained 40 mm Tris-HCl (pH 7.5), 50 mm KCl, 2.5 mm MgCl2, 0.1% Triton X-100, 10% glycerol, 2 μg of BSA, 1 mm DTT, 250 μm of each dNTP, 5 nm of primer-template, and 60 nm of exonuclease-proficient DNA polymerase. Reactions were incubated for 5 min at 37 °C, quenched with an equal volume of 95% formamide, 20 mm EDTA, heated to 95 °C for 5 min, and separated on a 14% polyacrylamide-urea gel.
Molecular DNA Combing
Iododeoxyuridine, chlorodeoxyuridine, and SDS were obtained from Sigma. Proteinase K was from Roche Diagnostics. Secondary antibodies for molecular combing were from Fisher. Cells were incubated with 50 μm iododeoxyuridine for 20 min followed by 50 μm chlorodeoxyuridine for an additional 20 min in the culture medium. Thymidine chase was then carried out at 1 mm for 1 h. Cells were embedded in agarose plugs and treated with 1 mg/ml proteinase K, 1% SDS in 0.5 m EDTA (pH 8.0) for 48 h. DNA was combed on silanized coverslips as described previously (22). Primary antibodies were described previously (23) except the anti-single-stranded DNA antibody was used at 1:100 (Chemicon Inc). Secondary antibodies were used at 1:50 (anti-mouse Alexa-488), 1:150 (anti-mouse Alexa-350), and 1:25 (anti-rat Alexa-594). Signals were captured and measured as described previously (23). Fork speed was calculated by dividing the median track size by the labeling time. Differences in fork speed between individual cell lines were examined using the Mann-Whitney test, which is a nonparametric equivalent of the t test and provides a measure of identity of two independent populations.
RESULTS
Purification of pol δ Active Site Mutants
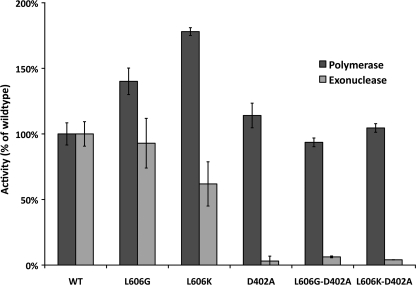
Four-subunit human pol δ variants were purified from E. coli by the method of Fazlieva et al. (17). The purification scheme involves transformation of E. coli with two plasmids: one plasmid expresses the 125-kDa catalytic subunit, and the other plasmid expresses the p66, p50, and p12 accessory subunits. A polyhistidine tag on the 125-kDa subunit allows the complex to be isolated on a nickel column; subsequent fractionation on a cation exchange column results in a nearly homogeneous preparation of a 4-subunit pol δ holoenzyme. We used this method to obtain 4-subunit wild type human pol δ as well as variants with amino acid substitutions at the Leu-606 position; human L606G and L606K pol δ correspond to the mouse L604G and L604K variants. Each of the three human pol δ variants was purified with exonuclease activity intact as well as with the exonuclease domain inactivated by the D402A point mutation. The mutant pol δ protein complexes all exhibited similar levels of purity, as assessed by SDS-PAGE and Coomassie Blue staining. The relative intensity of staining of the four pol δ subunits was similar for each of the purified variants (supplemental Fig. S1). Polymerase activity of each complex was assayed by measuring the incorporation of [α-32P]dTTP into an activated calf thymus DNA template in the presence of all four complementary dNTPs and Mg2+. All the pol δ variants exhibited DNA polymerase activity; both pol δG and pol δK had slightly greater specific activity than the wild type enzyme (Fig. 1). An increase in DNA polymerase activity has been reported for the Leu to Met mutation at the analogous residue in both S. cerevisiae pol δ (24) and human pol δ (18). The 3′ → 5′-exonuclease activity of the mutants was determined utilizing a linear single-stranded DNA substrate (25). Substitutions for Leu-606 did not substantially alter exonuclease activity (Fig. 1), although with each of the pol δ variants analyzed, the D402A substitution inactivated exonuclease activity by 95% or greater as expected.
FIGURE 1.
Polymerase and exonuclease activity of pol δ variants. Polymerase activity was assayed by incorporation of radioactive nucleotides into activated calf thymus DNA, and exonuclease activity was determined on a radioactively labeled single-stranded poly(dT) homopolymer. Assays were performed in triplicate and normalized to wild type pol δ; standard deviations are indicated.
Processivity
A decrease in processivity, which is defined as the number of nucleotides copied during each DNA-binding event, could result in impaired synthesis by mutant pol δ and therefore might contribute to the phenotype seen in mice expressing pol δK. To measure processivity, the pol δ variants were compared in their ability to extend a singly primed 7.2-kb DNA substrate. Limiting amounts of polymerase and excess DNA substrate were used to ensure that the detected extension events were due to a single polymerase-template interaction (26). Each pol δ variant was observed to extend the template by 1–10 nucleotides, similar to that reported for S. cerevisiae pol δ (24). Minor differences in site-specific termination probability were seen among the different mutants, but the overall length of product extension was similar (supplemental Fig. S2). Thus, the wild type, exonuclease-deficient, pol δG, and pol δK variants exhibit comparable processivity, and the phenotype seen in vivo upon expression of these alleles is unlikely to be due to an alteration in this property.
Stimulation by PCNA
pol δ interacts with the PCNA sliding clamp at the replication fork in vivo, with PCNA increasing pol δ activity by enhancing its ability to bind DNA (27). We measured the effect of PCNA on the activity of each pol δ variant when copying a primed DNA substrate (poly(dA)-oligo(dT)) (supplemental Fig. S3). On this template, in the absence of PCNA, wild type pol δ and pol δK had similar activity, whereas pol δG had somewhat increased activity, which could indicate an increased capability for pol δG to replicate through areas of secondary structure that are known to occur in homopolymeric templates (28). In the presence of PCNA, 4-subunit wild type pol δ, pol δG, and pol δK were all stimulated to comparable overall levels of synthesis. This result is consistent with a report indicating that the interaction between pol δ and PCNA is mediated predominantly by the p50 accessory subunit of pol δ and not the p125 catalytic subunit (29), which suggests that mutations in p125 would not be expected to alter the pol δ-PCNA interaction.
Single Nucleotide Kinetics
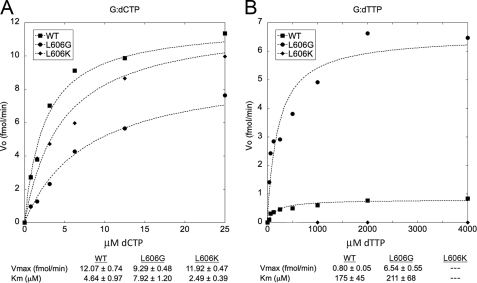
Nucleotide selection by the pol δG and pol δK mutants relative to wild type pol δ was analyzed by measuring the kinetics of incorporation of a single correct or incorrect nucleotide onto the 3′-primer terminus of a DNA substrate (21). To focus on base selection at the active site, exonuclease-deficient versions of the enzymes were utilized in the single nucleotide incorporation experiments. The rate of insertion of a single dCTP residue opposite template dG by pol δ is shown in Fig. 2. Wild type pol δ, pol δG, and pol δK catalyzed this insertion event with similar efficiency (Fig. 2A), consistent with the comparable activities of the three variants on activated calf thymus DNA. The rate of nucleotide mis-insertion was investigated by measuring incorporation of dTTP opposite template dG (Fig. 2B). Wild type pol δ catalyzed mis-insertion of dTTP at low but detectable levels, with a maximum reaction velocity of 0.80 fmol/min. pol δG catalyzed the mis-insertion event at a significantly higher rate of 6.5 fmol/min, indicating reduced accuracy of nucleotide selection at the polymerase active site. Although the Vmax for dG:dTTP insertion differed between wild type pol δ and pol δG, the Km values for this event were identical within experimental error. Under the steady-state conditions used in the assay, the Km approaches the nucleotide binding constant and can be considered to represent the residence time of nucleotides in the enzyme-DNA-dNTP complex (30). Thus, the comparable Km values we observed suggest that wild type pol δ and pol δG have comparable residence times for mismatched dNTPs at the dNTP-binding site. This observation is consistent with our previously published molecular modeling studies concerning the L604G and L604K variants of mouse pol δ (16), which suggested that the substitutions do not greatly alter the active site geometry and thus might not be expected to alter dNTP association/dissociation. The mechanism by which the motif A substitutions affect the fidelity of DNA synthesis may therefore involve an alteration in the conformational change that is thought to occur during nucleotide incorporation at the polymerase active site (31, 32).
FIGURE 2.
Nucleotide incorporation kinetics of pol δ mutant enzymes. Reactions were carried out under steady-state conditions (21) using a 5′-32P-labeled primer and a template with G at the first nucleotide position downstream from the 3′ terminus. The primer-template was extended by the indicated variants of exonuclease-deficient human pol δ holoenzyme in the presence of the indicated concentrations of either dCTP or dTTP. Band intensities were quantitated and the results fit to the Michaelis-Menten equation. Kinetic values are shown ± S.D. ■, wild type pol δ; ●, L606G; ♦, L606K. A, correct incorporation of dCTP opposite template dG. B, mis-incorporation of dTTP opposite template dG. Because L606K did not catalyze significant levels of dG:dTTP mis-incorporation under these conditions, kinetic values for this event were not determined.
pol δK did not catalyze detectable nucleotide mis-insertion, even with nucleotide concentrations as high as 4 mm. This lack of detectable mis-insertion by pol δK, in contrast to that observed with wild type pol δ and pol δG, suggests that the lysine substitution reduces incorporation of noncomplementary bases at the active site. Incorporation was insufficient to determine whether the reduced single nucleotide mis-insertion by pol δK is concordant with a change in Km or Vmax.
Fidelity of DNA Synthesis
To quantify the fidelity of each pol δ variant for all possible base substitution errors, we utilized the M13-lacZ gapped DNA forward mutation assay (33). This assay utilizes double-stranded M13 DNA containing a 407-nucleotide single-stranded gap in the sequence encoding the lacZ α-complementation fragment. The single-stranded region is copied in vitro by the polymerase. Complete gap filling is confirmed by agarose gel electrophoresis, and the products are transformed into α-complementation E. coli cells followed by plating on media containing X-gal and isopropyl 1-thio-β-d-galactopyranoside. Correct synthesis by the polymerase results in the production of β-galactosidase and the formation of dark blue plaques, whereas polymerase errors that inactivate the lacZα gene result in light blue or colorless plaques. The ratio of light blue or colorless plaques to dark blue plaques reflects the overall fidelity of the polymerase; sequencing of mutant plaques allows determination of the mutational signature of the polymerase within multiple sequence contexts.
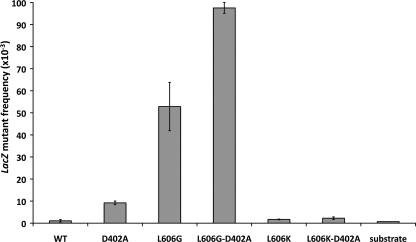
All of the pol δ mutants copied the entire gapped segment of the plasmid (data not shown). As reported previously (18), wild type pol δ replicates DNA with high accuracy, resulting in mutant plaques at a frequency of 1.1 × 10−3 ± 0.6 × 10−3 (average of duplicate experiments ± S.D.). This frequency is only slightly greater than the background lacZ mutant frequency of 0.7 × 10−3. The error frequency of the pol δG variant is 50-fold greater, with a mutant plaque frequency of 53 × 10−3 ± 11 × 10−3 (Fig. 3). Inactivation of proofreading further increases the mutant frequency by less than 2-fold to 98 × 10−3 ± 3 × 10−3. This observation implies that the mutator phenotype induced by the L606G mutation is primarily due to an alteration in base selection at the active site rather than altered proofreading or impaired polymerase-exonuclease domain switching (34), although these mechanisms may contribute to the inaccuracy of the L606G mutant as well. The elevated mutation frequency in the L606G,D402A double mutant additionally demonstrates that proofreading only partially compensates for the error prone mutant active site.
FIGURE 3.
lacZ mutant frequencies in the in vitro forward mutation assay. Each purified 4-subunit pol δ variant was used to copy a 407-nucleotide single-stranded gap in a double-stranded M13 DNA substrate. The single-stranded region of the substrate encodes the lacZ α-complementation fragment, such that accurate replication of the region yields a dark blue plaque in α-complementation E. coli, whereas polymerase errors result in a light blue or colorless plaque. The frequency of mutant plaques ± S.D. is shown for gapped DNA extended with each of the polymerase variants.
In sharp contrast, the pol δK variant yielded a lacZ mutant frequency of 1.7 × 10−3 ± 0.2 × 10−3, which is near the background of the assay and not substantially elevated relative to wild type pol δ. Moreover, when proofreading is inactivated in the L606K,D402A double mutant, the lacZ mutant frequency of 2.3 × 10−3 ± 0.6 × 10−3 is 4-fold lower than that seen with inactivation of proofreading alone (9.2 × 10−3 ± 0.8 × 10−3). The difference between pol δ L606K,D402A and pol δ D402A is significant (p < 1 × 10−4, Fisher's exact test). This observation suggests that the L606K substitution enhances the accuracy of DNA synthesis, which is consistent with the results seen in the single nucleotide incorporation assay.
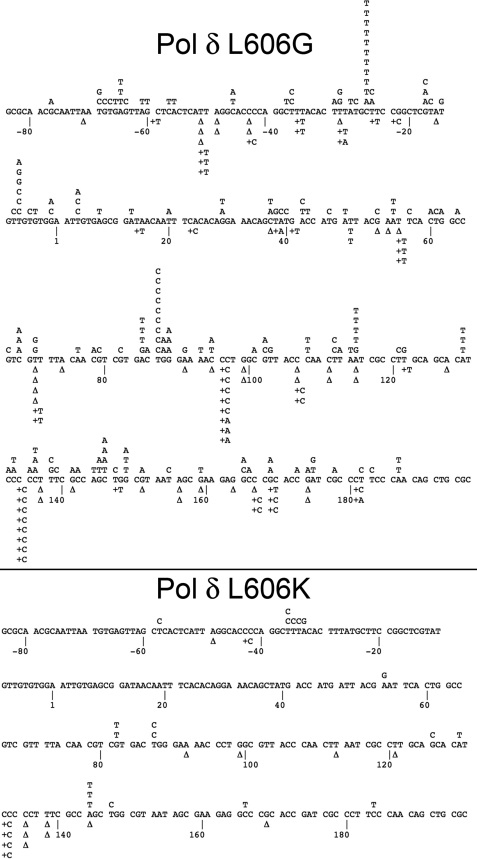
The types of mutations introduced by the exonuclease-proficient pol δ variants were determined by sequencing the copied segment of gapped DNA in the lacZ mutant plaques (for detailed mutation data, please see supplemental Table S1). pol δG introduced a large number of mutations throughout the lacZ target sequence (Fig. 4). We observed an increase in mispair frequency relative to wild type for every possible single-base substitution error other than dCMP mis-incorporation opposite template C. This indicates that the L606G mutation causes an overall reduction in the fidelity of DNA synthesis and that the lowered fidelity is expressed in multiple sequence contexts. In contrast, synthesis by pol δK results in a low frequency of errors throughout the lacZ target (Fig. 4). Both base substitution and single-nucleotide frameshifts are seen at a rate similar to that of wild type pol δ and approximately equal to the background of the assay; this implies that most or all mutations observed for pol δK are likely due to pre-existing mutations in the DNA substrate rather than being caused by errors during gap-filling synthesis. Although the frequency of deletions with pol δK was somewhat elevated relative to wild type in this in vitro assay, this difference was not significant (p = 0.13, Fisher's exact test).
FIGURE 4.
Mutation spectrum of pol δ mutants. The + strand coding sequence of the single-stranded portion of lacZα is shown, with point mutations indicated above the line and deletions (Δ) or insertions below. Top panel, pol δ L606G; bottom panel, pol δ L606K.
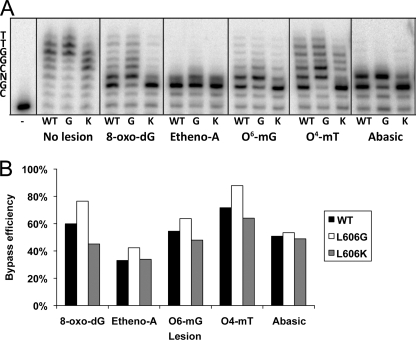
Lesion Bypass
The alterations in nucleotide selectivity seen in the Leu-606 mutant enzymes might affect the ability of the enzymes to copy DNA templates containing altered nucleotides. We measured the ability of the exonuclease-proficient pol δ variants to replicate a template containing a site-specific lesion at the +3 position (Fig. 5A). On a control template, slight differences in activity were seen among the wild type, L606G, and L606K variants, although all exhibited sufficient activity to extend beyond the +3 position. In the presence of a template lesion, the wild type pol δ stalls but is able to incorporate nucleotides opposite 8-oxoguanine, 1,N6-ethenoadenine, O6-methylguanine, O4-methylthymine, and a synthetic abasic site (tetrahydrofuran), with a partial ability to continue synthesis past the lesions. pol δG differs from wild type by exhibiting an increased ability to both incorporate opposite and continue extension beyond all the site-specific modifications. This observation is consistent with the reduced accuracy of nucleotide selection by pol δG, which is presumably due in part to relaxed geometry at the active site of the mutant polymerase during the nucleotide addition step. pol δK, on the other hand, was reduced in lesion bypass activity relative to wild type. Reactions with pol δK resulted in nearly complete termination of DNA synthesis at the position prior to the site-specific modification for each lesion analyzed. Quantification of the lesion bypass efficiency of each pol δ variant is shown in Fig. 5B; bypass efficiency of each variant was normalized to the extent of synthesis seen on the undamaged template. We observed that synthesis by pol δK was impaired most prominently when encountering 8-oxoguanine, which is a common lesion due to reactive oxygen species in vivo and is readily bypassed by most DNA polymerases (31); diminished synthesis by pol δK relative to wild type was also observed for the alkylated lesions O6-methylguanine and O4-methylthymine. Minimal extension was observed beyond ethenoadenine and abasic sites for all of the polymerases tested.
FIGURE 5.
Impaired lesion bypass capability of human pol δ L606K. A, reactions used a 5′-32P-labeled 28-nucleotide primer annealed to a 36-nucleotide template containing a site-specific lesion at the +3 position. The sequence of the 3′-segment of the template is indicated to the left of the figure, with N denoting the position of the modified nucleotide: deoxythymidine (no lesion), 8-oxoguanine (8-oxo-dG), 1,N6-ethenoadenine (etheno-A), O6-methylguanine (O6-mG), O4-methylthymine (O4mT), or an abasic site. B, lesion bypass was quantified by measuring the intensity of product bands at and beyond the position of the lesion and dividing by the total band intensity of the lane. The values were normalized to the extent of synthesis on the control template by dividing by the values measured in the control reactions.
Replication Fork Progression
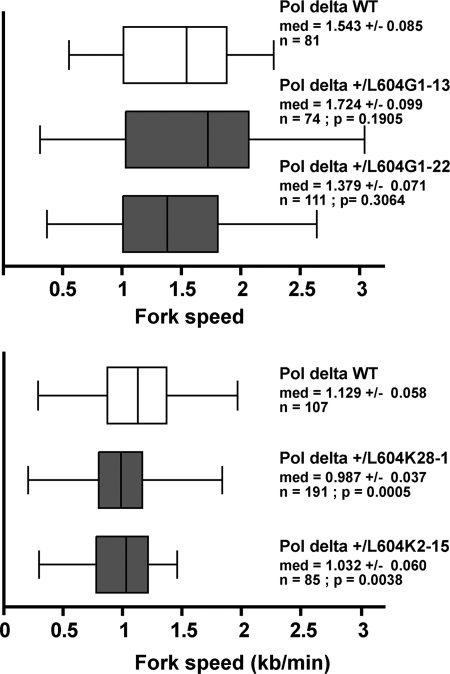
We hypothesized that a diminished ability of the pol δK polymerase to copy past altered nucleotides would be manifested in impaired progression of the replication fork in vivo due to stalled synthesis at endogenous sites of DNA damage. We probed this postulated mechanism by measuring replication fork progression on a genome-wide scale by molecular combing (22, 23). We sequentially labeled exponentially dividing mouse embryonic fibroblasts with the nucleotide analogs iododeoxyuridine and chlorodeoxyuridine, isolated total DNA, and stretched the DNA molecules on silanized coverslips. Neo-replicated tracks were revealed with fluorescent antibodies specific for the nucleotide analogs. Distributions of fork velocity values, as presented in Fig. 6, were obtained from track size analysis as described under “Experimental Procedures.” The fork velocity in cells expressing pol δG was unchanged in distribution relative to a lineage-matched control (p = 0.190 and p = 0.306, nonparametric Mann-Whitney test). In contrast, the distribution of fork velocity in cells expressing pol δK exhibited a significant shift toward reduced fork speeds relative to control cells (p = 5 × 10−4 and p = 3.8 × 10−3). These results imply that DNA synthesis in cells expressing pol δK is impaired, consistent with stalling of replication secondary to impaired bypass of endogenous lesions in DNA by the mutant pol δ.
FIGURE 6.
Replication fork progression is impaired in pol δ +/L604K mouse embryo fibroblasts. The distribution of replication fork velocities (kb/min) was measured at a single molecule level by molecular combing in wild type, pol δ +/L604G, and pol δ +/L604K mouse embryonic fibroblasts. The wild type cell line used in each experiment was derived from a cross of heterozygous animals. Box, 25–75 percentile range. Whiskers, minimum and maximum values. Med, median fork speed (kb) ± median absolute deviation; n, number of measurements; p, p value determined with nonparametric Mann-Whitney test.
DISCUSSION
The expression of the four-subunit complex of human pol δ in E. coli has facilitated biochemical studies of mutant mammalian pol δ (17, 18) and has allowed for analysis of the consequences of substitutions in the active site of a mammalian replicative DNA polymerase. Our studies indicate that both the L606G and L606K substitutions in human pol δ result in retained polymerase and exonuclease activity, unchanged processivity, and intact interaction with PCNA. However, the two variants differ greatly in their base selection accuracy; human pol δ L606G is extremely error prone, incorporating single noncomplementary nucleotides at a 50-fold elevated rate relative to wild type pol δ. In contrast, pol δ L606K is highly accurate, with base selection by the active site that is more faithful than that of wild type pol δ. To our knowledge, this study represents the first demonstration of a substitution in a mammalian polymerase that results in improved accuracy of base selection, a finding that suggests that base selection accuracy by DNA polymerases has not been optimized during evolution for maximal fidelity (35, 36). The enhanced base selection fidelity of pol δ L606K is associated with an apparent defect in bypass of DNA lesions in vitro, as well as impaired progression of the replication fork in mouse cells in vivo. Small differences in lesion bypass efficiency could have a major impact on DNA replication in vivo when one considers the large number of lesions that are likely to be encountered by a replicative polymerase, and we hypothesize that stalling of pol δ L606K at DNA lesions is responsible for the observed defect in replication fork progression.
The catalytic subunit of human pol δ is 94% conserved with mouse pol δ, and the sequence of motif A is identical between the two organisms. Thus, our results with mutated human pol δ in vitro may be relevant to understanding the phenotype seen in mice expressing the homologous variants in vivo. Heterozygous expression in mice of the highly error prone pol δG variant results in no change in life span and no discernable phenotype. In contrast, heterozygous expression of the highly accurate pol δK variant is associated with genomic instability, shortened life span, and accelerated carcinogenesis (16). Although we found that pol δG and pol δK exhibit substantial differences in base selection accuracy in vitro, in S. cerevisiae and in mice, both pol δG and pol δK exhibit a similar elevation of point mutations in vivo. Given the large difference in base selection accuracy between the two variants in vitro, it is likely the mutations occurring in vivo arise from distinct mechanisms in cells expressing pol δG versus pol δK. In the case of pol δG, which is highly error prone in vitro, point mutations in vivo are likely due to mis-incorporation by the polymerase active site. In contrast, the elevated frequency of point mutations in cells expressing pol δK, which is highly accurate in vitro, may be due to an increased role during nuclear DNA replication of the error prone bypass polymerases such as pol η and pol κ (37), secondary to the impaired lesion bypass capability of pol δK. In contrast to the differing phenotypes seen upon expression of the two variants in heterozygous mice, pol δG and pol δK are both embryonic lethal upon homozygous expression. The lethality of these polymerases in homozygous mice, despite the near-wild type catalytic activity of the purified enzymes in vitro, suggests the accumulation of a lethal burden of mutations that is not compatible with mammalian development (10).
The impaired replication fork progression that was seen in cells expressing pol δK in the in vivo replication fork progression assay, in conjunction with the impaired lesion bypass activity of this variant in vitro, is consistent with stalling of the mutant polymerase at endogenous sites of DNA damage. An alternative possibility is that pol δK replicates DNA in vivo at a similar rate to that of pol δG when utilizing undamaged nucleotides, but it is inhibited in synthesis in the molecular combing assay due to an impaired ability to insert the labeled nucleotide analogs that are used to track synthesis. A substantial fraction of the DNA damage that occurs in vivo is likely to occur to free nucleotides prior to their incorporation into DNA (38), and thus this interpretation is also consistent with the overall hypothesis that synthesis by pol δK is impaired in vivo due to a defect in the ability of the mutant polymerase to process DNA damage.
Notably, in our previous work with pol δ+/G and pol δ+/K mouse embryonic fibroblasts (16), we did not detect a difference between the variants in activation of the DNA damage response as assayed by serine 139 phosphorylation of histone H2AX. However, activation of the checkpoint response is likely to occur only in response to near-lethal levels of DNA damage, and thus the partial defect in lesion bypass synthesis we observed by pol δK in vitro may be sufficient to result in accumulation of deleterious DNA rearrangements over multiple cellular generations, without causing an absolute block to synthesis and thus without widespread activation of the checkpoint response. We hypothesize that cells expressing pol δK will exhibit a disproportionate increase in the induction of replication fork stalling and genomic rearrangements when lower levels of DNA damage are induced in cells. However, it is also possible that factors that were not studied in this work may also contribute to the observed in vivo defect in DNA synthesis by pol δK, such as sequence-specific stalling at endogenous replication barriers such as structured non-B DNA and common fragile sites (39, 40).
The ability of the replicative DNA polymerases to copy past limited amounts of damaged DNA without interruption is likely a requirement for efficient progression of the replication fork. In addition to DNA damage in cells induced by environmental processes, DNA is modified by endogenous reactive molecules generated by normal cellular metabolism, and in particular by reactive oxygen species (41). It has been estimated that 1% of oxygen metabolism results in reactive oxygen species (42) and that reactive oxygen species induce ∼20,000 damaged nucleotides in each cell per day (43). Although cells possess specialized repair processes that are induced in the presence of significant levels of damage, these processes are unlikely to be sufficient to remove the thousands of damaged bases that are likely present in most cells at the time of genome replication. In addition, failed DNA repair can result in intermediates that are more detrimental than the initial lesion (44, 45). During DNA replication, pol δ is likely to encounter many blocking lesions; although eukaryotic cells do possess lesion bypass polymerases that are able to take over for the replicative polymerases and replicate beyond sites of damage that have not yet undergone repair, the catalytic efficiency of the major replicative polymerases is much greater than that of the bypass polymerases, and as a result small alterations in bypass by pol δ may have profound effects in cells. Thus, mutations in pol δ (such as L606G) that enhance the ability to bypass lesions might be better tolerated than mutations in pol δ (such as L606K) that reduce bypass.
Overall, pol δG and pol δK exhibit a similar burden of point mutations in vivo, yet a cancer phenotype is seen only in the case of pol δK, which exhibits impaired lesion bypass in vitro as well as stalled replication and large genomic rearrangements in vivo. These observations may be relevant to understanding the relative contributions of point mutations versus genomic rearrangements in spontaneous carcinogenesis. DNA sequencing of human cancers has revealed large numbers of mutations in each tumor analyzed. Presently, every tumor reported has at least 50 clonal mutations that alter the coding sequence of genes (46); fully sequenced DNA from human tumor has been reported to contain ∼30,000 clonal somatic mutations (47). Moreover, there are no mutations that are invariantly present in any type of tumor, and there is little evidence for a series of mutations that are diagnostic of any particular type of tumor (46). Nearly all of the reported mutations in tumors are single-base substitutions, predominantly transitions (48, 49). An increase in transition mutations can be considered as a hallmark of mis-incorporation by replicative DNA polymerases α, δ, and ϵ (1, 50) but can also be the result of DNA damage by reactive oxygen species. Many mutations identified in tumors are likely to be passenger mutations that have arisen during the last round of clonal selection. Evidently cells are able to tolerate large numbers of single-base substitutions, even those that result in nonsynonymous amino acid changes.
In contrast to single-base substitutions, rearrangements can result in frameshift mutations and large deletions that excise multiple contiguous genes and regulatory regions of DNA. Large deletions can cause inactivation of the corresponding genes on the homologous chromosome resulting in loss of heterozygosity, which is a characteristic of many tumors (51). Thus, per mutagenic event, deletions would be predicted to have a more profound effect on cellular phenotype than would single-base substitutions. The relative importance of DNA deletions compared with point mutations is supported by investigations on the fidelity of mitochondrial DNA replication. Substantial circumstantial evidence links mitochondrial DNA mutations to the aging process (52). A 500-fold increase in single base substitutions in mouse mitochondria results in a far greater burden of point mutations than is seen in vivo, yet with no apparent phenotype (53). The high tolerance to elevated point mutations in the mitochondrial genome is particularly striking when one considers its small size, with only 16 kb of sequence, and its extremely high density of coding sequence. The nuclear genome, with its substantial prevalence of noncoding sequence, would thus be expected to have an even higher tolerance to random point mutations. In contrast, a modest increase in random mitochondrial DNA deletions is associated with a dramatic accelerated aging phenotype (54), suggesting that mitochondrial DNA deletion events may be a driver of mammalian aging. We propose that nuclear DNA rearrangements may play a similarly central role in cancer progression.
Large genomic rearrangements are a known hallmark of cancer. However, smaller insertions and deletions that are present in individual cells within tumors are difficult to detect by current DNA sequencing methods. Even though massive DNA sequencing has revolutionized the way we think about many disease processes, its application in studying cancer may be limited by the heterogeneity of cells within a tumor. Accurate quantitation of these alterations would require long read lengths or the sequencing of single molecules. Moreover, most rearrangements that arise during tumorigenesis would likely either be lethal or would be negatively selected during tumor progression. The rate at which rearrangements occur in tumors is likely to be much higher than sequencing would predict, and random insertions and deletions that are not detected by current technology may represent an important driving force in tumor initiation and progression.
Supplementary Material
Acknowledgments
We thank Erica Ellingson for excellent technical assistance, Etienne Schwob and Marjorie Drac from the Plateforme Peignage Moléculaire de Montpellier for providing silanized coverslips, and the members of the Loeb laboratory for critically reviewing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA102029, R01 CA115802, and P01 CA077852 (to L. A. L.) and F30 AG030314 and T32 GM007266 (to M. W. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- pol
- polymerase
- PCNA
- proliferating cell nuclear antigen.
REFERENCES
- 1.Kunkel T. A., Bebenek K. (2000) Annu. Rev. Biochem. 69, 497–529 [DOI] [PubMed] [Google Scholar]
- 2.Loeb L. A., Monnat R. J., Jr. (2008) Nat. Rev. Genet. 9, 594–604 [DOI] [PubMed] [Google Scholar]
- 3.Sweasy J. B., Lauper J. M., Eckert K. A. (2006) Radiat. Res. 166, 693–714 [DOI] [PubMed] [Google Scholar]
- 4.Shevelev I. V., Hübscher U. (2002) Nat. Rev. Mol. Cell Biol. 3, 364–376 [DOI] [PubMed] [Google Scholar]
- 5.Modrich P., Lahue R. (1996) Annu. Rev. Biochem. 65, 101–133 [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 7.Loeb L. A., Springgate C. F., Battula N. (1974) Cancer Res. 34, 2311–2321 [PubMed] [Google Scholar]
- 8.Beckman R. A., Loeb L. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14140–14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldsby R. E., Hays L. E., Chen X., Olmsted E. A., Slayton W. B., Spangrude G. J., Preston B. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15560–15565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albertson T. M., Ogawa M., Bugni J. M., Hays L. E., Chen Y., Wang Y., Treuting P. M., Heddle J. A., Goldsby R. E., Preston B. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17101–17104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Modrich P. (2006) J. Biol. Chem. 281, 30305–30309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Y. H., Garg P., Stith C. M., Al-Refai H., Sterling J. F., Murray L. J., Kunkel T. A., Resnick M. A., Burgers P. M., Gordenin D. A. (2005) Mol. Cell. Biol. 25, 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinkai A., Loeb L. A. (2001) J. Biol. Chem. 276, 46759–46764 [DOI] [PubMed] [Google Scholar]
- 14.Patel P. H., Kawate H., Adman E., Ashbach M., Loeb L. A. (2001) J. Biol. Chem. 276, 5044–5051 [DOI] [PubMed] [Google Scholar]
- 15.Venkatesan R. N., Hsu J. J., Lawrence N. A., Preston B. D., Loeb L. A. (2006) J. Biol. Chem. 281, 4486–4494 [DOI] [PubMed] [Google Scholar]
- 16.Venkatesan R. N., Treuting P. M., Fuller E. D., Goldsby R. E., Norwood T. H., Gooley T. A., Ladiges W. C., Preston B. D., Loeb L. A. (2007) Mol. Cell. Biol. 27, 7669–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fazlieva R., Spittle C. S., Morrissey D., Hayashi H., Yan H., Matsumoto Y. (2009) Nucleic Acids Res. 37, 2854–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt M. W., Matsumoto Y., Loeb L. A. (2009) Biochimie 91, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jónsson Z. O., Hindges R., Hübscher U. (1998) EMBO J. 17, 2412–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgers P. M., Gerik K. J. (1998) J. Biol. Chem. 273, 19756–19762 [DOI] [PubMed] [Google Scholar]
- 21.Creighton S., Bloom L. B., Goodman M. F. (1995) Methods Enzymol. 262, 232–256 [DOI] [PubMed] [Google Scholar]
- 22.Conti C., Saccà B., Herrick J., Lalou C., Pommier Y., Bensimon A. (2007) Mol. Biol. Cell 18, 3059–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillaire M. J., Betous R., Conti C., Czaplicki J., Pasero P., Bensimon A., Cazaux C., Hoffmann J. S. (2007) Cell Cycle 6, 471–477 [DOI] [PubMed] [Google Scholar]
- 24.Nick McElhinny S. A., Stith C. M., Burgers P. M., Kunkel T. A. (2007) J. Biol. Chem. 282, 2324–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maki H., Kornberg A. (1985) J. Biol. Chem. 260, 12987–12992 [PubMed] [Google Scholar]
- 26.Eckert K. A., Kunkel T. A. (1993) J. Biol. Chem. 268, 13462–13471 [PubMed] [Google Scholar]
- 27.Maga G., Hubscher U. (2003) J. Cell Sci. 116, 3051–3060 [DOI] [PubMed] [Google Scholar]
- 28.Klarmann G. J., Schauber C. A., Preston B. D. (1993) J. Biol. Chem. 268, 9793–9802 [PubMed] [Google Scholar]
- 29.Lu X., Tan C. K., Zhou J. Q., You M., Carastro L. M., Downey K. M., So A. G. (2002) J. Biol. Chem. 277, 24340–24345 [DOI] [PubMed] [Google Scholar]
- 30.Boosalis M. S., Petruska J., Goodman M. F. (1987) J. Biol. Chem. 262, 14689–14696 [PubMed] [Google Scholar]
- 31.Freisinger E., Grollman A. P., Miller H., Kisker C. (2004) EMBO J. 23, 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beard W. A., Wilson S. H. (2003) Structure 11, 489–496 [DOI] [PubMed] [Google Scholar]
- 33.Bebenek K., Kunkel T. A. (1995) Methods Enzymol. 262, 217–232 [DOI] [PubMed] [Google Scholar]
- 34.Stocki S. A., Nonay R. L., Reha-Krantz L. J. (1995) J. Mol. Biol. 254, 15–28 [DOI] [PubMed] [Google Scholar]
- 35.Loh E., Choe J., Loeb L. A. (2007) J. Biol. Chem. 282, 12201–12209 [DOI] [PubMed] [Google Scholar]
- 36.Loh E., Salk J. J., Loeb L. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1154–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo C., Kosarek-Stancel J. N., Tang T. S., Friedberg E. C. (2009) Cell. Mol. Life Sci. 66, 2363–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathews C. K. (2006) FASEB J. 20, 1300–1314 [DOI] [PubMed] [Google Scholar]
- 39.Weaver D. T., DePamphilis M. L. (1984) J. Mol. Biol. 180, 961–986 [DOI] [PubMed] [Google Scholar]
- 40.Shah S. N., Opresko P. L., Meng X., Lee M. Y., Eckert K. A. (2010) Nucleic Acids Res. 38, 1149–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barnes D. E., Lindahl T. (2004) Annu. Rev. Genet. 38, 445–476 [DOI] [PubMed] [Google Scholar]
- 42.Floyd R. A. (1995) in The Oxygen Paradox (Daview K. J., Ursini F. eds) pp. 89–103, CLEUP University Press, Padova, Italy [Google Scholar]
- 43.Beckman K. B., Ames B. N. (1997) J. Biol. Chem. 272, 19633–19636 [DOI] [PubMed] [Google Scholar]
- 44.Sung J. S., Demple B. (2006) FEBS J. 273, 1620–1629 [DOI] [PubMed] [Google Scholar]
- 45.Meira L. B., Moroski-Erkul C. A., Green S. L., Calvo J. A., Bronson R. T., Shah D., Samson L. D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 888–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fox E. J., Salk J. J., Loeb L. A. (2009) Cancer Res. 69, 4948–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pleasance E. D., Cheetham R. K., Stephens P. J., McBride D. J., Humphray S. J., Greenman C. D., Varela I., Lin M. L., Ordóñez G. R., Bignell G. R., Ye K., Alipaz J., Bauer M. J., Beare D., Butler A., Carter R. J., Chen L., Cox A. J., Edkins S., Kokko-Gonzales P. I., Gormley N. A., Grocock R. J., Haudenschild C. D., Hims M. M., James T., Jia M., Kingsbury Z., Leroy C., Marshall J., Menzies A., Mudie L. J., Ning Z., Royce T., Schulz-Trieglaff O. B., Spiridou A., Stebbings L. A., Szajkowski L., Teague J., Williamson D., Chin L., Ross M. T., Campbell P. J., Bentley D. R., Futreal P. A., Stratton M. R. (2010) Nature 463, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leary R. J., Lin J. C., Cummins J., Boca S., Wood L. D., Parsons D. W., Jones S., Sjöblom T., Park B. H., Parsons R., Willis J., Dawson D., Willson J. K., Nikolskaya T., Nikolsky Y., Kopelovich L., Papadopoulos N., Pennacchio L. A., Wang T. L., Markowitz S. D., Parmigiani G., Kinzler K. W., Vogelstein B., Velculescu V. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16224–16229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'Meara S., Vastrik I., Schmidt E. E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D. P., Louis D. N., Goldstraw P., Nicholson A. G., Brasseur F., Looijenga L., Weber B. L., Chiew Y. E., DeFazio A., Greaves M. F., Green A. R., Campbell P., Birney E., Easton D. F., Chenevix-Trench G., Tan M. H., Khoo S. K., Teh B. T., Yuen S. T., Leung S. Y., Wooster R., Futreal P. A., Stratton M. R. (2007) Nature 446, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bielas J. H., Loeb K. R., Rubin B. P., True L. D., Loeb L. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18238–18242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bielas J. H., Venkatesan R. N., Loeb L. A. (2007) Trends Genet. 23, 154–157 [DOI] [PubMed] [Google Scholar]
- 52.Greaves L. C., Turnbull D. M. (2009) Biochim. Biophys. Acta 1790, 1015–1020 [DOI] [PubMed] [Google Scholar]
- 53.Vermulst M., Bielas J. H., Kujoth G. C., Ladiges W. C., Rabinovitch P. S., Prolla T. A., Loeb L. A. (2007) Nat. Genet. 39, 540–543 [DOI] [PubMed] [Google Scholar]
- 54.Vermulst M., Wanagat J., Kujoth G. C., Bielas J. H., Rabinovitch P. S., Prolla T. A., Loeb L. A. (2008) Nat. Genet. 40, 392–394 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.