Abstract
Trypanosoma brucei is a parasitic protist that undergoes a complex life cycle during transmission from its mammalian host (bloodstream forms) to the midgut of its insect vector (procyclic form). In both parasitic forms, most glycolytic steps take place within specialized peroxisomes, called glycosomes. Here, we studied metabolic adaptations in procyclic trypanosome mutants affected in their maintenance of the glycosomal redox balance. T. brucei can theoretically use three strategies to maintain the glycosomal NAD+/NADH balance as follows: (i) the glycosomal succinic fermentation branch; (ii) the glycerol 3-phosphate (Gly-3-P)/dihydroxyacetone phosphate (DHAP) shuttle that transfers reducing equivalents to the mitochondrion; and (iii) the glycosomal glycerol production pathway. We showed a hierarchy in the use of these glycosomal NADH-consuming pathways by determining metabolic perturbations and adaptations in single and double mutant cell lines using a combination of NMR, ion chromatography-MS/MS, and HPLC approaches. Although functional, the Gly-3-P/DHAP shuttle is primarily used when the preferred succinate fermentation pathway is abolished in the Δpepck knock-out mutant cell line. In the absence of these two pathways (Δpepck/RNAiFAD-GPDH.i mutant), glycerol production is used but with a 16-fold reduced glycolytic flux. In addition, the Δpepck mutant cell line shows a 3.3-fold reduced glycolytic flux compensated by an increase of proline metabolism. The inability of the Δpepck mutant to maintain a high glycolytic flux demonstrates that the Gly-3-P/DHAP shuttle is not adapted to the procyclic trypanosome context. In contrast, this shuttle was shown earlier to be the only way used by the bloodstream forms of T. brucei to sustain their high glycolytic flux.
Keywords: Glucose Metabolism, Glycolysis, Metabolic Regulation, RNA Interference (RNAi), Trypanosome, Glycosomes, Metabolic Adaptation, Proline Metabolism, Redox Balance, Succinic Fermentation Pathway
Introduction
Trypanosomes of the Trypanosoma brucei group are the etiological agents of human African trypanosomiasis, a parasitic disease that affects over 36 countries in sub-Saharan Africa (1). T. brucei is a unicellular eukaryote, belonging to the protist order Kinetoplastida, that undergoes a complex life cycle during transmission from the bloodstream of a mammalian host (bloodstream stages of the parasite) to the alimentary tract (procyclic stage) and salivary glands (epimastigote and metacyclic stages) of a blood-feeding insect vector, the tsetse fly.
In the glucose-rich environment of the mammalian bloodstream, the parasite relies solely on glucose to produce energy (for review see Ref. 2). The procyclic insect stage of T. brucei, our experimental model in this analysis, develops a more elaborate energy metabolism based on different carbon sources, including glucose, proline, and threonine (3, 4). Although proline is the major component of the fly hemolymph (5), the parasite prefers glucose when this carbon source is available (6, 7).
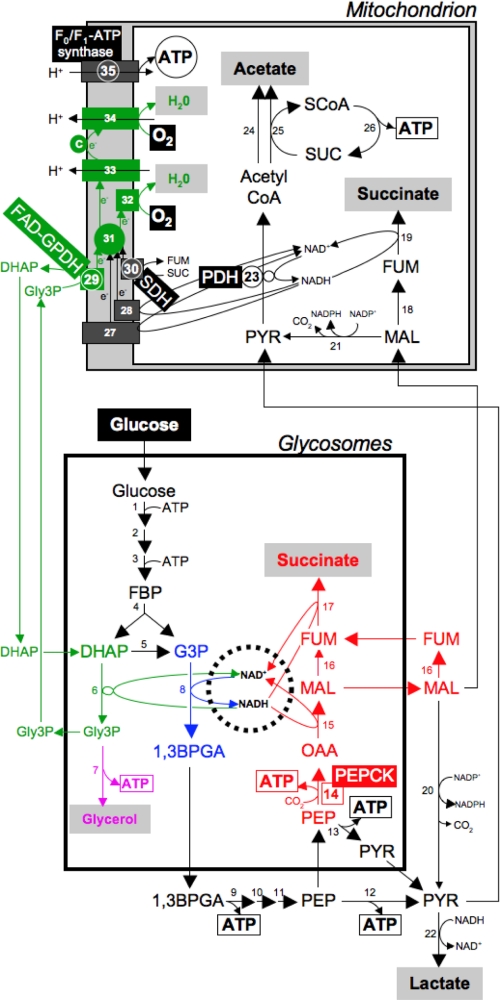
The procyclic trypanosomes convert glucose by aerobic fermentation into partially oxidized end products, such as succinate and acetate (Fig. 1) (for reviews see Refs. 8, 9). Most glycolysis takes place in specialized peroxisomes, called glycosomes (steps 1–5 and 8) (10). In the course of glycolysis, phosphoenolpyruvate is produced in the cytosol (steps 9–11), where it is located at a branching point. It can be converted into pyruvate (steps 12 and 13), which enters the mitochondrion to produce acetate (steps 23–25) (11, 12). Phosphoenolpyruvate can also reenter the glycosomes to be converted to succinate within that compartment (steps 14–17) or else, after conversion to malate, transferred to the mitochondrion to make succinate in that compartment (steps 18 and 19) (13, 14). Within the glycosomes, consumption and production of NADH are tightly balanced; NADH resulting from the reaction catalyzed by glyceraldehyde-3-phosphate dehydrogenase (step 8, colored in blue, Fig. 1) needs to be re-oxidized inside the organelle. It has been proposed that the glycosomal succinic fermentation pathway (steps in red, Fig. 1), which contains two NADH-dependent oxidoreductases (steps 15 and 17), is involved in this process (13). The glycosomal redox balance can also be theoretically maintained by the Gly-3-P3/DHAP shuttle (steps in green, Fig. 1) (15). This pathway involves the following: (i) a glycosomal NADH-dependent glycerol-3-phosphate dehydrogenase (NADH-GPDH, step 6), which produces Gly-3-P from DHAP; (ii) a putative glycosomal exchanger, which exchanges Gly-3-P for DHAP; and (iii) the mitochondrial FAD-dependent glycerol-3-phosphate dehydrogenase (FAD-GPDH, step 29), which regenerates DHAP from Gly-3-P. Electrons produced by FAD-GPDH are ultimately transferred to O2 via the mitochondrial respiratory chain (steps 31–34). Gly-3-P could also be converted into glycerol in the glycosomes (step 7, purple in Fig. 1), with a net production of one molecule of NAD+ per molecule of glycerol excreted.
FIGURE 1.
Schematic representation of glucose metabolism in the procyclic form of T. brucei. This figure describes the glycosomal NADH producing and consuming pathways, highlighted by a dashed circle and colored pathways. The glycosomal NADH-producing step is shown in blue; the glycosomal succinic fermentation pathways is shown in red; the Gly-3-P/DHAP shuttle and the associated complexes of the respiratory chain are shown in green; and the glycerol-producing step is shown in purple. Excreted end products from glucose metabolism are shown in black, red, green, or purple characters on a gray rectangle as background. ATP molecules produced by substrate level phosphorylation and oxidative phosphorylation are boxed and circled, respectively. Enzymatic steps targeted by RNAi are circled, and the PEPCK step, in which the gene has been deleted, is boxed; the name of the genetically manipulated enzymes is also indicated. Abbreviations used are as follows: 1,3BPGA, 1,3-bisphosphoglycerate; c, cytochrome c; e−, electrons; FBP, fructose 1,6-bisphosphate; FUM, fumarate; G3P, glyceraldehyde 3-phosphate; Gly3P, glycerol 3-phosphate; MAL, malate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PYR, pyruvate; SCoA, succinyl-CoA; SUC, succinate. Enzymes used are as follows: steps 1, hexokinase; 2, glucose-6-phosphate isomerase; 3, phosphofructokinase; 4, aldolase; 5, triose-phosphate isomerase; 6, NADH-dependent glycerol-3-phosphate dehydrogenase; 7, glycerol kinase; 8, glyceraldehyde-3-phosphate dehydrogenase; 9, phosphoglycerate kinase; 10, phosphoglycerate mutase; 11, enolase; 12, pyruvate kinase; 13, pyruvate phosphate dikinase; 14, phosphoenolpyruvate carboxykinase (PEPCK, Tb927.2.4210); 15, malate dehydrogenase; 16, cytosolic fumarase; 17, glycosomal NADH-dependent fumarate reductase; 18, mitochondrial fumarase; 19, mitochondrial NADH-dependent fumarate reductase; 20, cytosolic malic enzyme; 21, mitochondrial malic enzyme; 22, unknown enzyme; 23, pyruvate dehydrogenase complex (PDH-including the E2 subunit, PDH-E2, Tb927.10.7570); 24, unknown enzyme; 25, acetate:succinate CoA-transferase; 26, succinyl-CoA synthetase; 27, rotenone-sensitive NADH dehydrogenase (complex I of the respiratory chain); 28, rotenone-insensitive NADH dehydrogenase; 29, FAD-dependent glycerol-3-phosphate dehydrogenase (FAD-GPDH, Tb11.02.5280); 30, succinate dehydrogenase (SDH, complex II of the respiratory chain, Tb927.8.6580); 31, ubiquinone; 32, SHAM-sensitive alternative oxidase; 33, complex III of the respiratory chain; 34, complex IV of the respiratory chain; 35, F0F1-ATP synthase (ATPϵ- including the F1β subunit, ATPϵ-F1β, Tb927.3.1380).
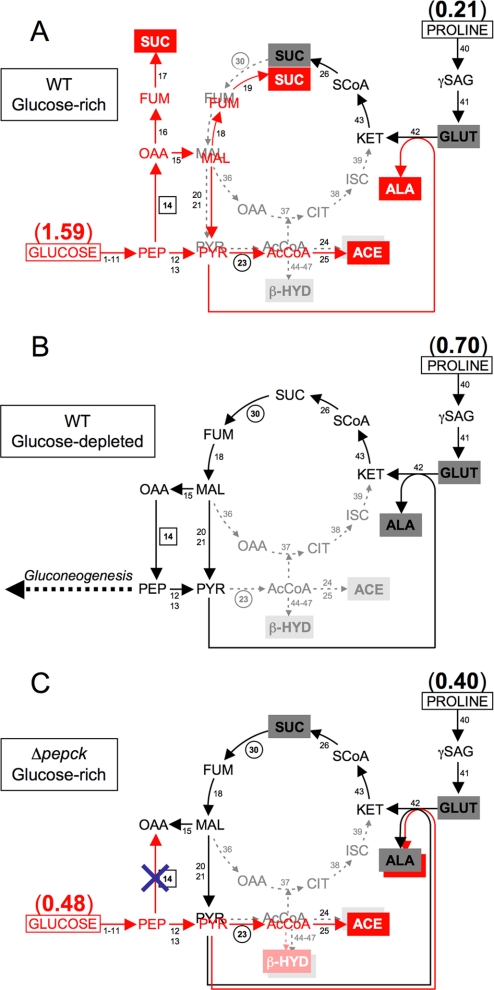
In a glucose-depleted environment, the procyclic trypanosomes modify their metabolism by increasing the rate of proline consumption compared with glucose-rich conditions. Succinate produced in the mitochondrion from proline metabolism is further converted into alanine (Fig. 2, A and B) (7, 16–18). The metabolic adaptation resulting from a repressive effect of glucose metabolism also affects the mode of ATP production, i.e. oxidative phosphorylation (step 35) versus substrate level phosphorylation (steps, 9, 12, and 26). In glucose-rich conditions, ATP is primarily produced by substrate level phosphorylation (4, 7, 19), whereas oxidative phosphorylation becomes essential in the absence of glucose (7, 20), although some differences may exist between strains (20).
FIGURE 2.
Schematic representation of proline and glucose metabolism in the wild type and Δpepck procyclic trypanosomes. A and B represent the wild type cells grown in glucose-rich and glucose-depleted conditions, respectively. C describes the switch to proline metabolism observed for the Δpepck mutant grown in glucose-rich conditions. Major and minor end products from proline metabolism are black characters on dark and light gray rectangles as backgrounds, respectively. End products from glucose metabolism are white characters on red rectangles. Dashed gray arrows indicate steps, which are considered to occur at a background level or not at all. The rate of proline and/or glucose consumption (μmol consumed/h/mg of protein) is indicated by parentheses. Deletion of the PEPCK gene is represented by a blue cross in C. Enzymatic steps targeted by RNAi are circled, and the PEPCK step, in which the gene has been deleted, is boxed. Abbreviations not used in Fig. 1 are as follows: ACE, acetate; AcCoA, acetyl-CoA; ALA, alanine; β-HYD, β-hydroxybutyrate; GLUT, glutamate; γSAG, glutamate γ-semialdehyde. Enzymes not shown in Fig. 1 are as follows: steps 36, mitochondrial malate dehydrogenase; 37, citrate synthase; 38, aconitase; 39, isocitrate dehydrogenase; 40, l-proline dehydrogenase; 41, pyrroline-5-carboxylate dehydrogenase; 42, l-alanine aminotransferase; 43, 2-ketoglutarate dehydrogenase complex; 44; acetyl-CoA acetyltransferase; 45, 3-hydroxy-3-methylglutaryl-CoA synthase; 46, 3-hydroxy-3-methylglutaryl-CoA lyase; 47, β-hydroxybutyrate dehydrogenase.
Here, we further analyze the metabolic flexibility of the procyclic trypanosomes by abolishing a main branch of glucose metabolism. Abolition of the succinate production pathway in the phosphoenolpyruvate carboxykinase (PEPCK) gene knock-out mutant (Δpepck) induced successful metabolic adaptations that allowed the parasite to maintain its growth rate, although its rate of glucose consumption is 3.3-fold reduced. To understand these adaptations, we have generated a number of RNAi mutant cell lines in the PEPCK null background, which were analyzed by global metabolomics approaches.
EXPERIMENTAL PROCEDURES
Growth and Maintenance of Trypanosomes
The procyclic form of T. brucei EATRO1125.T7T was cultured at 27 °C in SDM79 medium containing 10% (v/v) heat-inactivated fetal calf serum (FCS) and 2.5 mg/ml hemin (21) or in a glucose-depleted medium derived from SDM79, called SDM80 (6). The SDM80 medium is supplemented with 9% (v/v) heat-inactivated fetal calf serum dialyzed by ultrafiltration against 0.15 m NaCl (molecular mass cutoff: 10,000 Da; Sigma F0392; glucose concentration, 1 mm) and 1% (v/v) heat-inactivated FCS (glucose concentration, 5 mm). The glucose concentration in the glucose-depleted medium (SDM80) is 0.15 mm as compared with 6 mm in the glucose-rich media. We recently used a new FCS batch, which caused an increase of the procyclic cell doubling time grown in glucose-rich conditions (10.5 versus 13.5 h). For consistency, we always compared doubling times determined with the same FCS, such as described in Table 3. The FCS issue does not concern data obtained in glucose-depleted conditions, because no significant differences were observed with the different FCS batches.
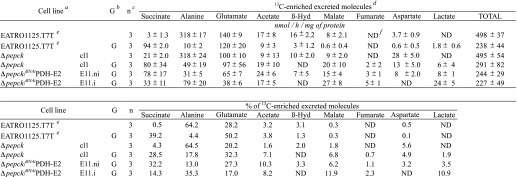
TABLE 3.
Excreted end products of d-[1-13C]glucose metabolism by procyclic T. brucei cell lines
β-Hyd means β-hydroxybutyrate.
a i means RNAi cell lines were tetracycline-induced during 6–10 days depending on the cell line and the experiments; ni means non-induced RNAi cell lines.
b n means number of experiments. When only two experiments have been analyzed per cell lines, the deviation of the mean (difference between the values divided by 2) is lower than 20% of the mean.
c The result of a single experiment or the means of 2–5 experiments are presented; the results are shown ± S.D. when more than two experiments have been performed per cell line.
d Glycerol and Gly-3-P peaks cannot be distinguished by this NMR analysis.
e Data are from Ref. 7.
f ND means not detectable.
Gene Knock-out of PEPCK
Replacement of the PEPCK gene (Tb927.2.4210) by the blasticidin and puromycin resistance markers via homologous recombination was performed with DNA fragments containing a resistance marker gene flanked by the PEPCK UTR sequences. Briefly, the pGEMt plasmid was used to clone an HpaI DNA fragment containing the blasticidin and puromycin resistance marker genes preceded by the PEPCK 5′-UTR fragment (514 bp) and followed by the PEPCK 3′-UTR fragment (597 bp). Primers used to produce PCR fragments are described in supplemental Fig. S1. The PEPCK knock-out was generated in the EATRO1125.T7T parental cell line, which constitutively expresses the T7 RNA polymerase gene and the tetracycline repressor under the control of a T7 RNA polymerase promoter for tetracycline inducible expression (22). Transfection and selection of drug-resistant clones were performed as reported previously (23). The first and second PEPCK alleles were replaced by blasticidin- and puromycin-resistant genes, respectively. Transfected cells were selected in glucose-rich SDM79 medium containing hygromycin B (25 μg/ml), neomycin (10 μg/ml), blasticidin (10 μg/ml), and puromycin (1 μg/ml).
Inhibition of Gene Expression by RNAi
Accession numbers of genes targeted by RNAi are as follows: F1β subunit of the F0/F1-ATP synthase (ATPϵ-F1β, Tb927.3.1380), FAD-GPDH (Tb11.02.5280), E2 subunit of the pyruvate dehydrogenase complex (PDH-E2, Tb927.10.7570), and succinate dehydrogenase (SDH, Tb927.8.6580). The inhibition by RNAi of gene expression in the procyclic form (24) was performed by expression of stem-loop “sense/antisense” RNA molecules of the targeted sequences (22) introduced in the pLew100 or pLew79 expression vectors (kindly provided by E. Wirtz and G. Cross) (25) or the p2T7Ti-177 vector (kindly provided by B. Wickstead and K. Gull) (26), as described previously. These three vectors contain the phleomycin resistance gene. Construction of the pLew-SDH-SAS, pLew-PDH-E2-SAS, and pLew-ATPϵ-F1β used to produce the double mutants Δpepck/RNAiSDH, Δpepck/RNAiPDH-E2, and Δpepck/RNAiATPϵ-F1β, respectively, have been described before (7). The RNAiSDH-A1, RNAiPDH-E2, and RNAiATPϵ-F1β cell lines have been generated before (7). The pLew-FAD-GPDH-SAS construct targets a fragment (from position 367 to 903 bp) of the FAD-GPDH gene. Briefly, a PCR-amplified 584-bp fragment, containing the antisense FAD-GPDH sequence with restriction sites added to the primers (supplemental Table S1), was inserted into the HindIII and BamHI restriction sites of the pLew100 plasmid. The separate PCR-amplified fragments containing the sense FAD-GPDH sequence were then inserted upstream of the antisense sequence, using HindIII and XhoI restriction sites (XhoI was introduced at the 3′-extremity of the antisense PCR fragment). The resulting plasmid (pLew-FAD-GPDH-SAS) contains a sense and antisense version of the targeted gene fragment, separated by a 50-bp fragment, under the control of the procyclic acidic repetitive protein promoter linked to a prokaryotic tetracycline operator.
The Δpepck null mutant and the EATRO1125.T7T parental cell line have been transformed with expression plasmids described above. The RNAi-harboring single mutant cell lines were selected in glucose-rich SDM79 medium containing hygromycin B (25 μg/ml), neomycin (10 μg/ml), and phleomycin (5 μg/ml). For transfection of the Δpepck cell line, blasticidin (10 μg/ml) and puromycin (1 μg/ml) were also included in the medium. Aliquots were frozen in liquid nitrogen to provide stocks of each line that had not been cultivated long term in medium. The selected cell lines were then adapted to the glucose-rich SDM80glu medium and the glucose-depleted SDM80 medium containing the same concentration of the five antibiotics.
Enzyme Assays
Sonicated (5 s at 4 °C) crude extracts of trypanosomes resuspended in cold hypotonic buffer (10 mm potassium phosphate, pH 7.8) were tested for mitochondrial FAD-GPDH (EC 1.1.99.5) (15) and malate dehydrogenase (MDH, EC 1.1.1.40) (27).
Immune Sera Production and Western Blot Analyses
For the production of PDH-E2 antibodies, a recombinant fragment corresponding to the full-length PDH-E2 gene preceded by an N-terminal histidine tag (10 histidine codons) was expressed in the Escherichia coli BL21, using the pET16b expression vector (Novagen). Cells were harvested by centrifugation, and recombinant proteins were purified by nickel chelation chromatography (Novagen) according to the manufacturer's instructions. Antisera were raised in rabbits or mice by three injections at 15-day intervals of 200 μg of PDH-E2-His recombinant nickel-purified proteins, electroeluted after separation on SDS-PAGE, and emulsified with complete (first injection) or incomplete Freund's adjuvant.
Total protein extracts of procyclic form T. brucei (5 × 106 cells) were separated by SDS-PAGE (10%) and immunoblotted on Immobilon-P filters (Millipore) (28). Immunodetection was performed as described previously (28, 29) using as primary antibodies the rat antiserum against PEPCK diluted 1:1000 (gift from T. Seebeck, Bern, Switzerland), the rabbit antiserum against the F1 moiety of the mitochondrial F0/F1-ATP synthase isolated from Crithidia fasciculata diluted 1:1000 (30) (gift from D. Speijer, Amsterdam, Netherlands), the heat shock protein 60 (hsp60) diluted 1:10,000 (31), or the E2 subunit of PDH diluted 1:500. Goat anti-rat, anti-rabbit, or anti-mouse Ig/peroxidase (1:10,000 dilution) was used as secondary antibody, and revelation was performed using ECLTM Western blotting detection reagents as described by the manufacturer (Amersham Biosciences).
Determination of Glucose and Proline Consumption
To determine the rate of glucose and proline consumption, cells (inoculated at 1–1.5 × 107 cells/ml) were grown in 10 ml of SDM79, SDM80glu (6 mm glucose), or SDM80 (0.15 mm glucose) medium. Aliquots of each growth medium (500 μl) were collected 0, 1, 6, 9, 10, 23, and 24 h after incubation at 27 °C. The quantity of glucose present in the medium was determined using the “Glucose GOD-PAP” kit (Biolabo SA). Proline concentration was determined with a colorimetric assay as described previously (32) after deproteinization of the samples by perchloric acid treatment.
Analysis of Excreted End Products from Glucose and Proline Metabolism
5–8 × 108 T. brucei procyclic cells were collected by centrifugation at 1400 × g for 10 min, washed once with phosphate-buffered saline (PBS), and incubated in 30 ml of PBS. For the analysis of glucose metabolism, the cells were maintained for 6 h at 27 °C in incubation buffer containing 110 μmol of d-[1-13C]glucose and 2 g/liter NaHCO3, pH 7.4. For the analysis of l-proline metabolism, the cells were maintained in PBS, pH 7.4 (without NaHCO3), containing 20 μmol of l-[4-13C] proline in the presence of 100 μmol of unenriched d-glucose. For NMR experiments, the supernatant was lyophilized, and 13C NMR spectra were collected at 125.77 MHz with a Bruker DPX500 spectrometer, as described before (7).
The supernatants analyzed by NMR were then lyophilized and dissolved in 1 ml of Milli-Q H2O for further analyses by high performance liquid chromatography (HPLC). The amount of glycerol in samples was determined by using an HPLC system (HP 1100 Series, Agilent, Santa Clara, CA) coupled to a Shodex RI-101 refractive index detector. The analytical column (Aminex® HPX-87H, 300 × 7.8 mm, 9 μm) was maintained at 48 °C. The binary pump was operated isocratically with 5 mm H2SO4 at a flow rate of 0.5 ml/min for 40 min. The injection volume was 50 μl.
Analysis of Intracellular Metabolites
T. brucei procyclic cells grown in glucose-rich conditions were sampled by fast filtration (33). Briefly, 2 × 107 cells were taken from the culture media, directly filtered on a vacuum (Whatman glass microfiber filters), and washed with 500 μl of culture media diluted 1:10 in PBS. The filters on which the cells were deposited were directly wrapped in aluminum paper and quenched in liquid nitrogen. Total sampling time was below 8 s. The filters were then transferred into 5 ml of boiling water. After 30 s of incubation, 200 μl of a uniformly 13C-labeled extract of E. coli was added as a quantification standard, and the solution was vortexed for 2 s. The extracts were immediately filtered (0.2 μm), chilled with liquid nitrogen, lyophilized, and dissolved in 200 μl of Milli-Q water prior to analysis. Three replicates were taken from each culture media, sampled, and analyzed separately.
The ion chromatography system (ICS 2000, Dionex, Sunnyvale, CA) was equipped with an EG50 potassium hydroxide eluent generator and a 2-mm ASRS-ULTRA II suppressor. Intracellular metabolites were separated at 29 °C on an IonPac AS11 analytical column (250 × 2 mm, Dionex) as described previously (34).
Ion chromatography was coupled to a 4000 QTRAP triple quadrupole linear ion trap mass spectrometer (Applied Biosystem, Foster City, CA) operating in the negative ion mode. Multiple reaction monitoring was used to detect selective fragments of the precursor ions with a dwell time of 25 ms. The acquisition parameters are described in more detail in supplemental Tables S2 and S3. The method allowed the potential detection of 43 intracellular metabolites, including most intermediates from glycolysis and pentose-phosphate pathway, organic acids, and nucleotides (supplemental Table S2). In the trypanosome extracts, 31 metabolites showed signal-to-noise ratios consistent with accurate quantification of the metabolite pools (supplemental Table S3).
Cell Permeabilization and Measurement of Oxygen Consumption
A total of 3 × 108 cells were collected from exponential phase growth, centrifuged at 10,000 rpm, and washed twice in buffer, pH 7.2, containing 240 mm mannitol, 100 mm KCl, 1 mm EGTA, 20 mm MgCl2, and 10 mm KH2PO4. Oxidation rates were determined polarographically with a Clark-type oxygen electrode (Rank Brothers) at 25 °C in a glass vessel (final volume 2 ml). The Clark-type oxygen electrode response was calibrated in air-equilibrated buffer in the absence (100% O2) and presence (0% O2) of sodium dithionite. 100% O2 concentration was taken as 240 μm (35). Permeabilization of whole cells was performed by the addition of 160 μg of digitonin to dilute cellular metabolites and consequently arrest the initial oxygen consumption, leaving the integrity of glycosomal and mitochondrial membranes intact (36). Respiratory chain response to substrate was tested by the consecutive addition of 12.5 mm Gly-3-P and 12.5 mm succinate, followed by KCN (6.25 mm) and SHAM (1.56 mm) to completely inhibit mitochondrial respiration.
RESULTS
Deletion of the PEPCK Gene Stimulates Growth of the Parasite
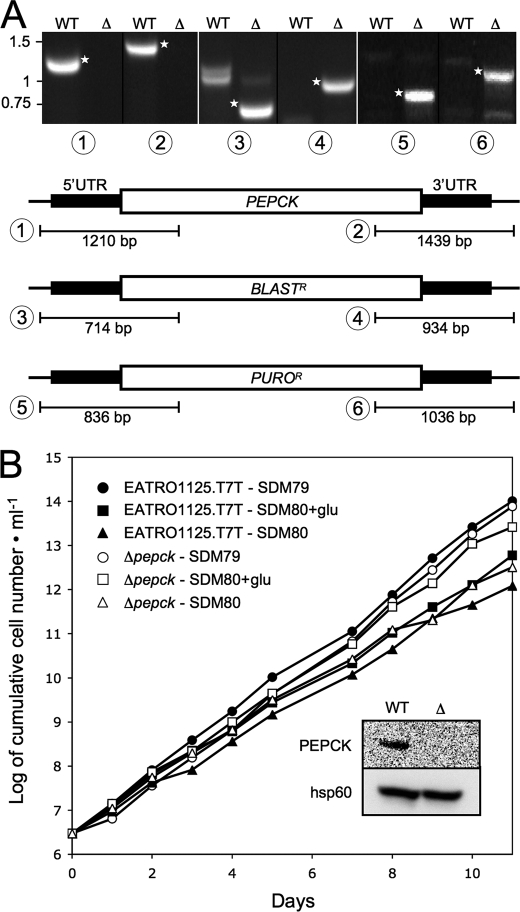
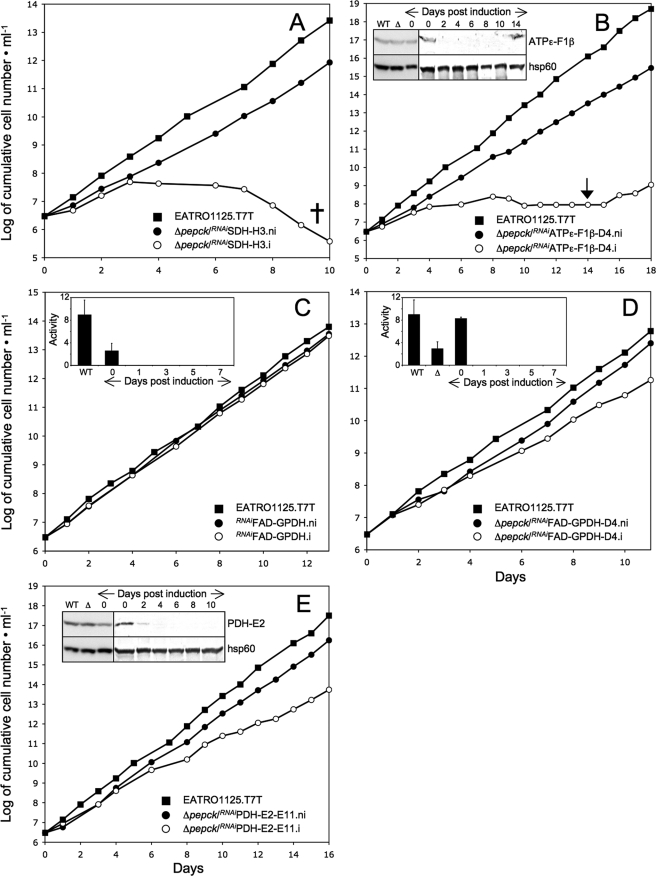
We previously observed that knockdown of the PEPCK gene (Tb927.2.4210) by RNAi only slightly affected procyclic trypanosomes (7), although PEPCK activity has been considered crucial to maintain both the glycosomal ATP/ADP and redox balances. To study metabolic adaptations caused by the absence of PEPCK activity, we have generated three independent PEPCK knock-out mutant cell lines (Δpepck::BLA/Δpepck::PURO, named Δpepck) by replacing both PEPCK alleles by selectable markers in the procyclic EATRO1125.T7T cell line, which express the tetracycline repressor and T7 RNA polymerase. Deletion of both alleles was confirmed by PCR and by Western blot with an anti-PEPCK immune serum (Fig. 3 for the Δpepck-cl1 cell line). The three freshly selected Δpepck mutant cell lines grew faster compared with the EATRO1125.T7T wild type cell line (11% decrease of the doubling time). However, after a few weeks of growth in standard SDM79 medium, the doubling time of the Δpepck-cl1 cell line returned back to wild type levels (Fig. 3B and Table 1) (not done for the two other Δpepck clones). The same observation was made when the Δpepck-cl1 cell line was grown in glucose-depleted conditions (SDM80). Because the three analyzed clones showed the same genotype and initial growth phenotype, only the Δpepck-cl1 cell line will be further described here.
FIGURE 3.
Analysis of the Δpepck mutant cell line. A shows a PCR analysis of genomic DNA isolated from the parental EATRO1125.T7T (WT) and Δpepck (Δ) cell lines. Amplifications were performed with primers based on sequences that flank the 5′UTR and 3′UTR fragments used to target the PEPCK gene depletion (black boxes) and internal sequences from the PEPCK gene (PCR products 1 and 2), the blasticidin resistance gene (BLASTR, PCR products 3 and 4), or the puromycin resistance gene (PUROR, PCR products 5 and 6). As expected, PCR amplification using primers derived from the PEPCK gene and drug-resistant genes were only observed for the parental EATRO1125.T7T (WT) and Δpepck (Δ) cell lines, respectively (DNA bands labeled with a star). B shows the growth curve of the EATRO1125.T7T and Δpepck cell lines incubated in SDM79 and the SDM80 medium either containing (SDM80glu) or lacking (SDM80) glucose. Cells were maintained in the exponential growth phase (between 106 and 107 cells/ml), and cumulative cell numbers reflect normalization for dilution during cultivation. The inset shows a Western blot analysis of the parental (WT) and Δpepck (Δ) cell lines with the anti-PEPCK and anti-hsp60 immune sera.
TABLE 1.
Effect of enzyme depletions on the doubling time of procyclic T. brucei cell lines
| Step(s) | Cell linea | Vector | ni/ib | Activityc | Proteind | Doubling timee |
|
|---|---|---|---|---|---|---|---|
| Glu-rich | Glu-depleted | ||||||
| % | h | ||||||
| EATRO1125.T7T | 10.5 (13.5)f | 13.9 | |||||
| 14 | Δpepck (cl1) | NDg | ND | WT | WT | ||
| 30 | RNAiSDH (A1)h | plew100 | ni | 80 | ndi | WTf | WT |
| i | ND | nd | WTf | †j | |||
| 14, 30 | Δpepck/RNAiSDH (H3) | plew100 | ni | nd | nd | 13.4 | 30.1 |
| i | nd | nd | † | † | |||
| 35 | RNAiATPϵ-F1βh | p2T7Ti-177 | ni | nd | +++ | 17.6f | 40.1 |
| i | nd | ND | 23.2f | † | |||
| 14, 35 | Δpepck/RNAiATPϵ-F1β (D4) | plew100 | ni | nd | +++ | 14.3 | 20.5 |
| i | nd | ND | † | † | |||
| 29 | RNAiFAD-GPDH | plew100 | ni | 30 | nd | WT | nd |
| i | ND | nd | WT | nd | |||
| 14, 29 | Δpepck/RNAiFAD-GPDH (D4) | plew100 | ni | 90 | nd | WT | nd |
| i | ND | nd | 13.1 | nd | |||
| 23 | RNAiPDH-E2h | plew79 | ni | 100 | +++ | WTe | WT |
| i | ND | ND | WTe | WT | |||
| 14, 23 | Δpepck/RNAiPDH-E2 (E11) | plew79 | ni | nd | +++ | WT | nd |
| i | nd | ND | 15.8 | nd | |||
a The clone number is indicated in parentheses.
b ni means noninduced; i means tetracycline-induced.
c Relative enzymatic activities of the targeted protein(s) in the RNAi-harboring cell lines are expressed as a percentage of the activity detected in the parental EATRO1125.T7T cell line.
d Relative amount of the targeted protein(s) in the RNAi-harboring cell lines are expressed as a relative amount of the protein(s) detected by Western blotting in the parental EATRO1125.T7T cell line.
e WT means that the doubling time of the mutant cell line is similar to that of the parental cell line (EATRO1125.T7T).
f Introduction of a new FCS batch caused a decrease of the EATRO1225.T7T doubling time (from 13.5 to 10.5 h). Cell lines described in 7 were grown with the old FCS.
g ND means not detectable.
h Data are from Ref. 7.
i nd means not determined.
j † means not viable.
The next two sections describe the metabolic adaptations occurring in the Δpepck-cl1 cell line to compensate for the loss of the succinate fermentation pathway. The metabolic fate of glucose and proline in the Δpepck-cl1 cell line was investigated, before addressing the question of the glycosomal redox balance.
Proline Metabolism Becomes More Active in the Δpepck Cell Line
The Δpepck cell line maintained in a glucose-rich environment showed a reduced rate of glucose consumption (3.3-fold) associated with an increased rate of l-proline consumption (2-fold) in comparison with the wild type cell line (Table 2). This suggests that the lack of PEPCK caused a reduced glycolytic flux that could be compensated by the activation of proline utilization. To further investigate this metabolic adaptation, 13C labeling experiments were carried out to detail the metabolism of glucose and proline, separately. The Δpepck procyclic cell line was maintained in PBS containing either 4 mm d-[1-13C]glucose or 1 mm l-[4-13C]proline, and the incubation medium was analyzed by 13C NMR. This approach allowed determination of the nature of the metabolic end products, as well as the total amount of 13C label recovered in these compounds (7).
TABLE 2.
Glucose and proline consumption in procyclic T. brucei cell lines
| Cell linea | Rate of glucose consumption | Rate of proline consumption |
|---|---|---|
| mmol/h/mg protein | ||
| EATRO1125.T7T | 1.59 ± 0.20 | 0.21 ± 0.05 |
| EATRO1125.T7T (−Glu) | NAb | 0.70 ± 0.20 |
| Δpepck | 0.48 ± 0.23 | 0.40 ± 0.12 |
| Δpepck (−Glu) | NA | 0.67 ± 0.15 |
| RNAiFAD-GPDH.ni | 0.86 ± 0.05 | 0.27 ± 0.09 |
| RNAiFAD-GPDH.i | 0.79 ± 0.15 | 0.19 ± 0.08 |
| Δpepck/RNAiFAD-GPDH.ni | 0.29 ± 0.01 | 0.34 ± 0.04 |
| Δpepck/RNAiFAD-GPDH.i | 0.10 ± 0.01 | 0.53 ± 0.07 |
| RNAiPDH-E2.nic | 1.50 ± 0.20 | 0.18 ± 0.05 |
| RNAiPDH-E2.ic | 1.60 ± 0.10 | 0.20 ± 0.04 |
| Δpepck/RNAiPDH-E2.ni | 0.68 ± 0.19 | 0.26 ± 0.11 |
| Δpepck/RNAiPDH-E2.i | 0.03 ± 0.03 | 0.50 ± 0.14 |
a i means tetracycline-induced; ni means noninduced.
b NA means not applicable.
c Cell line grown in SDM80glu.
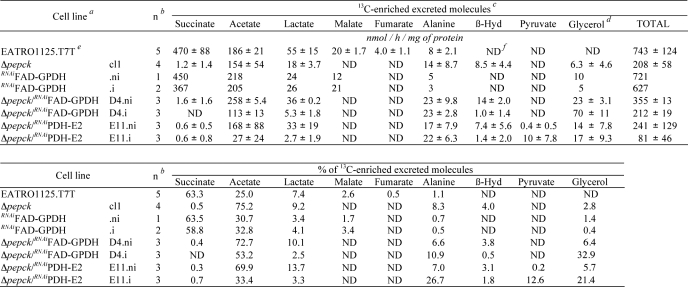
The PBS medium collected after 6 h of incubation with d-[1-13C]glucose was analyzed by 13C NMR spectroscopy to quantify the excretion of 13C-labeled products. In such conditions, a total of 743 nmol of 13C label was recovered in the supernatant of wild type cells (Table 3), with most of the label being detected in succinate (63.3%), acetate (25.0%), and lactate (7.4%). In comparison, much less label was recovered in the medium of the Δpepck mutant cells (208 nmol of 13C-enriched excreted molecules/h/mg of protein, see Table 3). This 3-fold reduction in 13C label recovery is consistent with the 3.3-fold reduction in glucose utilization in the Δpepck cell line. The spectrum of 13C-labeled products is also modified in the mutant cell line. In the parental EATRO1125.T7T cell, the major end product of glucose metabolism is succinate, which was barely detectable in the Δpepck mutant. This was also true for the downstream metabolites of succinate fermentation, namely malate and fumarate (Fig. 4, see Table 3). These data confirm that conversion of glucose into succinate proceeds uniquely via the PEPCK pathway in procyclic T. brucei. In addition, the NMR analyses showed increased signals of glycerol/Gly-3-P (the two compounds overlap in 13C NMR spectra) and β-hydroxybutyrate in the Δpepck cell line. This suggests a redistribution of flux toward the Gly-3-P/DHAP shuttle and acetate metabolism, respectively, when no glycosomal succinic fermentation is occurring (see below).
FIGURE 4.
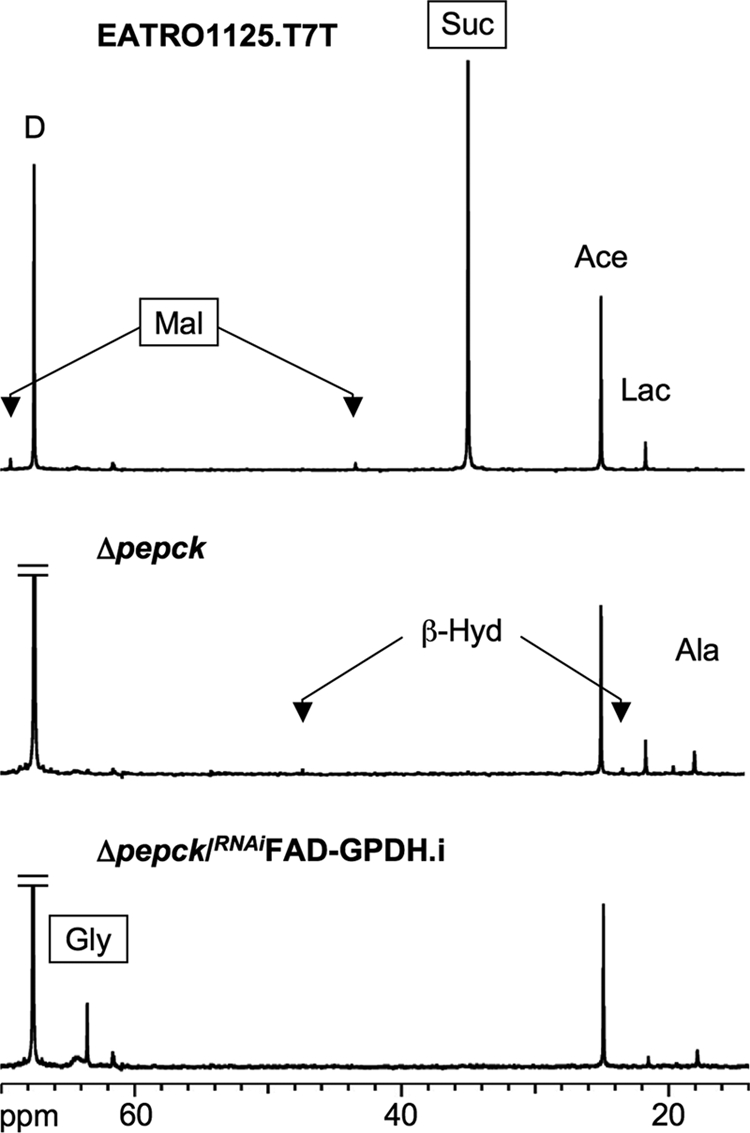
Carbon-13 NMR spectra of metabolic end products excreted by procyclic cell lines incubated with d-[1-13C]glucose. For these NMR analyses, the parental EATRO1125.T7T, the Δpepck, and tetracycline-induced Δpepck/RNAiFAD-GPDH.i cell lines were incubated with 4 mm d-[1-13C]glucose in PBS/NaHCO3 buffer. The NMR spectra were obtained after addition of 15 μl of dioxane. Each spectrum corresponds to one representative experiment from a set of at least three. The boxed metabolites (succinate, malate, and glycerol) are excreted metabolites produced in the glycosomes. The resonances were assigned as follows: Ala, alanine; Ace, acetate; β-Hyd, β-hydroxybutyrate; D, dioxane; Gly, glycerol; Lac, lactate; Mal, malate; Suc, succinate.
The metabolism of proline in the wild type EATRO1125.T7T procyclic cells is known to depend highly on the presence of glucose (7). Accordingly, in the absence of glucose, the parasite excretes mainly alanine (64.2% of label recovery from l-[4-13C]proline) and glutamate (28.2%), with only small amounts of acetate (3.2%), β-hydroxybutyrate (3.1%), and succinate (0.5%) (Table 4 and Fig. 2A). In the presence of glucose, proline utilization was significantly reduced, and its metabolic fate was also modified, i.e. succinate became a major end product, and alanine production was 30-fold reduced (Table 4 and Fig. 2B). In the same glucose-rich conditions, the Δpepck cell line produced similar amounts of alanine and succinate (Table 4 and Fig. 2C). The alanine/succinate ratio observed in these conditions was 0.62. This was 5-fold higher than observed with wild type cells in the absence of glucose (0.11) but 7-fold lower than observed for the wild type cells in glucose-rich conditions (106). In conclusion, the Δpepck cell line shows, in the presence of glucose, an intermediary state between glucose-depleted and glucose-rich conditions in the wild type cells.
TABLE 4.
Excreted end products of l-[4-13C]proline metabolism by procyclic T. brucei cell lines
a i means tetracycline-induced; ni means noninduced.
b Cells were grown in SDM79 and incubated in PBS/l-[4- 13C]proline containing (G) or not ( ) 3.3 mm d-glucose.
c Number of experiments for each cell line are shown.
d The results are shown ± S.D.
e Data from Ref. 7.
f ND means not detectable.
The intracellular metabolite content was analyzed by ion chromatography-MS/MS. In glucose-rich conditions, the levels of fumarate, malate, and succinate were significantly lower in the Δpepck mutant, compared with the wild type cells (supplemental Fig. S1). But the fact that significant extents of these metabolites could be detected in the Δpepck mutant, where the conversion of glucose into succinate no longer occurs, indicates that the utilization of proline could replenish the pool of dicarboxylic acids (fumarate, malate, and succinate) (Fig. 2C).
To further ascertain the activation of proline metabolism when PEPCK is lacking, RNAi knockdowns of each of succinate dehydrogenase (SDH, step 30) and the F0/F1-ATP synthase (step 35) were performed in the Δpepck cell line. These two enzymatic steps are dispensable in glucose-rich conditions. However, in glucose-depleted conditions, SDH is a key step in the conversion of proline into alanine (see Fig. 2B), and the F1β (ATPϵ-F1β) is essential for ATP production by oxidative phosphorylation (7). Considering that the increase in proline metabolism plays a critical role in the Δpepck cell line, the down-regulation of these two genes in the PEPCK null background was expected to strongly affect the cellular viability (see Fig. 2C). Indeed, both double mutants showed a strong growth phenotype in glucose-rich conditions. After tetracycline induction, the Δpepck/RNAiSDH cell line (Δpepck/RNAiSDH.i) died within 8 days, whereas the uninduced mutant (Δpepck/RNAiSDH.ni) showed only a moderate growth phenotype (Fig. 5A and Table 1). Three different Δpepck/RNAiATPϵ-F1β.i cell lines were analyzed, all of which showed a cessation of growth in glucose-rich medium, before a reversion to uninduced doubling time levels (Fig. 5B). This reversion was concomitant with the re-emergence of the ATPϵ-F1β protein in Western blots of cell extracts (Fig. 5B, inset). Altogether, these data confirmed that the Δpepck cell line switched to proline metabolism even in the presence of glucose.
FIGURE 5.
Analysis of mutant cell lines. A–E show the growth curve of the Δpepck/RNAiSDH, Δpepck/RNAiATPϵ-F1β, RNAiFAD-GPDH, Δpepck/RNAiFAD-GPDH, and Δpepck/RNAiPDH-E2 cell lines, respectively, incubated in SDM79 in the presence (.i, ○) or in the absence (.ni, ●) of tetracycline. Cells were maintained in the exponential growth phase (between 106 and 107 cells/ml), and cumulative cell numbers reflect normalization for dilution during cultivation. The cross indicates that further incubation led to the death of the whole population (A), and the arrow indicates the observed reexpression of the ATPϵ-F1β protein (B). The insets in B and E show Western blot analyses of the EATRO1125.T7T (WT) and Δpepck (Δ) cell lines, plus the Δpepck/RNAiATPϵ-F1β (B) or Δpepck/RNAiPDH-E2 (E) cell lines upon tetracycline addition (lanes 0–10/14). The name of the proteins recognized by the immune sera is indicated in the right margin. The insets in C and D show the FAD-GPDH activity (milliunits/mg of protein) of the EATRO1125.T7T parental cell line (WT) plus the RNAiFAD-GPDH (C) or Δpepck/RNAiFAD-GPDH (D) cell lines upon tetracycline addition (lanes 0–7). The inset of D also shows the FAD-GPDH activity of the Δpepck mutant (Δ). The FAD-GPDH activities are normalized with the malate dehydrogenase activity measured in the same samples.
Redox Balance within the Glycosomes
Current schemes of redox potential within the glycosomes show a tightly regulated balance of NADH production and usage, where the NADH produced by the glyceraldehyde-3-phosphate dehydrogenase is re-oxidized within the succinate fermentation pathway (2). In the Δpepck null mutant, where the latter pathway is no longer active, alternative process(es) must operate to maintain the glycosomal redox balance. Two processes could potentially play this role, namely the Gly-3-P/DHAP shuttle and glycerol fermentation. To address the role of the Gly-3-P/DHAP shuttle, we compared knockdown effects of the FAD-GPDH gene in the wild type and Δpepck null backgrounds. Both the tetracycline-induced RNAiFAD-GPDH.i and Δpepck/RNAiFAD-GPDH.i mutant cell lines showed a stable loss of the FAD-GPDH enzyme activity up to 7 days, even after 24 h of induction with tetracycline (Fig. 5, C and D, inset).
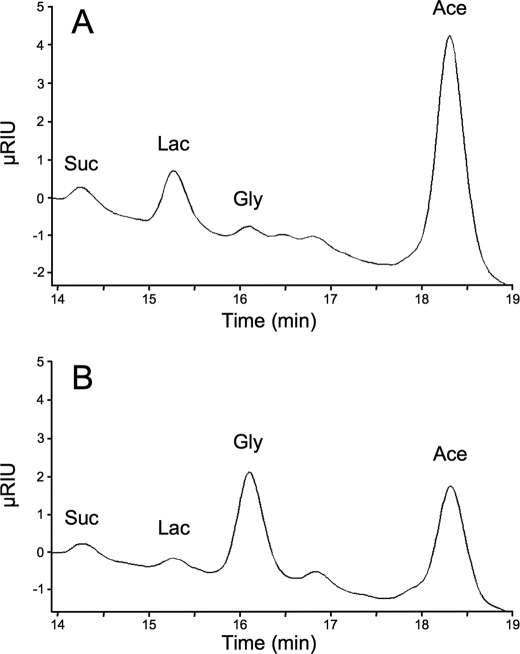
Although the Δpepck/RNAiFAD-GPDH.i double mutant showed a moderate growth phenotype upon tetracycline induction (Fig. 5D), the loss of FAD-GPDH in the Δpepck null background caused a number of metabolic alterations. First, glucose consumption was drastically reduced in the double mutant (16- and 5-fold, as compared with wild type and Δpepck cell lines, respectively). This was associated with an increase in proline consumption (Table 2). Second, the NMR analysis of incubation supernatants showed a reduced conversion of d-[1-13C]glucose into β-hydroxybutyrate, most likely as a result of the overall reduction in glycolytic flux (Fig. 4 and Table 3). Third, HPLC measurements of incubation supernatants showed that glycerol production was increased 14.4-fold in the Δpepck/RNAiFAD-GPDH.i mutant upon tetracycline induction (Fig. 6). Glycerol was not detected in the wild type, Δpepck, RNAiFAD-GPDH.ni, and RNAiFAD-GPDH.i cell lines (data not shown). Finally, the ion chromatography-MS/MS analysis of intracellular metabolites showed that the intracellular amount of Gly-3-P was ∼2.5-fold increased in the Δpepck/RNAiFAD-GPDH.i mutant compared with the uninduced cells (supplemental Fig. S1). These data indicate the concomitant accumulation of intracellular Gly-3-P and excretion of glycerol, which are, respectively, the substrate and product of glycerol kinase (step 7), in the Δpepck/RNAiFAD-GPDH.i mutant. Altogether, this shows that glycerol fermentation was increased in the PEPCK null background only when the FAD-GPDH was no longer active. This means that in the Δpepck mutant, reoxidation of glycosomal NADH mainly proceeds via the Gly-3-P/DHAP shuttle. These data also highlight the production of NAD+ by the glycosomal NADH-GPDH (step 6) in the absence of the glycosomal succinate fermentation branch.
FIGURE 6.
HPLC analysis of metabolic end products excreted by procyclic cell lines incubated with d-[1-13C]glucose. Metabolites contained in incubation medium of uninduced (A) and tetracycline-induced (B) Δpepck/RNAiFAD-GPDH cells were separated by HPLC and analyzed by refractometry. The same samples were analyzed by NMR (Table 3 and Fig. 4). Abbreviations used are as follows: Ace, acetate; Gly, glycerol; Lac, lactate; Suc, succinate; RIU, refractive index unit.
In contrast, none of the metabolic perturbations found in the Δpepck/RNAiFAD-GPDH.i double mutant were observed in the RNAiFAD-GPDH.i single mutant cell line, indicating that the Gly-3-P/DHAP shuttle is not critical for glycolysis when the PEPCK enzyme is active (Tables 2 and 3, Fig. 5D, and supplemental Fig. S1). In conclusion, these data demonstrate that the Gly-3-P/DHAP shuttle is primarily used in the absence of glycosomal succinic fermentation to maintain the glycosomal redox balance.
Functional Analysis of FAD-GPDH
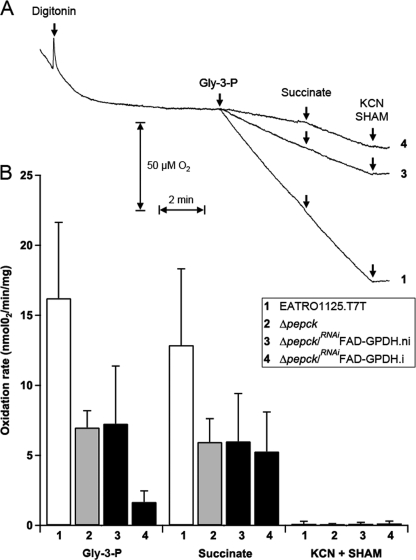
FAD-GPDH is the first step of the respiratory chain involved in electron transfer from Gly-3-P to molecular oxygen (see Fig. 1). To further confirm the involvement of FAD-GPDH in the Δpepck null background, it was important to demonstrate that knockdown of FAD-GPDH by RNAi significantly affected oxygen consumption from Gly-3-P. Thus, we compared oxygen consumption of wild type and mutant cell lines in the presence of Gly-3-P. Intact trypanosome cells are unable to use Gly-3-P as a respiratory substrate, because this compound cannot cross the plasma membrane. Therefore, we selectively permeabilized the plasma membrane by a mild digitonin treatment, without affecting mitochondrial integrity, as described previously (36). The wild type cells incubated in the absence of respiratory substrates showed a high rate of endogenous respiration, which dramatically decreased in the presence of digitonin (Fig. 7A). This suggests that the consumption of an unknown endogenous carbon source was abolished when the subcellular compartments were disconnected. In these conditions, the plasma membrane has been successfully permeabilized, because addition of 12.5 mm Gly-3-P restored oxygen consumption. A subsequent addition of 12.5 mm succinate did not further stimulate oxygen consumption. Succinate is one of the preferred respiratory substrates of procyclic trypanosomes (37), which is oxidized by complex II of the respiratory chain (SDH, step 30), indicating that Gly-3-P is also a good respiratory substrate in these experimental conditions. As expected, addition of both 6.25 mm KCN (inhibitor of the terminal oxidase, step 34) and 1.56 mm SHAM (inhibitor of the alternative oxidase, step 32) completely inhibited oxygen consumption (Fig. 7A). The Δpepck and uninduced Δpepck/RNAiFAD-GPDH.ni cell lines showed the same behavior, although the rate of oxygen consumption was ∼2-fold lower compared with the parental cell line (Fig. 7A). In contrast, the tetracycline-induced Δpepck/RNAiFAD-GPDH.i mutant showed a 4-fold reduction of the rate of oxygen consumption in the presence of Gly-3-P, compared with uninduced cells. The addition of succinate increased the respiration rate to levels comparable with that of the Δpepck and uninduced Δpepck/RNAiFAD-GPDH.ni cells. This result shows that in the induced double mutant, the reduced oxygen consumption observed in the presence of Gly-3-P was not due to a reduced capacity of oxygen consumption but the direct result of the loss of FAD-GPDH. In conclusion, these experiments demonstrate that, as expected, the FAD-GPDH function was considerably reduced upon tetracycline induction of the Δpepck/RNAiFAD-GPDH cell line.
FIGURE 7.
Oxygen consumption in permeabilized procyclic T. brucei cells. A, trace of respiration rates in 3 × 108 EATRO1125.T7T (1), Δpepck/RNAiFAD-GPDH.ni (3), and Δpepck/RNAiFAD-GPDH.i (4) cells, upon the addition of digitonin (60 μg), Gly-3-P (12.5 mm), succinate (12.5 mm), and KCN/SHAM (6.25/1.56 mm). B, rate of oxygen consumption for the EATRO1125.T7T (1), Δpepck (2), Δpepck/RNAiFAD-GPDH.ni (3), and Δpepck/RNAiFAD-GPDH.i (4) cell lines (4 < n < 6, error bars are ± S.E.).
Inhibition of Both the Succinate and Acetate Branches Is Detrimental for Glucose Metabolism
Glucose metabolism in procyclic trypanosomes mainly results in succinate and acetate production. To further alter glucose metabolism, the production of both end products was abolished by generating a RNAi mutant targeting the pyruvate dehydrogenase (PDH, step 23) in the PEPCK null background. In glucose-rich conditions, the RNAiPDH-E2 single mutant showed no growth phenotype upon tetracycline induction (7). The Δpepck/RNAiPDH-E2.i cell line survived tetracycline induction, with only a moderate increase (1.5-fold) of its doubling time (Table 1 and Fig. 5E). However, the Δpepck/RNAiPDH-E2.i double mutant showed important metabolic differences compared with wild type cells and the Δpepck mutant. First, glucose was no longer the main carbon source used in the Δpepck/RNAiPDH-E2.i cell line, because only a residual consumption was detected in glucose-rich conditions (2 and 6% of the wild type and Δpepck rates of consumption, respectively) (Table 2). Second, the incapability of the Δpepck/RNAiPDH-E2.i cell line to metabolize glucose was confirmed by NMR. The conversion of d-[1-13C]glucose into 13C-labeled end products was decreased 3- and 9-fold compared with the Δpepck and wild type cell lines, respectively (Table 3). Third, pyruvate, the PDH substrate, became a major excreted end product of glucose metabolism (accounting for 12.6% of total end products, with a 25-fold increase after tetracycline induction). Acetate was still detectable because of the impossibility to completely switch off gene expression by RNAi. Finally, as observed for the Δpepck mutant, the Δpepck/RNAiPDH-E2.i cell line switched to proline metabolism as shown by the increased rate of proline consumption (Table 2) and the increased amount of alanine produced from l-[4-13C]proline (Table 4). Altogether, these data show that the inhibition of acetate production in the absence of the succinate branches induced the arrest of glycolysis, compensated by a switch to proline (as observed for wild type procyclic cells grown in glucose-depleted conditions).
DISCUSSION
Kinetoplastids, including trypanosomatids and bodonids, are the only known organisms performing glycolysis in the specialized peroxisome organelles, called glycosomes. Within glycosomes, NAD+ consumed by the glyceraldehyde-3-phosphate dehydrogenase (step 8) needs to be regenerated by NADH-dependent oxidoreductases, to maintain the organellar redox (NAD+/NADH) balance. The procyclic form of T. brucei contains three different glycosomal pathways for re-oxidation of NADH into NAD+, i.e. succinic fermentation (steps 14–17), glycerol production (steps 6 and 7), and the Gly-3-P/DHAP shuttle (steps 6, 29, and 31–34).
Succinate Fermentation Is the Preferred Route for Glycosomal Reoxidation of NADH
Our data demonstrated that the contribution of the Gly-3-P/DHAP shuttle in the wild type environment is low (and possibly not significant). The RNAi knockdown of FAD-GPDH (step 29) resulted in no distinct phenotype discernable from the wild type cells, in terms of growth rate, rates of proline/glucose consumption, or metabolome analyses using NMR, HPLC, or ion chromatography-MS/MS. Similarly, the wild type parasites and the RNAiFAD-GPDH.i cell line did not use the glycerol production pathway, because glycerol is not detectable in the extracellular medium of cells incubated in the presence of d-[1-13C]glucose. Consequently, glycosomal succinic fermentation alone is sufficient for maintaining the glycosomal redox balance in wild type procyclic cells, as proposed previously (13). This contrasts with the bloodstream forms of T. brucei grown in aerobic conditions, which use only the Gly-3-P/DHAP shuttle to maintain the glycosomal redox balance.
Abolition of the Glycosomal Succinic Fermentation Stimulated Growth of the Parasite
This surprising behavior was observed in three independent Δpepck cell lines. This clearly indicates that, in the absence of this key glycosomal metabolic branch, the parasites develop successful metabolic alternatives. We have identified the following three main adaptations in response to the PEPCK gene deletion, (i) use of the Gly-3-P/DHAP shuttle to maintain the glycosomal redox balance; (ii) down-regulation of glucose metabolism, and (iii) switch to proline metabolism. Each of these adaptations will be individually addressed below.
The growth stimulation observed in the Δpepck cell lines is probably a consequence of these metabolic adaptations. Indeed, an increase of proline metabolism, although glucose is still consumed (albeit with a reduced rate), may stimulate the biosynthetic pathways by increasing the production of both ATP and biosynthetic precursors. This is consistent with the growth stimulation observed for different wild type procyclic strains grown in glucose-rich medium supplemented with N-acetyl-d-glucosamine (a glucose transport inhibitor), which also induces a switch to proline metabolism (38, 39). However, after a couple of weeks in the glucose-rich medium, the Δpepck cell line analyzed here reverted to normal growth, suggesting that the mutant reached a more stable metabolic steady state by reducing its metabolic activity.
Maintenance of the Glycosomal Redox Balance
Our data demonstrate a hierarchy between the three different glycosomal pathways for the regeneration of NAD+. As already mentioned, succinate fermentation is the first NAD+-production pathway used by the wild type procyclics to maintain the redox balance. When succinate fermentation is inactivated (Δpepck), the Gly-3-P/DHAP shuttle is primarily used instead. Glycerol production, which is not detectable in wild type cells, is also increased in the Δpepck mutant, but its contribution remains marginal (2.8% of the excreted metabolites). When both the Gly-3-P/DHAP shuttle and succinate fermentation were blocked (Δpepck/RNAiFAD-GPDH.i), glycerol became a major end product of glucose metabolism (21.4% of excreted metabolites), indicating that glycerol production is significantly used only in the absence of the two other alternatives.
Down-regulation of Glucose Metabolism in PEPCK Null Environment
The consumption of glucose was significantly reduced (3.3-fold) in the Δpepck cell line, compared with wild type cells. In parallel, the intracellular amounts of the glycolytic intermediates glucose 6-phosphate and fructose 6-phosphate were also reduced in the Δpepck cell line, which is consistent with a reduced glycolytic rate (supplemental Fig. S1). The reduced glucose consumption in the Δpepck cell line is not due to the inability of the Gly-3-P/DHAP shuttle to maintain the redox balance, because both the glycosomal and mitochondrial parts of the shuttle are functional in procyclic cells (15, 40). In parallel, we confirmed the activity of the shuttle in the PEPCK null environment by showing that the rate of oxygen consumption in the presence of Gly-3-P is ∼10-fold reduced in the Δpepck/RNAiFAD-GPDH.i mutant, compared with the wild type cells (Fig. 7). Altogether, these data suggest that, although the Gly-3-P/DHAP shuttle is functional, it is not well adapted to sustain a relatively high glycolytic flux in the procyclic trypanosome metabolic context.
Glycerol production is definitively not a good alternative for maintaining the glycosomal balance in procyclic trypanosomes, because glucose metabolism is almost abolished when both succinate fermentation and the Gly-3-P/DHAP shuttle are inactivated (16-fold reduction in the Δpepck/RNAiFAD-GPDH.i mutant compared with wild type cells). This may be related to the kinetic properties of the glycerol kinase. The conversion of Gly-3-P to glycerol is thermodynamically unfavorable and is only feasible at high Gly-3-P concentrations and a high ADP/ATP ratio (41). However, in the experimental conditions used for NMR investigations, the Δpepck/RNAiFAD-GPDH.i mutant incubated with glucose as the sole carbon source was forced to consume glucose at a rate similar to that observed for the Δpepck cell line (212 versus 208 nmol of 13C-enriched molecules excreted/h/mg of protein) (Table 3). In these conditions, the double mutant mainly converts glucose into acetate and glycerol (53.2 and 32.9% of the excreted end products, respectively) (Table 3). Interestingly, the nature of glucose metabolism in the Δpepck/RNAiFAD-GPDH.i cell line is highly similar to that observed in the bloodstream forms grown anaerobically, except that the procyclic mutant further converts pyruvate into acetate. Although this double mutant cell line still has the capability to perform glycolysis, it switches to proline metabolism when placed in glucose-rich conditions, with almost no glucose consumed.
Activation of Proline Metabolism in the Δpepck Cell Line Grown in Glucose-rich Conditions
The absence of a growth phenotype in the Δpepck cell line is primarily due to the capacity of the parasite to switch to proline metabolism, as observed previously for wild type cells grown in glucose-depleted conditions (6, 7). This means that the metabolism becomes more oxidative in the Δpepck mutant. Accordingly, the Δpepck/RNAiATPϵ-F1β.i and Δpepck/RNAiSDH.i double mutants were not able to grow in glucose-rich conditions, although such a phenotype was observed for the RNAiATPϵ-F1β.i and RNAiSDH.i single mutants only in glucose-depleted conditions (7). However, the metabolism of the Δpepck mutant in glucose-rich conditions does not closely mimic that of the wild type cells in glucose-depleted conditions. First, proline is consumed at a lower rate by the Δpepck mutant (0.40 versus 0.70 μmol consumed/h/mg of protein, respectively). Second, the ratio of alanine/succinate excreted from proline metabolism is considerably larger in glucose-depleted conditions (wild type and Δpepck mutant cell lines) compared with glucose-rich conditions (wild type cells) (106–15.1 versus 0.11). Again, the Δpepck cell line grown in glucose-rich conditions shows an intermediate value (0.65). This incomplete switch to proline metabolism is probably related to the remaining consumption of glucose observed in the Δpepck cell line. Interestingly, the Δpepck/RNAiPDH-E2.i double mutant expresses similar metabolic parameters (0.50 μmol of proline consumed/h/mg of protein, with alanine/succinate = 2.4), although its glucose consumption is almost abolished. This raises a number of questions such as the following. What is the maximum glycolytic flux compatible with the maintenance of the highest rate of proline consumption or how much glycolytic flux is required to fully repress proline metabolism? It has been shown previously that the metabolic adaptation is a graded response rather than a binary switch and that the repressive effect of glucose on proline metabolism seems to be related to the overall metabolic status of the cells rather than any allosteric action of glucose on its own (39). Alternatively, the presence of high concentrations of extracellular glucose (6 mm) might influence proline metabolism, even in the absence of a significant glycolytic flux. The two first steps of the proline metabolic pathway (proline uptake and dehydrogenation) are up-regulated upon glucose depletion (6), suggesting that their activity is controlled by glucose. However, the precise nature of this control is unknown, and further investigations would be needed to fully take apart the components of proline regulation.
Acetate Production in the PEPCK Null Environment
The production of acetate is similar in the Δpepck and wild type cell lines (154 versus 186 nmol of 13C-enriched molecules excreted/h/mg of protein, respectively). Acetate is produced from glucose, the consumption of which is ∼3.5-fold higher in the wild type cells compared with the Δpepck mutant. These data suggest that acetate production could be at its maximal capacity in the wild type parasite, because the higher glycolytic flux in this strain does not result in significant increase in acetate production. This hypothesis is consistent with the observation that glucose is converted into β-hydroxybutyrate in all cell lines lacking the PEPCK gene (Table 3). β-Hydroxybutyrate is usually produced in conditions where acetyl-CoA accumulates. Its production by Δpepck mutants may result from a redistribution of carbon fluxes toward the acetate branch, which would become saturated, as a consequence of a limiting capacity. The absence of β-hydroxybutyrate production in the wild type cells is consistent with this model, because the succinic fermentation pathways redirect part of carbon flow toward succinate production and thus may prevent acetyl-CoA accumulation. Considering this flux redistribution, one would anticipate the observed abolition of glucose consumption when the acetate branch is inactivated in the PEPCK null background (Δpepck/RNAiPDH-E2.i) (Table 2).
In conclusion, we show that the succinate fermentation, used by the procyclic trypanosomes to maintain the glycosomal redox and ATP/ADP balances, is not essential for the growth of the parasite. However, it is required to maintain a relatively high glycolytic flux. The other glycosomal NADH-consuming pathways (Gly-3-P/DHAP shuttle and glycerol production) are functional but not adapted to sustain a glycolysis-based metabolism in the procyclic cells. The flexibility provided by the redundant pathways might prove important under certain biological conditions that the parasite encounters in its complicated life cycle, such as during the migration from one tissue or an organ to another. In contrast, acetate production, the other major branch of glucose metabolism, is essential for the procyclic trypanosomes to generate ATP in the mitochondrion (11, 12) as well as to feed the cytosolic fatty acid biosynthesis through the so-called “acetate shuttle” (42). At least two different enzymatic activities account for the last step of the acetate branch (12). Identification of the acetate production enzymes will certainly prove helpful to fully understand all the roles played by this key pathway.
Supplementary Material
Acknowledgments
We particularly thank D. Speijer (Amsterdam, Netherlands) for providing us with the immune serum against the F1 subunit of the F0/F1-ATP synthase and T. Seebeck (Bern, Switzerland) for providing us with the anti-PEPCK antibody.
This work was supported by the CNRS, the Université Victor Segalen Bordeaux 2, the Fondation pour la Recherche Médicale, and the Agence Nationale de la Recherche (ANR) Program Grants METABOTRYP (ANR-MIME2007)l and SysTryp (ANR-BioSys2007).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S3.
- Gly-3-P
- glycerol-3-phosphate
- DHAP
- dihydroxyacetone phosphate
- FAD-GPDH
- FAD-dependent glycerol-3-phosphate dehydrogenase
- NADH-GPDH
- NADH-dependent glycerol-3-phosphate dehydrogenase
- PDH
- pyruvate dehydrogenase
- PEPCK
- phosphoenolpyruvate carboxykinase
- SDH
- succinate dehydrogenase
- SHAM
- salicylhydroxamic acid.
REFERENCES
- 1.Barrett M. P., Burchmore R. J., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., Krishna S. (2003) Lancet 362, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 2.Michels P. A., Bringaud F., Herman M., Hannaert V. (2006) Biochim. Biophys. Acta 1763, 1463–1477 [DOI] [PubMed] [Google Scholar]
- 3.Cross G. A., Klein R. A., Linstead D. J. (1975) Parasitology 71, 311–326 [DOI] [PubMed] [Google Scholar]
- 4.Coustou V., Besteiro S., Biran M., Diolez P., Bouchaud V., Voisin P., Michels P. A., Canioni P., Baltz T., Bringaud F. (2003) J. Biol. Chem. 278, 49625–49635 [DOI] [PubMed] [Google Scholar]
- 5.Fairlamb A. H., Opperdoes F. R. (1986) in Carbohydrate Metabolism in Cultured Cells (Morgan M. J. ed) pp. 183–224, Plenum Publishing Corp., New York [Google Scholar]
- 6.Lamour N., Rivière L., Coustou V., Coombs G. H., Barrett M. P., Bringaud F. (2005) J. Biol. Chem. 280, 11902–11910 [DOI] [PubMed] [Google Scholar]
- 7.Coustou V., Biran M., Breton M., Guegan F., Rivière L., Plazolles N., Nolan D., Barrett M. P., Franconi J. M., Bringaud F. (2008) J. Biol. Chem. 283, 16342–16354 [DOI] [PubMed] [Google Scholar]
- 8.Cazzulo J. J. (1992) FASEB J. 6, 3153–3161 [DOI] [PubMed] [Google Scholar]
- 9.Bringaud F., Rivière L., Coustou V. (2006) Mol. Biochem. Parasitol. 149, 1–9 [DOI] [PubMed] [Google Scholar]
- 10.Opperdoes F. R., Borst P., Spits H. (1977) Eur. J. Biochem. 76, 21–28 [DOI] [PubMed] [Google Scholar]
- 11.Van Hellemond J. J., Opperdoes F. R., Tielens A. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 3036–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivière L., van Weelden S. W., Glass P., Vegh P., Coustou V., Biran M., van Hellemond J. J., Bringaud F., Tielens A. G., Boshart M. (2004) J. Biol. Chem. 279, 45337–45346 [DOI] [PubMed] [Google Scholar]
- 13.Besteiro S., Biran M., Biteau N., Coustou V., Baltz T., Canioni P., Bringaud F. (2002) J. Biol. Chem. 277, 38001–38012 [DOI] [PubMed] [Google Scholar]
- 14.Coustou V., Besteiro S., Rivière L., Biran M., Biteau N., Franconi J. M., Boshart M., Baltz T., Bringaud F. (2005) J. Biol. Chem. 280, 16559–16570 [DOI] [PubMed] [Google Scholar]
- 15.Guerra D. G., Decottignies A., Bakker B. M., Michels P. A. (2006) Mol. Biochem. Parasitol. 149, 155–169 [DOI] [PubMed] [Google Scholar]
- 16.Obungu V. H., Kiaira J. K., Olembo N. K., Njogu M. R. (1999) Indian J. Biochem. Biophys. 36, 305–311 [PubMed] [Google Scholar]
- 17.van Weelden S. W., Fast B., Vogt A., van der Meer P., Saas J., van Hellemond J. J., Tielens A. G., Boshart M. (2003) J. Biol. Chem. 278, 12854–12863 [DOI] [PubMed] [Google Scholar]
- 18.van Weelden S. W., van Hellemond J. J., Opperdoes F. R., Tielens A. G. (2005) J. Biol. Chem. 280, 12451–12460 [DOI] [PubMed] [Google Scholar]
- 19.Bochud-Allemann N., Schneider A. (2002) J. Biol. Chem. 277, 32849–32854 [DOI] [PubMed] [Google Scholar]
- 20.Zíková A., Schnaufer A., Dalley R. A., Panigrahi A. K., Stuart K. D. (2009) PLoS Pathog. 5, e1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brun R., Schönenberger M. (1979) Acta Trop. 36, 289–292 [PubMed] [Google Scholar]
- 22.Bringaud F., Robinson D. R., Barradeau S., Biteau N., Baltz D., Baltz T. (2000) Mol. Biochem. Parasitol. 111, 283–297 [DOI] [PubMed] [Google Scholar]
- 23.Bringaud F., Baltz D., Baltz T. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7963–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngô H., Tschudi C., Gull K., Ullu E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 26.Wickstead B., Ersfeld K., Gull K. (2002) Mol. Biochem. Parasitol. 125, 211–216 [DOI] [PubMed] [Google Scholar]
- 27.Opperdoes F. R., Markoŝ A., Steiger R. F. (1981) Mol. Biochem. Parasitol. 4, 291–309 [DOI] [PubMed] [Google Scholar]
- 28.Harlow E., Lane D. (eds) (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29.Sambrook J., Fritsch E. F., Maniatis T. (eds) (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30.Speijer D., Breek C. K., Muijsers A. O., Hartog A. F., Berden J. A., Albracht S. P., Samyn B., Van Beeumen J., Benne R. (1997) Mol. Biochem. Parasitol. 85, 171–186 [DOI] [PubMed] [Google Scholar]
- 31.Bringaud F., Peyruchaud S., Baltz D., Giroud C., Simpson L., Baltz T. (1995) Mol. Biochem. Parasitol. 74, 119–123 [DOI] [PubMed] [Google Scholar]
- 32.Bates L. S., Waldren R. P., Teare I. D. (1973) Plant Soil 39, 205–207 [Google Scholar]
- 33.Wittmann C., Krömer J. O., Kiefer P., Binz T., Heinzle E. (2004) Anal. Biochem. 327, 135–139 [DOI] [PubMed] [Google Scholar]
- 34.Kiefer P., Nicolas C., Letisse F., Portais J. C. (2007) Anal. Biochem. 360, 182–188 [DOI] [PubMed] [Google Scholar]
- 35.Dufour S., Rousse N., Canioni P., Diolez P. (1996) Biochem. J. 314, 743–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vercesi A. E., Bernardes C. F., Hoffmann M. E., Gadelha F. R., Docampo R. (1991) J. Biol. Chem. 266, 14431–14434 [PubMed] [Google Scholar]
- 37.Turrens J. F. (1989) Biochem. J. 259, 363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peacock L., Ferris V., Bailey M., Gibson W. (2006) Parasitology 132, 651–658 [DOI] [PubMed] [Google Scholar]
- 39.Ebikeme C. E., Peacock L., Coustou V., Riviere L., Bringaud F., Gibson W. C., Barrett M. P. (2008) Parasitology 135, 585–594 [DOI] [PubMed] [Google Scholar]
- 40.Broman K., Knupfer A. L., Ropars M., Deshusses J. (1983) Mol. Biochem. Parasitol. 8, 79–87 [DOI] [PubMed] [Google Scholar]
- 41.Králová I., Rigden D. J., Opperdoes F. R., Michels P. A. (2000) Eur. J. Biochem. 267, 2323–2333 [DOI] [PubMed] [Google Scholar]
- 42.Rivière L., Moreau P., Allmann S., Hahn M., Biran M., Plazolles N., Franconi J. M., Boshart M., Bringaud F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12694–12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.