Abstract
We present a body of ultrastructural, biochemical, and genetic evidence that demonstrates the oligomerization of virulence-associated autotransporter proteins EspC or EspP produced by deadly human pathogens enterohemorrhagic and enteropathogenic Escherichia coli into novel macroscopic rope-like structures (>1 cm long). The rope-like structures showed high aggregation and insolubility, stability to anionic detergents and high temperature, and binding to Congo Red and thioflavin T dyes. These are properties also exhibited by human amyloidogenic proteins. These macroscopic ropes were not observed in cultures of nonpathogenic Escherichia coli or isogenic espP or espC deletion mutants of enterohemorrhagic or enteropathogenic Escherichia coli but were produced by an Escherichia coli K-12 strain carrying a plasmid expressing espP. Purified recombinant EspP monomers were able to self-assemble into macroscopic ropes upon incubation, suggesting that no other protein was required for assembly. The ropes bound to and showed cytopathic effects on cultured epithelial cells, served as a substratum for bacterial adherence and biofilm formation, and protected bacteria from antimicrobial compounds. We hypothesize that these ropes play a biologically significant role in the survival and pathogenic scheme of these organisms.
Keywords: Adhesion, Amyloid, Bacterial Toxins, Cell Adhesion, Serine Protease, Adherence, Autotransporters, E. coli O157:H7, EspP
Introduction
Pathogenic bacteria produce a prodigious array of virulence factors in the form of adhesins, toxins, cytoskeleton-manipulating effectors, enzymes, and mediators of motility that are usually secreted across the bacterial cell surface by a variety of simple or complex highly organized secretion mechanisms (1). In doing so, infectious bacteria ensure their ability to survive and multiply within their host. Among the different protein secretion mechanisms described in Gram-negative bacteria, the type 5 protein secretion system is perhaps the simplest of all because it does not require the repertoire of components and sophisticated structures associated with other systems (1–4). Proteins secreted via this pathway are called autotransporters (AT)2 because their export from the cytosol across the double membrane is a self-mediated process attributed to the presence of defined domains with specific functions required for secretion. The N terminus of AT proteins contains the signal peptide that leads the protein toward the Sec-dependent pathway. The C terminus contains the β-domain, which forms a β-barrel pore-like structure in the outer membrane through which the passenger domain, corresponding to the mid-portion of the protein, is secreted and can remain associated to the cell surface or be released to the milieu (2). Members of the family of SPATE (serine protease ATs of the Enterobacteriacea) are proteins from Escherichia coli (E. coli) and Shigella spp., which possess a consensus serine protease motif (2). SPATEs are generally large proteins that display an array of distinct biological activities such as adhesins, hemagglutinins, cytotoxins, or enzymes with different substrate specificities (2).
Enterohemorrhagic E. coli (EHEC) O157:H7 is a food-borne pathogen implicated as a major cause of diarrheal illness in developed countries. The production of Shiga toxins and colonization of the large intestine are central aspects of the pathogenicity of EHEC strains (5). Cattle are natural hosts of EHEC O157:H7, and they constitute the primary source of infection for humans (5). Enteropathogenic E. coli (EPEC) is a cause of potentially fatal infantile diarrhea in developing countries (5). EPEC and EHEC possess a pathogenicity island termed the locus of enterocyte effacement, which contains a repertoire of genes encoding regulators, the adhesin intimin and its receptor Tir, a type 3 secretion system (T3SS) machinery, and T3 secreted effector molecules, that together with non-locus of enterocyte effacement encoded effectors (Nles), act in concert to confer on the bacteria the ability to adhere and inflict damage to the gut mucosa (5). In addition to the outer membrane adhesin intimin (5), several other pili or non-pili adhesins have been described in EPEC and EHEC O157:H7 (6–10).
EspP and EspC are SPATE proteins (>100 kDa) secreted by EHEC and EPEC, respectively, that share significant similarity at their amino acid sequence level and are two of the predominant secreted proteins found in liquid culture supernatants of these organisms (2). The EspC protein of EPEC shows considerable homology with the IgA protease of Neisseria gonorrhoeae and Haemophilus influenza, although it does not display this activity (2, 11). Enterotoxin and cytotoxin activities on rat jejunal segments and cultured epithelial cells have been demonstrated for EspC (12, 13). Cytotoxicity is more efficient when EspC is translocated during contact of EPEC with epithelial cells (14), a process that apparently also requires the participation of the T3SS (15). Recently, it was also found that EspC conferred enhanced lysozyme resistance to EPEC (16). EspC and EspP display enzymatic activities, and although EspC cleaves spectrin (also called fodrin) and hemoglobin, EspP of EHEC cleaves pepsin A, and human coagulation factor V (2, 11, 14, 17–19). The EspP protein was also shown to influence gut colonization in calves and contributes to adherence of EHEC to bovine primary rectal cells (20). Recently, EspP was found to be directly involved in biofilm formation and adherence of EHEC to T84 intestinal epithelial cells (21). In this study, we provide compelling evidence that EspC and EspP proteins oligomerize into macroscopic rope-like structures that possess cell adhesiveness, exert enzymatic and cytopathic effects, and serve as a substratum for bacterial biofilm formation sheltering the bacteria from external foes.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
The bacterial strains and plasmids used in this study are listed in supplemental Table S1. The bacteria were cultured in Luria-Bertani broth or in DMEM (Invitrogen) at 37 °C with shaking, unless otherwise stated. When required, antibiotics were added to the medium at the following concentrations: kanamycin at 50 μg/ml, ampicillin at 200 μg/ml, gentamicin at 50 μg/ml, and chloramphenicol at 30 μg/ml.
Ultrastructural Analysis
For scanning electron microscopy (SEM) and ultra thin sectioning the ropes were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in PBS and processed for SEM and transmission electron microscopy. Ultrathin longitudinal and transversal sections of ropes were used for structural analysis and immunodetection of EspP with primary rabbit anti-EspP antibodies (produced in this study against purified EHEC EspP) or preimmune sera and anti-rabbit IgG gold-labeled conjugates as previously described (9).
Purification of Ropes, Analysis by SDS-PAGE, and Immunoblotting
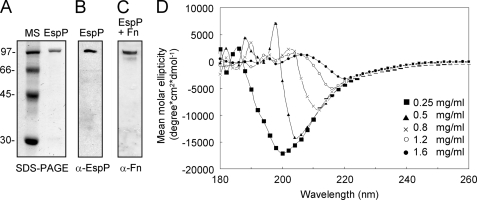
Ropes produced by E. coli in liquid (DMEM) cultures were pulled out and extensively washed by vigorous and repeated vortexing and centrifugation in distilled water to remove bacteria. The ropes were analyzed by conventional SDS-PAGE using 10% polyacrylamide gels or immunoblotting using rabbit anti-EspP serum as primary antibody (1:2,000) and anti-rabbit IgG horseradish peroxidase-conjugate (1:20,000). A 104-kDa protein identified in the EHEC ropes was excised from polyacrylamide gels and subjected to mass spectrometry analysis after digestion with trypsin. To demonstrate host protein-EspP binding, the immobilized EspP protein derived from the ropes was incubated with fibronectin (5 μg/ml) and reacted with rabbit anti-fibronectin serum (Sigma) (1:20,000) and secondary antibody. Immunoblots were developed with HyGLO chemiluminescent HRP antibody detection reagent (Denville) (9).
Biochemical Characterization of Ropes
Ropes produced by E. coli K-12 DH5α(pB9-5) were subjected to different physical and chemical treatments and then analyzed visually for stability and dissociation. The ropes were treated with proteolytic enzymes proteinase K (0.2–0.8 μg/ml) and trypsin (50 μg/ml), 1–4 m urea, 2–20% SDS, and 0.05–0.5 m Triton X-100 or heated at 40 or 100 °C for 2 h in PBS.
Determination of Cytotoxicity on HeLa Cells
We sought to compare the cytotoxic activity of ropes versus EspP monomer. Purification of EspP monomers and ropes is described in the supplemental text. One hundred μg of ropes or purified EspP were incubated with HeLa cell monolayers in high glucose DMEM containing 1% BSA for 2, 4, 6, and 24 h at 37 °C. The supernatants were removed from the wells, and cell lysis was quantified by measuring lactate dehydrogenase (LDH) release as outlined by the manufacturer (Roche Applied Science). HeLa cells in high glucose DMEM and 1% BSA were used as low control (spontaneous LDH release). Maximum LDH release (high control) was determined by adding 100 μl/well of 2% Triton X-100. For immunofluorescence microscopy, anti-EspP antibodies were added to 2% formalin-fixed samples for 1 h in 10% horse serum followed by the Alexa Fluor-conjugated secondary antibody and then visualized using an Axio Imager1.0 Zeiss microscope as previously described (8, 9). The cytopathic effect exerted by ropes or EspP monomers were compared visually counting the number of rounded HeLa cells under fluorescence microscopy.
Binding of EspP versus Ropes to HeLa Cells
Different concentrations (two serial fold dilutions starting at 100 μg/ml) of EspP or ropes were incubated for 6 h with monolayers of HeLa cells in a black sterile polystyrene 96-well microplate (Packard). Bound ropes or EspP were detected with anti-EspP antibodies and a secondary antibody labeled with Alexa Flour 488. The fluorescence was read at an excitation of 485 ± 9 nm.
Cleavage of Pepsin A by Ropes and EspP
To determine whether the ropes shared the enzymatic activity shown by EspP, 30 μg of porcine pepsin A (Sigma) were mixed with 5 μg of EspP or ropes in a total volume of 10 μl of 150 mm PBS (pH 7.6) and incubated at 37 °C for 18 h as previously described (17). The digestion samples were analyzed by 16% SDS-PAGE and Coomassie Blue staining.
Antibiotics Protection Assay
Ropes obtained from cultures of EHEC, EPEC, or DH5α(pB9-5) were rinsed gently to remove any loosely associated bacteria and then incubated for 2 h with kanamycin, ampicillin, or gentamicin in triplicate on three different days. The ropes were washed three times with PBS to remove residual antibiotics and then incubated with 1 ml of 0.1% Triton X-100 for 30 min. Serial dilutions were plated out onto agar plates, and the colony-forming units (1 × 103) were recorded. All of the strains used are sensitive to the antibiotics employed, except for DH5α(pB9-5), which is resistant to kanamycin.
RESULTS
Discovery of Novel Macroscopic Structures in Pathogenic E. coli
We found that macroscopic fibrillar structures are formed by EPEC strain E2348/69 (O127:H6) and EHEC O157:H7 strains EDL933, 86-24, and 85-170 growing with shaking overnight at 37 °C in tissue culture DMEM (supplemental Fig. S1 and Table S1). These fibers could be visualized by the naked eye and pulled out for analysis. When extended on a glass slide they measured up to 2 cm in length and were several microns wide (Fig. 1A). These dimensions are well beyond those of other bacterial filamentous superstructures such as flagella (20 nm wide, 5–10 μm long) (22), type 4 pili (20 μm long), or T3SS needles (12 nm wide and 600 nm to several μm long) (7, 9, 23, 24). We were able to reproducibly see these highly flexible and sticky fibers, albeit of different dimensions on a day-to-day basis (supplemental Fig. S1).
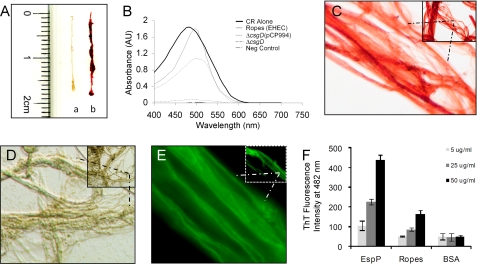
FIGURE 1.
Binding of CR dye to E. coli macroscopic ropes. A, measurement of bacterial macroscopic fibers (lane a) and staining with CR (lane b) on a glass slide. B, CR absorbance spectrum between 400 and 700 nm of EHEC ropes (thin solid line), CR alone (bold solid line), ropes without CR used as negative control (dashed line). As positive and negative controls of CR binding, we used the E2348/69ΔcsgD(pCP994) strain, which overproduces curli and binds CR (dotted line), and the curli mutant E2348/69ΔcsgD strain (dashed and dotted line), respectively. C and D, photomicrographs (taken at 100×) of CR-stained ropes or unstained ropes observed under bright field microscopy. E, binding of ThT to ropes visualized by fluorescence microscopy (micrographs taken at 100×). F, binding of 5–50 μg/ml of EspP (monomer), ropes, and BSA to 5 μm ThT and absorbance measured after excitation at 450 nm in a Synergy MX fluorometer (Bio-Tek Instruments, Inc.). Background values of ThT alone were subtracted from the sample values.
SEM analysis at low magnification of the fibers revealed a morphology resembling ropes, which appeared to be composed of hundreds of parallel long flexible filaments, which bundled up to increase the thickness of the ropes (Fig. 2). The bundle of filaments twisted together yielding rope-like structures that torqued forming loops at different sites. The helix handedness of the ropes examined was unambiguously determined by observation of the orientation of the filaments, which appeared to torque in a left-handed orientation forming coiled and supercoiled structures (Fig. 2, A and B). The particular handedness was not due to the rotation orientation (counterclockwise) of our shaking incubator because static growth of large cultures of EPEC or EHEC also produced ropes.
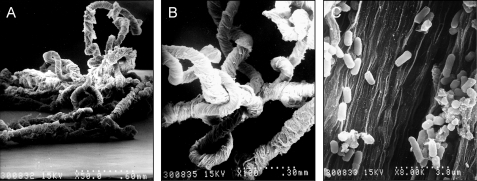
FIGURE 2.
E. coli macroscopic fibers visualized by SEM. A–C, EPEC macrofibers observed at 50×, 100×, and 8,000×, respectively.
Congo Red (CR) and thioflavin T (ThT) dye binding is a general property of highly aggregative and insoluble human amyloid fibers and of a few bacterial amyloidogenic proteins such as curli fibers (25–29). These dyes are believed to specifically interact in some unknown way with the crossed-β-sheet structure common to amyloid structures (28, 30–32). We found that, like human amyloids, the ropes bound CR dye (Fig. 1A). We compared the absorbance pattern of CR alone versus CR in the presence of ropes between 400 and 700 nm. We found a shift in maximal optical absorbance from 475 (CR alone) to 500 (CR plus ropes) nm (Fig. 1B). This pattern is in agreement with that exhibited by the E. coli curli fibers in the presence of CR (25). For these experiments, an EPEC curli mutant (E2348/69ΔcsgD) and its derivative complemented strain (E2348/69ΔcsgD(pCP994)), which overproduces curli (33), were used as negative and positive controls, respectively. Observation of unstained or CR-stained bacterial ropes under light microscopy also revealed their fibrillar nature (Fig. 1, C and D). We also found that the fluorescent ThT dye associated with ropes in a protein concentration-dependent manner (Fig. 1, E and F). After excitation at 450 nm, ThT exhibits a constant fluorescence emission (28, 34). When EspP or ropes were mixed with ThT in suspension, both mixtures exhibited an enhanced fluorescence emission at 482 nm. However, a higher intensity was obtained with the EspP monomers (Fig. 1F). In all, these data indicate that β-sheet structures are present in both the EspP monomers and the ropes.
The Ropes Are Composed of a Major Secreted Protein
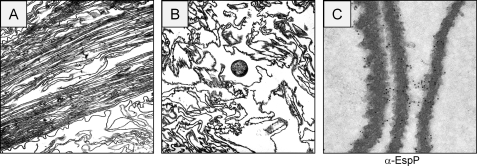
Regarding the nature of the ropes, our first thought was that they were formed by chains of bacteria in tandem. Previous work reported that Bacillus subtilis could form macrofibers composed of biologically active strands of chained bacteria (35). Also, E. coli colonizing the urinary tract can form filamentous chains of bacteria that represent a source of infectious units (36). In the present study, analysis by high magnification SEM inside the crevices of the EPEC ropes showed a highly organized filamentous substratum with scores of bacteria adhering to it (Fig. 2C). Analysis of longitudinal and transversal thin sections of bacterial ropes by transmission electron microscopy confirmed their stranded and sheeted architecture (Fig. 3, A and B) and that they are not composed of bacteria joined in tandem as in the case of the B. subtilis macrofibers or the filamentous uropathogenic E. coli. The presence of bacteria inside the ropes supports the provoking thought that the ropes could serve also as a biofilm substratum. It was necessary to discriminate the presence of other bacterial superstructures such as flagella, pili, or T3SS needles as components of the newly discovered ropes. Isogenic deletion mutants of EPEC in fliC (flagellin gene), bfpA (type 4 pilin gene), escN (T3SS ATPase gene), espB (T3SS pore-forming protein gene), or espA (T3SS translocon sheath gene), and EHEC EDL933 mutants in fliC and escN retained the ability to form ropes, indicating that rope formation was independent of pili, flagella, EspA or EspB secreted proteins, or T3S (supplemental Table S1).
FIGURE 3.
Thin sectioning of EPEC ropes. A, longitudinal sectioning. B, transversal sectioning. Note the presence of one bacterium. C, immunogold labeling of sectioned rope filament with anti-EspP antibody.
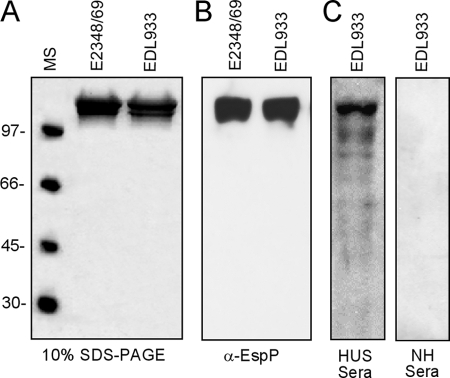
SDS-PAGE analysis of individual EPEC and EHEC ropes showed that these fibers were largely composed in each case of a doublet protein band of 104 kDa (Fig. 4A). The EHEC EspP protein was analyzed by trypsin digestion and mass spectrometry yielding peptide amino acid sequences (KTGEGLVILGAEKTF, VAGMQNTEADAVKQNGNAY, and IDLHAGKNITGDGF) that matched the predicted product of the espP gene. In agreement, the doublet band reacted with anti-EspP antibody by immunoblotting (Fig. 4B). To investigate whether HUS patients develop anti-EspP antibodies, we examined a pool of sera obtained from patients who suffered HUS. The HUS serum clearly reacted with EspP, whereas no reaction was observed with a pool of normal human sera (Fig. 4C). Immuno-EM experiments using rabbit anti-EspP antibody showed labeling along the length of thin sections of the EPEC macrofilaments (Fig. 3C). To provide genetic evidence of the role of EspP or EspC in rope formation, we constructed isogenic deletion mutants in EHEC espP or EPEC espC genes. Ropes were not observed in cultures of EPEC espC or EHEC (EDL933, 86-24, and 85-170) espP mutants. Laboratory strains E. coli K-12 DH5α or HB101, both naturally lacking espC or espP genes, did not produce ropes (supplemental Table S1). Further genetic proof of the nature of the ropes was provided by experiments demonstrating rope formation in DH5α transformed with espP harbored on plasmid pB9-5 (17) and in the EPEC espC and EHEC espP mutants complemented with pB9-5 (supplemental Table S1). We also found that Citrobacter rodentium, another attaching and effacing organism pathogenic for mice (37), produced ropes, whereas the C. rodentium espC mutant was not able to assemble ropes (supplemental Table S1). Rope formation was studied in the presence of anti-EspP antibodies in filtered supernatants containing EspP monomers obtained from overnight cultures of E. coli K-12 DH5α(pB9-5), EHEC EDL933, or EPEC E2348/69 incubated overnight with several dilutions of anti-EspP or preimmune sera. As expected, rope formation was abolished in a dose-dependent manner only in the samples containing anti-EspP antibody and not by the preimmune serum, suggesting that the antibody opsonized the soluble protein monomers blocking rope formation (supplemental Table S2).
FIGURE 4.
Biochemical characterization of ropes. A, analysis of EPEC (E2348/69) and EHEC (EDL933) ropes by SDS-PAGE. B, immunoblotting of EPEC and EHEC ropes with rabbit anti-EspP antibody. C, immunoreactivity of EHEC ropes with pool sera from HUS patients and normal human (NH) sera.
Biochemical and Physical Properties of the E. coli Ropes
CD employing different concentrations of purified EspP monomers (Fig. 5A) was used to evaluate the stability of the EspP protein at pH 8.0 and 20 °C using an Aviv 62DS spectropolarimeter. A distinctive CD spectra in the far ultraviolet region between 180 and 260 nm was obtained (Fig. 5D). Low concentrations of EspP (between 0.25 and 0.5 mg/ml) showed one peak at 200–205 nm, suggesting that the protein is present in a random coil structure. However, at higher protein concentrations (between 1.2 and 1.6 mg/ml), a mean molar ellipticity was achieved at 217 and 220 nm, respectively, suggesting conformational changes from a random coil structure to a β-sheet structure, which, as in the case of amyloidogenic proteins, could be important for the structural stability of the protein and for rope assembly. The secondary structure of the EspP passenger domain is predicted to be composed of 57% β-strands, which are likely to form β-sheet-rich intermediate fibrils that assemble into the complex quaternary rope structures. Experiments to determine the stability and resistance of the ropes to different physical and chemical treatments were performed. Similar to human amyloid fibris and curli, the ropes were highly aggregative, sticky, insoluble, and stable in aqueous solutions (including most common buffers at neutral pH) and resistant to vigorous mechanical shearing and disruption (e.g. vortexing or manual shaking). Bacterial ropes proved to be highly resistant to anionic detergents because only 75% dissociation of ropes was observed in 20% SDS, and Triton X-100 had no effect on the ropes (supplemental Table S3). Urea totally dissociated the ropes when used at 4 m, indicating the presence of noncovalent bonds between the monomeric subunits (supplemental Table S3). The ropes were sensitive to trypsin at 150 μg/ml and were highly sensitive to treatment with low concentrations of proteinase K, further proving their proteinaceous nature. The addition of 0.5 m EDTA did not destabilize the ropes; however, the presence of EDTA inhibited rope formation in EspP-containing supernatants, suggesting a role of divalent cations in oligomerization of the EspP subunits into ropes. Finally, the ropes were demonstrated to be thermo-resistant because incubation at 100 °C for 2 h did not disaggregate the structures (supplemental Table S3).
FIGURE 5.
Purification of monomeric EspP and circular dichroism. A, purified EspP monomer obtained from liquid cultures of DH5α(pB9-5) after ammonium sulfate precipitation and visualization by SDS-PAGE and Coomassie Blue staining. B, the immobilized protein was reacted with rabbit anti-EspP antibody by immunoblotting. C, EspP was incubated with a fibronectin (Fn) solution (0.5 μg/ml) and then reacted with anti-fibronectin antibody. D, circular dicroism performed in a range of 180–270 nm with different concentrations of EspP monomer.
Biological Properties Associated with EspP Ropes
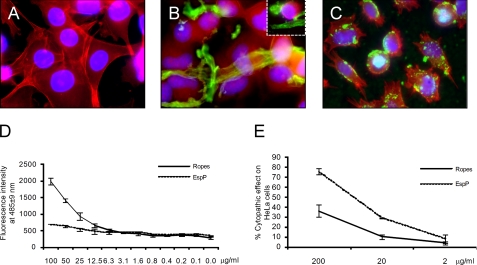
Determining whether or not the ropes retained the biological activities (cytotoxic or enzymatic) of the monomeric protein constituents was an important question to address. In agreement with a previous report suggesting that EspP is required for intestinal colonization in calves (20), we found that EspP interacts with fibronectin, indicating that EspP possesses lectin properties (Fig. 5C) and may recognize host proteins. In addition, we tested the ability of rope fragments and EspP to bind to cultured epithelial cells. After 6 h of incubation, isolated ropes and EspP monomers bound clearly in a dose-dependent manner to cultured epithelial cells, strongly suggesting that these structures possess cell adhesiveness (Fig. 6D).
FIGURE 6.
Binding of EspP-containing ropes to HeLa Cells. A, HeLa cells (red) alone. B and C, HeLa cells incubated with ropes (B) or EspP (C) (green) obtained from DH5α(pB9-5) after 24 h of incubation. In both cases note the loss of stress fibers and retraction of cell bodies indicating cytopathic activity. Photomicrographs (A–C) were taken at 60×. D, kinetics of adherence of EspP versus ropes to HeLa cells. E, cytopathic effects of EspP or ropes on HeLa cells were quantified using immunofluorescence microscopy counting the number of cells with morphological changes (e.g. cell rounding and loss of plasmic membrane) compared with the control without EspP or ropes.
EspP (also known as PssA) is considered by some authors to be a cytotoxin because it causes cytotoxic effects on Vero cells (38). We found that incubation of HeLa cell monolayers with bacteria-free ropes or EspP monomers for 24 h led to cell rounding (Fig. 6, B and C). Cytopathic effects were more evident with EspP at 200 μg/ml, whereas the ropes showed 50% fewer cytophatic effects (Fig. 6E). Quantification of the cytotoxic potential of EspP and ropes was done measuring LDH released from HeLa cells (39, 40). Neither EspP nor ropes were able to significantly increase LDH release compared with the medium alone (supplemental Fig. S2A). Thus, the data suggest that EspP and ropes exhibit a cytopathic, but not cytotoxic, effect on host cells.
To determine whether the ropes retain the enzymatic activity exhibited by EspP, pepsin A was used as target substrate as previously described (17). The ropes did not show the same level of proteolytic activity on pepsin A as that seen with EspP (supplemental Fig. 2B), suggesting that the enzymatic motifs in EspP are hidden in the rope structure.
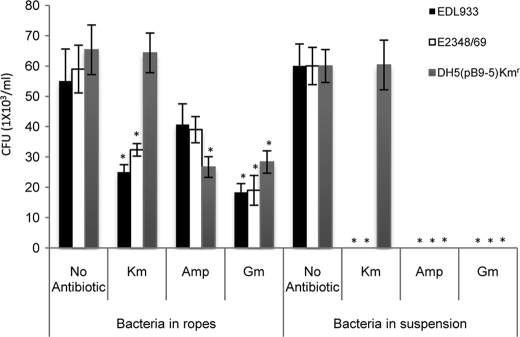
In agreement with our hypothesis that EspP ropes could serve as a biofilm substrate, we found that exogenously added EPEC, EHEC, and E. coli K-12 bacteria bound to bacteria-free ropes (supplemental Fig. 3). The presence of the bacteria within the ropes as revealed by ultrastructural studies (Fig. 2) also led us to hypothesize that the bacteria are protected within the megastructure, presumably from host or environmental factors. Thus, to test this hypothesis we performed experiments in which ropes from EPEC, EHEC, and DH5α(pB9-5) containing bacteria were incubated with or without several antibiotics. It was found that bacteria associated with the ropes were protected to a significant extent from killing by the antibacterial drugs. We were able to recover bacteria from the ropes incubated with antibiotics, albeit less than in the control samples without antibiotics (Fig. 7). Based on these data, it is tempting to speculate that the ropes might serve to protect bacteria from other external foes within the host (e.g. immune system factors) or in the environment.
FIGURE 7.
Ropes provide protection to the bacteria against antibiotics. The ropes obtained from EDL933, E2348/69, and DH5α(pB9-5) were incubated with different antibiotics for 2 h. As controls, Luria broth bacterial cultures were incubated with antibiotics. The bacteria within the ropes were protected against total killing by the indicated antibiotics. DH5α(pB9-5) (Kmr) survived the treatment with Km because of the presence of the pB9-5 plasmid. Otherwise, all of the cultures died in the presence of the antibiotics. *, p < 0.05 with respect to the wild-type strain without antibiotic.
DISCUSSION
In this study we provide the first observation that AT proteins EspP and EspC oligomerize to form coiled-coil megastructures that possess adhesive and cytopathic activities on host epithelial cells. Isogenic EPEC and C. rodentium espC and EHEC espP mutants do not produce ropes, and recombinant EspP expressed in E. coli K-12 led to rope formation. Further, purified EspP protein assembled into ropes upon incubation at 37 °C. To date, no other proteinaceous filamentous structure (e.g. pili, flagella, and T3SS needles) in pathogenic E. coli strains manifests such a macroscopic nature (7, 9, 23, 24). Striking similarities, in terms of structure, biochemical and biophysical properties, exist between the E. coli ropes and human amyloidogenic proteins implicated in human diseases such as Alzheimer, Parkinson, cystic fibrosis, and prion diseases. It has been reported that curli fibers of uropathogenic E. coli are involved in the polymerization of amyloid-like fibers, which were suggested to participate in neurodegenerative and amyloid-related diseases (25). The E. coli ropes showed high insolubility in common neutral buffer, high aggregation in aqueous solutions, unusual stability, and binding to ThT and CR dyes. These properties are reminiscent of human amyloid fibers. The E. coli ropes showed high resistance to mechanical forces and boiling and to solubilization and disruption at high concentrations of anionic detergents, urea, and mercaptoethanol. CD experiments supported the notion that EspP monomers aggregate in a concentration-dependent manner and that formation of ropes involves the transition of a random coil structure into a more stable, highly organized β-sheet structure. Furthermore, we found that cationic ions might be important for rope formation and stability because of the presence of EDTA inhibited rope formation, although the ropes did not dissociate in the presence of EDTA.
The domains within the large AT proteins that allow self-export across the bacterial envelope have been identified (2). However, the specific amino acid sequences in EspC or EspP that might allow monomer-monomer interaction leading to rope formation are unknown. Analysis of the primary sequence of EspP suggests that the region between amino acids 1000 and 1050 could potentially be responsible for oligomerization of the monomers, but supporting experimental data are missing.
ATs are the most abundant secreted proteins found in the supernatants of Gram-negative bacterial pathogens (2). It is presumably unlikely that pathogenic bacteria would devote considerable amounts of energy to produce and secrete ATs into the milieu to form these megastructures without a biological purpose. Our data indicate that the ropes display several biological activities that could be important for the survival of the organism within or out of the host. For example, the ropes were shown to bind to host epithelial cells in culture and to host proteins such as fibronectin, to serve as a substratum for biofilm formation, to protect the bacteria within the ropes from antibiotics, and lastly to exhibit cytopathic effects on cell monolayers. We did not find any supportive evidence of cytotoxicity by the ropes or EspP using the LDH determination. Published data indicate that EspP binds to T84 cells and that it promotes gut colonization in calves (20, 21). Rope formation does not appear to be accidental, because several enteric pathogens, in addition to EPEC, EHEC, and C. rodentium, produce macrofilaments. Macromolecular structures consisting of extended sheets associated with enhanced T3S secretion were seen in Shigella flexneri (41); however, their role in in vivo pathogenesis is unclear. Yersiniae enterocolitica assembles seven Yop secreted proteins aggregated into large filaments, with unknown function (42).
People naturally or experimentally infected with EPEC or EHEC develop antibodies against EspC or EspP, respectively (38, 43). We found that a pool of sera obtained from HUS patients reacted with the rope-forming EspP protein, whereas normal human sera did not show any reactivity. These data suggest that EspP-containing ropes could be produced during infections. However, after an effort to explore a role for the ropes in vivo, we could not find differences in terms of colonization between C. rodentium versus an isogenic espC mutant in a murine model of infection. Determining whether the ropes are produced in vivo (e.g. in the lumen of the gut) represents a technically challenging task because of the lack of a relevant animal model for EHEC infections and because of the lower density of bacteria expected to be present on colonic tissue, in contrast to growth in liquid cultures. Although host protein degradation and cytopathic/cytotoxic activities have been attributed to EspC and EspP, they only constitute one of the players of the multi-factorial pathogenic scheme of these pathogens. Based on the cell damage caused by EspP ropes on epithelial cells shown here, we cannot rule out the possibility that the ropes might contribute to the effacement and destruction of the intestinal linen seen upon EPEC and EHEC infections (5). That attaching and effacing pathogens, but not commensal E. coli, are able to assemble ropes is a remarkable phenomenon that suggests a virulence trait that must play a significant yet undefined role in the biology and disease-causing scheme of these pathogenic bacteria. The novel ropes may represent a sheltering mechanism for bacteria to avoid clearance in biological systems (e.g. by escaping the innate and adaptive immunity mechanisms) or a substratum for adherence, persistence, and survival in the animal and human hosts or the environment.
Supplementary Material
Acknowledgments
We thank María Alexandra Ledesma and Alejandra Vázquez for technical assistance; Maribel Vargas for processing and analysis of samples by electron microscopy, Ben Fane for analysis of EspP/EspC amino acid sequence; Helge Karch for kindly providing plasmid pB9-5; James B. Kaper for support; and Rob and Mavis McKenna, Herman Gordon, S. Padmanabhan, and Neil Mendelson for valuable insights.
This work was supported, in whole or in part, by National Institutes of Health Grants AI66012 and AI63211 (to J. A. G.). This work was also supported by DGAPA Grant IN201703-3 and CONACyT Grant 42918Q (to J. L. P.) and grants from Canadian Institutes of Health Research and Howard Hughes Medical Institute to BBF.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables S1–S3, and Figs. S1–S3.
- AT
- autotransporter
- EHEC
- enterohemorrhagic E. coli
- EPEC
- enteropathogenic E. coli
- T3SS
- type 3 secretion system
- SEM
- scanning electron microscopy
- LDH
- lactate dehydrogenase
- CR
- Congo Red
- ThT
- thioflavin T
- HUS
- hemolytic uremic syndrome.
REFERENCES
- 1.Coburn B., Sekirov I., Finlay B. B. (2007) Clin. Microbiol. Rev. 20, 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson I. R., Navarro-Garcia F., Desvaux M., Fernandez R. C., Ala'Aldeen D. (2004) Microbiol. Mol. Biol. Rev. 68, 692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E. (2005) Annu Rev. Microbiol. 59, 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaper J. B., Nataro J. P., Mobley H. L. (2004) Nat. Rev. Microbiol. 2, 123–140 [DOI] [PubMed] [Google Scholar]
- 6.Erdem A. L., Avelino F., Xicohtencatl-Cortes J., Girón J. A. (2007) J. Bacteriol. 189, 7426–7435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girón J. A., Ho A. S., Schoolnik G. K. (1991) Science 254, 710–713 [DOI] [PubMed] [Google Scholar]
- 8.Rendón M. A., Saldaña Z., Erdem A. L., Monteiro-Neto V., Vázquez A., Kaper J. B., Puente J. L., Girón J. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xicohtencatl-Cortes J., Monteiro-Neto V., Ledesma M. A., Jordan D. M., Francetic O., Kaper J. B., Puente J. L., Girón J. A. (2007) J. Clin. Invest. 117, 3519–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Low A. S., Dziva F., Torres A. G., Martinez J. L., Rosser T., Naylor S., Spears K., Holden N., Mahajan A., Findlay J., Sales J., Smith D. G., Low J. C., Stevens M. P., Gally D. L. (2006) Infect. Immun. 74, 2233–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein M., Kenny B., Stein M. A., Finlay B. B. (1996) J. Bacteriol. 178, 6546–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mellies J. L., Navarro-Garcia F., Okeke I., Frederickson J., Nataro J. P., Kaper J. B. (2001) Infect. Immun. 69, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navarro-García F., Canizalez-Roman A., Sui B. Q., Nataro J. P., Azamar Y. (2004) Infect. Immun. 72, 3609–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidal J. E., Navarro-García F. (2006) Infect. Immun. 74, 2293–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidal J. E., Navarro-García F. (2008) Cell Microbiol. 10, 1975–1986 [DOI] [PubMed] [Google Scholar]
- 16.Salinger N., Kokona B., Fairman R., Okeke I. N. (2009) Appl. Environ. Microbiol. 75, 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunder W., Schmidt H., Karch H. (1997) Mol. Microbiol. 24, 767–778 [DOI] [PubMed] [Google Scholar]
- 18.Drago-Serrano M. E., Parra S. G., Manjarrez-Hernández H. A. (2006) FEMS Microbiol. Lett. 265, 35–40 [DOI] [PubMed] [Google Scholar]
- 19.Dutta P. R., Cappello R., Navarro-García F., Nataro J. P. (2002) Infect. Immun. 70, 7105–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dziva F., Mahajan A., Cameron P., Currie C., McKendrick I. J., Wallis T. S., Smith D. G., Stevens M. P. (2007) FEMS Microbiol. Lett. 271, 258–264 [DOI] [PubMed] [Google Scholar]
- 21.Puttamreddy S., Cornick N. A., Minion F. C. (2010) Infect. Immun. 78, 2377–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giron J. A. (2005) in Colonization of Mucosal Surfaces (Nataro J. P. ed) pp. 213–236, ASM Press, Herndon, VA [Google Scholar]
- 23.Cornelis G. R. (2006) Nat. Rev. Microbiol. 4, 811–825 [DOI] [PubMed] [Google Scholar]
- 24.Ogino T., Ohno R., Sekiya K., Kuwae A., Matsuzawa T., Nonaka T., Fukuda H., Imajoh-Ohmi S., Abe A. (2006) J. Bacteriol. 188, 2801–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman M. R., Robinson L. S., Pinkner J. S., Roth R., Heuser J., Hammar M., Normark S., Hultgren S. J. (2002) Science 295, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fowler D. M., Koulov A. V., Balch W. E., Kelly J. W. (2007) Trends Biochem. Sci. 32, 217–224 [DOI] [PubMed] [Google Scholar]
- 27.Frid P., Anisimov S. V., Popovic N. (2007) Brain Res. Rev. 53, 135–160 [DOI] [PubMed] [Google Scholar]
- 28.Naiki H., Higuchi K., Hosokawa M., Takeda T. (1989) Anal. Biochem. 177, 244–249 [DOI] [PubMed] [Google Scholar]
- 29.LeVine H., 3rd. (1999) Methods Enzymol. 309, 274–284 [DOI] [PubMed] [Google Scholar]
- 30.Naiki H., Higuchi K., Matsushima K., Shimada A., Chen W. H., Hosokawa M., Takeda T. (1990) Lab. Invest. 62, 768–773 [PubMed] [Google Scholar]
- 31.Naiki H., Higuchi K., Nakakuki K., Takeda T. (1991) Lab. Invest. 65, 104–110 [PubMed] [Google Scholar]
- 32.Krebs M. R., Bromley E. H., Donald A. M. (2005) J. Struct. Biol. 149, 30–37 [DOI] [PubMed] [Google Scholar]
- 33.Saldaña Z., Xicohtencatl-Cortes J., Avelino F., Phillips A. D., Kaper J. B., Puente J. L., Girón J. A. (2009) Environ. Microbiol. 11, 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeVine H., 3rd. (1993) Protein. Sci. 2, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendelson N. H. (1982) J. Bacteriol. 151, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Justice S. S., Hunstad D. A., Seed P. C., Hultgren S. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19884–19889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng W., Puente J. L., Gruenheid S., Li Y., Vallance B. A., Vázquez A., Barba J., Ibarra J. A., O'Donnell P., Metalnikov P., Ashman K., Lee S., Goode D., Pawson T., Finlay B. B. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djafari S., Ebel F., Deibel C., Krämer S., Hudel M., Chakraborty T. (1997) Mol. Microbiol. 25, 771–784 [DOI] [PubMed] [Google Scholar]
- 39.Guyer D. M., Radulovic S., Jones F. E., Mobley H. L. (2002) Infect. Immun. 70, 4539–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts P. H., Davis K. C., Garstka W. R., Bhunia A. K. (2001) J. Microbiol. Methods 43, 171–181 [DOI] [PubMed] [Google Scholar]
- 41.Parsot C., Ménard R., Gounon P., Sansonetti P. J. (1995) Mol. Microbiol. 16, 291–300 [DOI] [PubMed] [Google Scholar]
- 42.Michiels T., Wattiau P., Brasseur R., Ruysschaert J. M., Cornelis G. (1990) Infect. Immun. 58, 2840–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarvis K. G., Kaper J. B. (1996) Infect. Immun. 64, 4826–4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.