Abstract
The human cytomegalovirus (HCMV) protein US2 hijacks the endoplasmic reticulum (ER)-associated degradation machinery to dispose of MHC class I heavy chain (HC) at the ER. This process requires retrotranslocation of newly synthesized HC molecules from the ER membrane into the cytosol, but the mechanism underlying the dislocation reaction has been elusive. Here we establish an in vitro permeabilized cell assay that recapitulates the retrotranslocation of MHC HC in US2-expressing cells. Using this assay, we demonstrate that the dislocation process requires ATP and ubiquitin, as expected. The retrotranslocation also involves the p97 ATPase. However, the mechanism by which p97 dislocates MHC class I HC in US2 cells is distinct from that in US11 cells: the dislocation reaction in US2 cells is independent of the p97 cofactor Ufd1-Npl4. Our results suggest that different retrotranslocation mechanisms can employ distinct p97 ATPase complexes to dislocate substrates.
Keywords: ATPases, Endoplasmic Reticulum(ER), Membrane Proteins, Protein Degradation, Protein Translocation, ERAD/Retrotranslocation, US2, Ufd1-Npl4, p97/cdc48, Ubiquitin
Introduction
The endoplasmic reticulum (ER)2 is a major site of protein synthesis for secretory and membrane proteins. To ensure that only properly folded polypeptides and correctly assembled protein complexes enter the secretory pathway to reach their final destinations, eukaryotic cells have evolved a conserved quality control mechanism. This quality control program selectively retains polypeptides that fail to reach native conformation and export them from the ER into the cytosol for degradation by the 26 S proteasome. This process was termed ER-associated protein degradation (ERAD) or retrotranslocation (1–3). Efficient degradation of misfolded ER proteins requires concerted actions by the p97 ATPase and the ubiquitin proteasome system (4). Polypeptides emerging into the cytosol via one or more protein conducting channels are polyubiquitinated by an ER membrane-associated ubiquitin ligase (5, 6). The p97 ATPase complex, which comprises of the AAA (ATPase associated with various cellular activities) ATPase p97 and the dimeric adaptor complex Ufd1-Npl4, then use a dual recognition mode to recognize both polypeptide chain and the attached polyubiquitin conjugates, leading to the extraction of the polypeptide from the ER membrane (7, 8). p97 subsequently delivers the substrate to the proteasome for degradation with the assistance from a collection of shuttling factors that are capable of interacting with both p97 and the proteasome (9–11).
The retrotranslocation pathway can be hijacked by viruses to facilitate viral evasion of host immune defense. The murine γ-herpesvirus 68 encodes a type III membrane protein named mK3, which down-regulates newly synthesized major histocompatibility complex (MHC) class I heavy chain (HC) by co-opting the ERAD mechanism (12). Two HCMV-encoded proteins, US2 and US11, can each target MHC class I HC to the ERAD pathway for degradation. In US11- or US2-expressing cells, MHC class I HC, a type I transmembrane protein carrying a single glycan, is rapidly moved back into the cytosol. Once emerging into the cytosol, HC molecules are modified with polyubiquitin chains, whereas the N-glycan is cleaved off. The polyubiquitinated polypeptide chain is subsequently degraded by the 26 S proteasome (13, 14). The elimination of the antigen presenting MHC HC molecules from the ER membrane apparently allows viruses to escape detection by the host immune system.
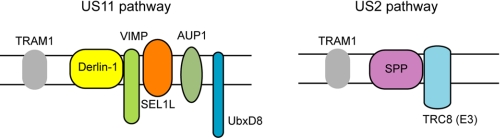
Both US11 and US2 directly bind MHC class I HC to initiate the retrotranslocation process, and both pathways involve ubiquitin, the 26 S proteasome, and a translocating chain-associated membrane protein-1 (15–19). Nonetheless, these two processes each posses its own unique features, indicating that these two viral proteins may use different retrotranslocation routes to dislocate MHC class I HC from the ER membrane. For example, on the ER lumen side, although the Hsp70 family protein BiP has been implicated in both pathways (20), the ER chaperone protein-disulfide isomerase is required only for US2-mediated retrotranslocation (21). The difference between the two pathways is most drastic when the membrane components involved are compared (Fig. 1). In the ER membrane, US11 and US2 proteins engage almost completely different sets of membrane proteins to accomplish the dislocation reaction. US2 targets MHC HC to a protein complex comprised of the signal peptide peptidase and an ER-membrane bound ubiquitin ligase termed TRC8 for subsequent export into the cytoplasm (22, 23). US11 does not interact with these proteins. Instead, it targets HC to a large protein complex containing Derlin-1, VIMP, UbxD8, SEL1L, and AUP1 for retrotranslocation (24–27) (Fig. 1). On the cytosolic side, previous studies have demonstrated the involvement of the p97-Ufd1-Npl4 complex in US11-mediatd retrotranslocation (7, 28). However, few studies have examined the cytosolic requirement for US2-induced retrotranslocation. In particular, it is unclear whether or not the p97 ATPase participates in this process, and if so, how it can cooperate with such distinct membrane retrotranslocation complexes.
FIGURE 1.
US11 and US2 use distinct membrane complexes to dislocate MHC class I heavy chain. The schemes show the known membrane components required for the degradation of MHC class I HC in US11 and US2 cells, respectively. TRAM1, translocating chain associated membrane protein-1; VIMP, VCP interacting protein.
In this report, we established an in vitro permeabilized cell assay that recapitulates important features of the dislocation reaction in intact US2-expressing cells (referred thereafter as US2 cells). Using this system, we demonstrate that the retrotranslocation of HC induced by US2 expression requires ubiquitin and the p97 ATPase. Surprisingly, the canonical adaptor complex Ufd1-Npl4 implicated in retrotranslocation of most ERAD substrates studied to date is dispensable for US2-induced retrotranslocation. We propose that adaptor switch may allow the p97 ATPase to cooperate with distinct retrotranslocation machineries in the ER membrane to serve different substrates.
MATERIALS AND METHODS
Constructs, Antibodies, Protein, and Chemicals
The pLNCX2-US2 plasmid was constructed in two steps. First, a DNA fragment comprised of the coding sequence for the signaling sequence (SS) of the prolactin gene and the FLAG tag (MDSKGSSQKGSRLLLLLVVSNLLLCQGVVSTPVDYKDDDDK) was amplified by PCR and inserted into the BglII and NotI sites of the pLNCX2 vector (Clontech, Mountain View, CA) to make pLNCX2-SS-FLAG. The US2 coding sequence was then amplified by PCR and cloned in the NotI and SalI sites of the pLNCX2-SS-FLAG. The sequences of all the constructs were confirmed by sequencing. The ON-TARGETplus SMARTpool siRNA targeting VCP/p97 (L-008727-00-0050) and the control siRNA (D-001810-10-20) were purchased from Thermo Scientific (Waltham, MA). The anti-Ufd1 siRNA were purchased from Ambion (Austin, TX). The targeting sequence is 5′-CCAACUCAGCCGACUUAAC. Bovine ubiquitin was purchased from Sigma. MG132 was purchased from Calbiochem. DeoxyBigCHAP was purchase from Dojindo (Rockville, MD). MHC HC and p97 antibodies were described previously (24). The construct expressing GST-tagged p97 and the anti-Ufd1 antibody were generously provided by Dr. Hemmo Meyer (University of Duisburg-Essen, Germany).
Cell Lines, Transfection, and Immunoblotting
293T was purchased from ATCC and maintained according to the standard protocol. Transfection was done with the Lipofectamine2000 reagent (Invitrogen) following the protocol provided by the manufacturer. Immunoblotting was performed according to standard protocol. Fluorescence-labeled secondary antibodies were used for detection. The fluorescent blots were imaged using a LI-COR Odyssey infrared imager. Protein bands were quantified using the Odyssey 2.1. Astrocytoma or 293T cells stably expressing FLAG-US2 were generated using the pLNCX2-based retroviral system as described previously (29). 293T cell stably expressing YFP tagged T-cell receptor α chain and astrocytoma cells stably expressing US11 were described previously (28, 29).
Preparation of Cow Liver Cytosol
Fresh bovine liver tissue was cut into small pieces to remove blood vessels and connective tissue. The resulting tissue (∼300 g) was thoroughly rinsed in ice-cold homogenization buffer (50 mm HEPES, pH 7.5, 80 mm KCl, 15 mm NaCl, 3 mm MgCl2, 250 mm sucrose, 1 mm dithiothreitol (DTT), 0.5 mm phenylmethylsulfonyl fluoride (PMSF)). Homogenization buffer (∼300 ml) containing extra protease inhibitors was added. The tissue was homogenized in a Polytron blender followed by further homogenization using a Potter homogenizer rotating at ∼1000 rpm. The homogenate was centrifuged at 9000 × g in a Beckman JA-10 rotor for 15 min. The supernatant was filtered through eight layers of cheesecloth, re-centrifuged, and filtered through cheesecloth a second time. The supernatant was then centrifuged in a Beckman Ti45 rotor at 45,000 × g for 3 h. The cytosol supernatant was saved carefully. The protein concentration of the cytosol was 20–30 mg/ml, as measured with the use of the Micro BCA Protein Assay (Pierce).
Protein Purification and Biochemical Depletion Experiments
GST-Ube2B C88S and GST-p97 proteins were purified from Escherichia coli as previously described (30). Purified proteins were further fractionated by size exclusion chromatography on Superdex 200 and Superose 6 columns, respectively, in 50 mm Tris-HCl, pH 8.0, 150 mm potassium chloride, 5% glycerol, and 2 mm magnesium chloride. Cow liver cytosol was purified as described previously (16). To deplete Ufd1-Npl4 from cytosol, GST-p97 protein was immobilized on glutathione beads. The beads were washed once with a buffer containing 50 mm Tris-HCl, pH 7.5, and 150 mm sodium chloride. 40 mg of cow liver cytosol was subjected to two rounds of depletion, each with glutathione beads containing 125 μg of GST-p97 protein. The beads were removed by centrifugation. To deplete ubiquitin from cow liver cytosol, cow liver cytosol (40 mg) was incubated with 3 mg of GST-Ubc2m in the presence of GST-Uba1 (1 μg) and an ATP regenerating system (16). To monitor the depletion efficiency, fluorescein-labeled ubiquitin (0.25 μg) was included and the reaction was incubated at 37 °C for 15 min. Ubiquitin aldehyde was added to a final concentration of 1 μm. GST-Ubc2m·ubiquitin complex was then removed by glutathione beads. The depletion efficiency was monitored by comparing the fluorescence intensity prior to and after the depletion reaction.
Permeabilized Cell Assay
Pulse-chase and permeabilization experiments were adapted from a previously described protocol (28, 31). US2-expressing U373-MG astrocytoma cells (32) were starved in a Met/Cys-free DMEM for 40 min and incubated in a medium containing [35S]Met/Cys for 10 min. Where indicated, the proteasome inhibitor MG132 (20 μm) was present in the starvation and pulse labeling medium. To permeabilize the plasma membrane, cells were washed with PBS supplemented with 0.9 mm CaCl2 and then incubated at 3 × 107 cells/ml in PB buffer (25 mm HEPES, pH 7.2, 115 mm potassium acetate, 5 mm sodium acetate, 2.5 mm MgCl2, 0.5 mm EGTA) containing 0.028% digitonin (Calbiochem, Gibbstown, NH) and a protease inhibitor mixture comprised of leupeptin (10 μg/ml), chymestatin (5 μg/ml), elastatinal (3 μg/ml), and pepstatin (1 μg/ml) at 4 °C for 5 min. For some experiments, an ATP regenerating system (ARS) containing 1 mm ATP, 7.8 mg/ml of phosphocretine (Sigma), and 0.5 mg/ml of creatine kinase (Roche Applied Science) was added to permeabilized US2 cells together with bovine ubiquitin (10 μm) and permeabilized cells were then incubated at 37 °C to initiate retrotranslocation. Alternatively, permeabilized US2 cells were first subjected to centrifugation at 20,000 × g for 5 min to remove the supernatant cytosol fraction. The membrane pellet was washed twice with PB buffer containing 270 mm NaCl. The salt-treated membrane pellet was resuspended in cow liver cytosol containing 10 μm bovine ubiquitin and ARS. The reaction mixture was then incubated at 37 °C. Samples taken at different time points were either analyzed directly by immunoprecipitation in a buffer containing 0.5% Nonidet P-40 (11), or first fractionated into a supernatant and a pellet fraction. These fractions were extracted by the Nonidet P-40 lysis buffer and then subjected to immunoprecipitation with an anti-HC antibody.
Co-immunoprecipitation Experiment
US2-expressing U373-MG astrocytoma cells were starved in a Met/Cys-free medium containing 20 μm MG132 for 1 h and incubated in medium containing [35S]Met/Cys for 30 min in the presence of MG132. Cells were washed with ice-cold PBS and lysed using DeoxyBigCHAP (1% DeoxyBigCHAP, 30 mm Tris-HCl, pH 7.4, 150 mm potassium acetate, 4 mm magnesium acetate and protease inhibitors) or Nonidet P-40 lysis buffer (11). Cells were subjected to centrifugation at 20,000 × g for 5 min and cell extract was pre-cleared using Staph A washed with the corresponding lysis buffer. Cell extract was then divided into 3 parts; 10% subjected to immunoprecipitation with an anti-HC antibody, 45% subjected to immunoprecipitation with a control antibody, and 45% subjected to immunoprecipitation with an anti-p97 antibody. 75% of the precipitated material from control and anti-p97 immunoprecipitations were resuspended in 2% SDS, 5 mm DTT and heated at 90 °C for 5 min. The eluted material was diluted with the Nonidet P-40 lysis buffer, and subjected to a second immunoprecipitation with an anti-HC antibody.
RESULTS
An in Vitro Assay for US2-mediated Retrotranslocation
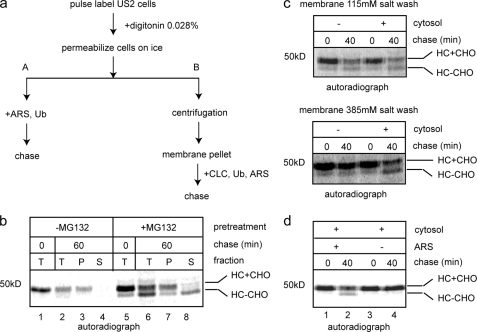
To study the mechanism of US2-mediated retrotranslocation, we set up a permeabilized cell system. A similar assay was previously established to dissect the mechanism of US11-dependent retrotranslocation of MHC class I HC (31). To this end, astrocytoma cells stably expressing US2 protein were labeled with [35S]methionine and permeabilized with a buffer containing a low concentration of the detergent digitonin. Digitonin-permeabilized cells were incubated with an ATP regenerating system to initiate retrotranslocation (Fig. 2a, scheme A). Samples taken at different time points were either analyzed directly by solubilization in a Nonidet P-40-containing extraction buffer followed by immunoprecipitation with an anti-HC antibody or first fractionated into pellet (containing the ER membrane) and supernatant (containing the cytosol) fractions before immunoprecipitation analysis. Incubation of permeabilized US2 cells with an ARS decreased the level of MHC class I HC due to retrotranslocation and proteasomal degradation (Fig. 2b, lane 1 versus 2). When US2 cells were treated with a proteasome inhibitor MG132 for 1 h prior to pulse labeling, the degradation of HC was inhibited, leading to the accumulation of a fraction of retrotranslocated MHC class I HC during the labeling process, which had the N-glycan cleaved off by a cytosolic N-glycanase (Fig. 2b, lane 5) (33). The majority of retrotranslocated HC also had the polyubiquitin chains removed, which was likely due to deubiquitination by cellular deubiquitinating enzymes, as reported for US11-mediated retrotranslocation and other ERAD substrates (11, 31, 34, 35). After incubation, we observed that the amount of dislocated HC product was increased with a concurrent reduction in glycosylated retrotranslocation precursor (Fig. 2b, lane 5 versus 6). Fractionation experiments showed that the majority of deglycosylated HC was indeed present in the cytosol fraction, whereas the glycosylated form was mostly associated with the membrane (Fig. 2b, lane 8 versus 7), consistent with previous studies using intact US2 cells. Thus, our in vitro assay recapitulates a key feature of the in vivo reaction that is the release of HC into the cytosol in a deglycosylated form upon inhibition of the proteasome.
FIGURE 2.
A permeabilized cell assay for US2-mediated retrotranslocation. a, experimental schemes for the permeabilized cell assay. After cells are permeabilized, they were either incubated with an ARS and ubiquitin (Ub) directly to initiate retrotranslocation (scheme A) or subjected to centrifugation to remove cytosol (scheme B). In the latter case, the membrane pellet fraction was washed with salt before the addition of CLC, Ub, and ARS. b, US2 cells were either untreated or treated with the proteasome inhibitor MG132 (20 μm, 1 h), radiolabeled, and permeabilized. Cells were then incubated in the presence of ARS for either 0 or 60 min at 37 °C. A portion of the sample (T) was directly analyzed, whereas the other was fractionated into supernatant (S) and membrane pellet (P). The extracts were subjected to immunoprecipitation with αHC. HC+CHO, glycosylated HC; HC-CHO, deglycosylated HC. c, MG132-treated US2 cells were radiolabeled and permeabilized. The membrane pellet was washed with a buffer containing the indicated concentrations of salt before being incubated with CLC or a buffer at 37 °C. Samples taken at the indicated time points were directly analyzed by immunoprecipitation with αHC. d, the membrane fraction from radiolabeled, MG132-treated US2 cells were salt treated and incubated with CLC either in the absence or presence of ARS before being subjected to immunoprecipitation with αHC. The image is a representative gel from three independent experiments.
We next tested whether the dislocation of HC in permeabilized US2 cells requires any cytosolic proteins. We introduced a centrifugation step following the permeabilization of cells, which removed cytosolic factors (31). The resulting membrane pellets were washed with a buffer containing 115 mm salt before being incubated with either buffer or cow liver cytosol (CLC) (Fig. 2a, scheme B). Interestingly, retrotranslocation of HC occurred even in the absence of cytosol. The addition of cytosol did not improve the dislocation efficiency (Fig. 2c, top panel). However, when membranes from permeabilized US2 cells were treated with a buffer containing 385 mm salt, the dislocation reaction became dependent on the addition of cytosol (Fig. 2c, bottom panel). These results suggest that US2-dependent dislocation likely involves a factor that is present both in the cytosol and on the ER membrane. This is in contrast to the dislocation reaction in US11 cells, which requires few cytosolic proteins (36). Most of the factors involved in US11-induced retrotranslocation are exclusively associated with the ER membrane either as anchored or peripheral proteins. These observations further consolidate the notion that the two viral proteins induce MHC class I HC degradation via distinct mechanisms. As expected, the dislocation of MHC class I HC in permeabilized US2 cells also required ATP as its omission completely blocked the appearance of the deglycosylated HC (Fig. 2d, lane 2 versus 4).
Ubiquitin Is Required for US2-dependent Retrotranslocation
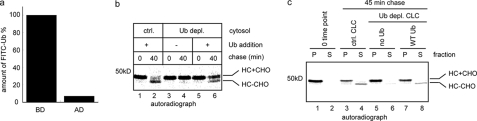
We tested whether retrotranslocation of MHC class I HC in permeabilized US2 cells requires ubiquitin. We used a previously established method to deplete ubiquitin from cytosol (16). This method used a GST-tagged mutant ubiquitin-conjugating enzyme Ube2B C88S, which has the catalytic cysteine converted to serine. As a result, it forms an ester bond with free ubiquitin molecules and the E2-ubiquitin complex is much more stable than wild type E2-Ub thioester complex. The GST-tagged E2-Ub complex could then be removed using glutathione beads (see “Materials and Methods”). This approach allowed us to reproducibly deplete >90% of ubiquitin from the cytosol, as indicated by efficient removal of a fluorescence labeled ubiquitin tracer (Fig. 3a). Consistent with previous results, cytosol depleted of ubiquitin did not support US11-dependent dislocation (Fig. 3b, lane 2 versus 4) and addition of purified bovine ubiquitin to ubiquitin-depleted CLC restored the dislocation activity, as a fraction of glycosylated HC was converted to dislocated deglycosylated species (Fig. 3b, lane 6). When the membranes from permeabilized US2 cells were incubated with either mock-depleted or ubiquitin-depleted CLC, dislocation of MHC class I HC was observed in the presence of control CLC, but not with ubiquitin-depleted CLC (Fig. 3c, lanes 5 and 6 versus lanes 3 and 4). Like US11 cells, re-introducing ubiquitin to ubiquitin-depleted cytosol restored the dislocation activity, causing the accumulation of deglycosylated HC in the cytosol fraction (Fig. 3c, lane 8). Together, these observations suggest that dislocation of MHC class I HC in permeabilized US2 cells also involves ubiquitination in agreement with previous studies in intact cells, which showed that polyubiquitinated MHC class I HC is present in US2 cells treated with a proteasome inhibitor (17), and that removal of potential ubiquitin conjugation sites in MHC class I HC abolished its dislocation (18).
FIGURE 3.
Ubiquitin is required for US2-mediated retrotranslocation. a, a small amount of FITC-labeled ubiquitin was added to CLC to monitor the depletion efficiency. Fluorescence intensity in CLC was measured before (BD) and after (AD) depletion. b, MG132-treated US11-expressing cells were radiolabeled, permeabilized. The membrane pellet fraction was salt treated and incubated with either control (ctrl.) cytosol or ubiquitin-depleted CLC. Where indicated, bovine ubiquitin was added back to ubiquitin-depleted CLC. Samples taken at the indicated time points were analyzed directly by immunoprecipitation with αHC. c, as in b, except that US2 cells were used and samples taken at the indicated time points were fractionated into membrane pellet (P) and supernatant (S) fractions before immunoprecipitation analyses. The image shows a representative gel from two independent experiments.
The Differential Effect of p97 Inhibitors on US11- and US2-induced Dislocation
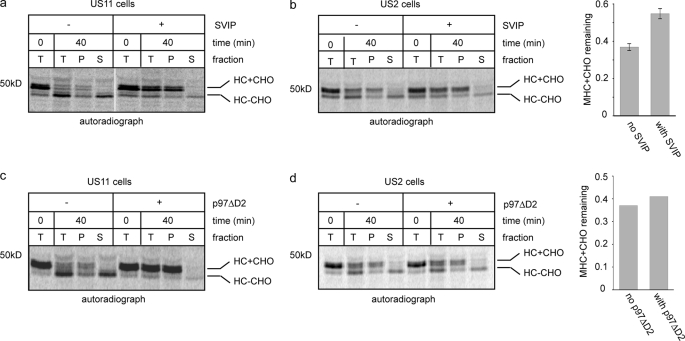
Previous studies show that polyubiquitin acts as a recognition signal to engage a p97 ATPase complex (8), which utilizes the energy generated by ATP hydrolysis to dislocate polypeptides from the ER membrane (28). We therefore tested whether US2-induced retrotranslocation involves p97. Because a significant fraction of p97 is tightly associated with the ER membrane, which is resistant to high salt treatment (28), we used a protein termed SVIP to inhibit the p97 function. SVIP is a small polypeptide containing a p97 interaction motif. Overexpression of SVIP inhibits p97-dependent protein degradation at the ER membrane (37). Addition of SVIP protein to permeabilized US11 cells inhibits US11-dependent retrotranslocation (Fig. 4a). In a parallel experiment, we also treated permeabilized US2 cells with the same dose of SVIP. However, we found that SVIP only had a marginal effect on US2-mediated retrotranslocation, leading to a small but reproducible reduction in the level of dislocated deglycosylated HC and a corresponding increase in the membrane-associated glycosylated HC precursors (Fig. 4b).
FIGURE 4.
Differential effects of p97 inhibitors on US2- and US11-mediated retrotranslocation. a, MG132-treated US11 cells were radiolabeled, permeabilized, and incubated as shown in the scheme A in Fig. 1a. Where indicated, recombinant SVIP protein (4 μm) was included in the chase reaction. Samples taken at the indicated time points were either analyzed directly (T) by immunoprecipitation with αHC or fractionated into a pellet (P) and supernatant (S) fraction before immunoprecipitation. b, as in a, except that US2 cells were used. Graph shows the quantification of results from two independent experiments. c, as in a, except that a dominant-negative form of p97 (p97ΔD2, 2 μm) was added to the reaction mixture before chase. d, as in c, except that US2 cells were used. The graph shows the quantification of the experiment.
To further examine the role of p97 in US2-mediated dislocation, we incubated permeabilized US2 cells with a p97 mutant lacking the second ATPase domain (ΔD2). This mutant was shown to block retrotranslocation of MHC class I HC in US11 cells (7) (Fig. 4c). Interestingly, addition of this mutant to permeabilized US2 cells had no significant impact on HC dislocation under the same condition (Fig. 4d). Together, these results suggest that US2-induced retrotranslocation either does not require p97 or employs a p97 complex that is different from the one used in US11 cells, so it cannot be efficiently inhibited by these inhibitory proteins.
Interaction of p97 with a Retrotranslocation Intermediate in US2 Cells
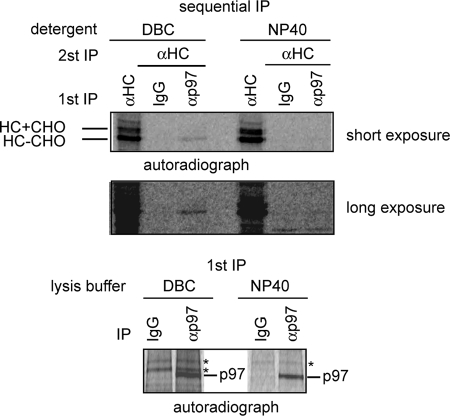
To gain more insights on the role of p97 in US2-mediated retrotranslocation, we examined the potential interaction of p97 with MHC class I HC undergoing retrotranslocation in US2 cells. Because the retrotranslocation and degradation of MHC class I HC in US2 cells occurs rapidly (t½ < 5 min), the interaction between p97 and the substrate HC, if existing, must be transient in nature. To capture this transient complex, we treated cells with a proteasome inhibitor for 1 h before labeling cells with [35S]methionine. Cell extracts were prepared using either the mild detergent DeoxyBigCHAP or a stronger detergent (Nonidet P-40). p97 was immunoprecipitated from cell extract (Fig. 5, bottom panel) and the associated proteins were subjected to a second round of immunoprecipitation with an anti-MHC HC antibody (Fig. 5, upper panels). The result showed that a fraction of MHC class I HC molecules associated with p97 only when DeoxyBigCHAP was used to prepare cell extract, consistent with our hypothesis that the p97-MHC class I HC interaction is unstable. Interestingly, unlike US11 cells in which p97 associated with both the glycosylated retrotranslocation precursor and the dislocated deglycosylated MHC molecules (38), p97 preferentially associated with deglycosylated MHC class I HC in US2 cells (Fig. 5). This observation indicates that p97 is still involved in US2-mediated retrotranslocation, but it appears to operate by a mechanism distinct from that used in US11 cells.
FIGURE 5.
Interaction of p97 with a heavy chain retrotranslocation intermediate in US2 cells. MG132-treated US2 cells were radiolabeled. Cells were lysed in buffers containing either 1% DeoxyBigCHAP (DBC) or 0.5% Nonidet P-40. Cell extracts were subjected to immunoprecipitation with either control IgG or αp97. A portion of the precipitated material was directly analyzed by SDS-PAGE and autoradiography (bottom panel). The remaining material was subjected to a second round of immunoprecipitation with αHC. Where indicated, a fraction of cell extract was subjected to immunoprecipitation with αHC antibody. Shown is a representative gel from two independent experiments.
Ufd1 Is Dispensable for US2-mediated Retrotranslocation
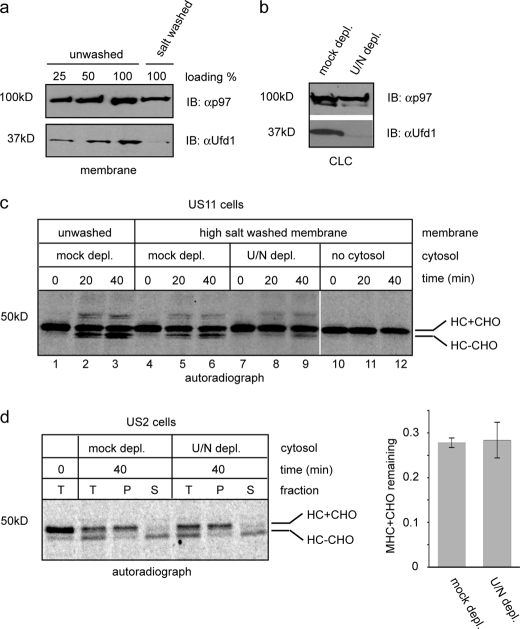
We then tested whether the retrotranslocation of MHC class I HC in US2 cells requires the Ufd1-Npl4 cofactor complex. Ufd1 and Npl4 are present in cells solely as a dimeric complex (39) and depletion of one subunit always causes the simultaneous loss of the other, as reported in several studies (40, 41). The Ufd1-Npl4 complex is mostly in the cytosol, but a fraction of the complex is also associated with the ER membrane. However, the interaction of the Ufd1-Npl4 complex with the membrane can be disrupted by salt (350 mm) (Fig. 6a). Because our in vitro assay includes a washing step with a buffer containing ∼400 mm salt, the only source of Ufd1-Npl4 in the in vitro retrotranslocation reactions is the cytosol. We therefore depleted the Ufd1-Npl4 complex from the cytosol using recombinant GST-p97. Consistent with previous studies, this approach efficiently removed the cofactor complex from the cytosol, as confirmed by immunoblotting with an anti-Ufd1 antibody (Fig. 6b). We first tested the effect of Ufd1 depletion on US11-mediated dislocation. When US11 cells were permeabilized and washed with salt, retrotranslocation of HC was partially inhibited, as indicated by the reduced level of deglycosylated HC during the chase period (Fig. 6c, lanes 5 and 6 versus 2 and 3). This was at least in part due to the removal of the Ufd1 complex from the membrane as depletion of Ufd1 from CLC further impaired HC retrotranslocation (Fig. 6c, lanes 8 and 9). In contrast, when US2-induced dislocation assay was performed using either the cofactor-depleted cytosol or a mock depleted cytosol, we observed no difference in their ability to support the retrotranslocation of HC (Fig. 6d). Thus, we conclude that unlike US11 cells, US2-mediated retrotranslocation likely does not require Ufd1-Npl4.
FIGURE 6.
US2-mediated retrotranslocation is independent of the Ufd1-Npl4 complex in vitro. a, the membrane fraction from digitonin-permeabilized astrocytoma cells was either untreated or treated with a buffer containing 350 mm salt. The membrane was then extracted with the Nonidet P-40 lysis buffer and the protein extract was analyzed by immunoblotting with the indicated antibodies. b, control CLC or Ufd1-Npl4-depleted CLC were analyzed by immunoblotting (IB) with the indicated antibodies. c, MG132-treated US11 cells were radiolabeled and permeabilized. The membrane pellet fraction was either untreated or salt treated and then incubated with the indicated CLCs. Samples taken at the indicated time points were analyzed by immunoprecipitation with αHC antibody. ctrl., control. d, MG132-treated US2 cells were radiolabeled and permeabilized. The membrane pellet fraction was incubated with the indicated CLCs to initiate retrotranslocation. Samples taken at the indicated time points were either analyzed directly by immunoprecipitation with αHC (T) or fractionated into membrane pellet (P) and supernatant (S) fractions before immunoprecipitation analysis. Shown is a representative gel from two independent experiments. The graph shows the quantification of the results. Error bars indicate the mean of two independent experiments.
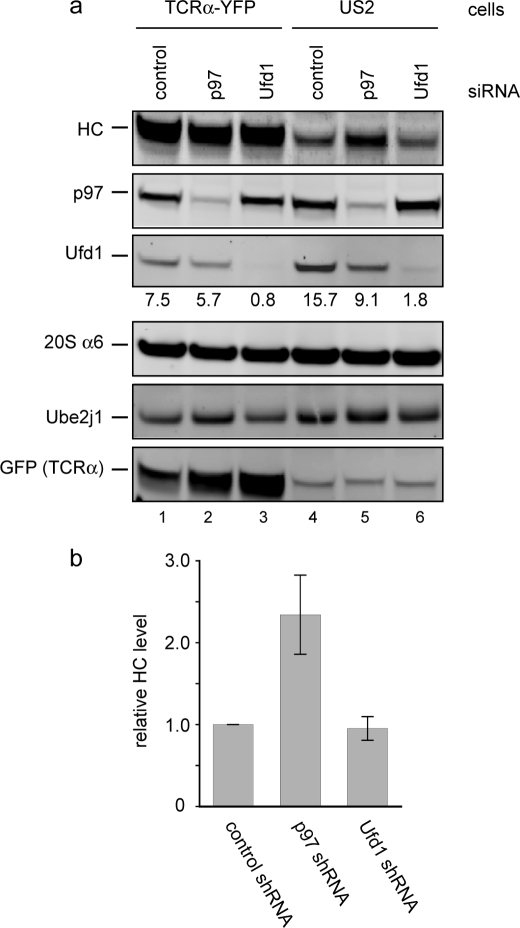
We finally confirmed our findings using intact 293T cells stably expressing US2. We knocked down both p97 and Ufd1 using a siRNA approach. The depletion of these proteins from cells could be verified by immunoblotting (Fig. 7a). The depletion of either p97 or Ufd1 significantly impaired the ERAD pathway, as indicated by the increased stability of the classical ERAD substrate TCRα (Fig. 7a, lanes 1–3). Interestingly, in US2-expressing 293T cells, depletion of p97 stabilized endogenous MHC class I HC, whereas knockdown of Ufd1 had no effect on the degradation of MHC class I HC (Fig. 7a, lanes 4-6, and b). These results confirm that US2-induced retrotranslocation proceeds via a p97-dependent but Ufd1-independent mechanism.
FIGURE 7.
US2-mediated retrotranslocation is independent of Ufd1. a, 293T cells stably expressing TCRα or US2 were transfected with either control siRNA or siRNA targeting p97 or Ufd1. Whole cell extracts were analyzed by immunoblotting with antibodies against the indicated proteins. The number indicates the relative intensity of the Ufd1 band. Note that the GFP antibody also detects a nonspecific protein in US2 cell extract, which migrates at the same position as TCRα-YFP. b, shown are normalized HC levels in US2 cells transfected with the indicated siRNAs from three independent experiments. Error bars represent S.E. (n = 3).
DISCUSSION
The HCMV protein US2 hijacks the ERAD pathway to dispose of newly synthesized MHC class I HC. It has been shown previously that in US2-expressing cells, MHC class I HC molecules undergo polyubiquitination and are dislocated into the cytosol prior to proteasomal degradation (13). Removal of ubiquitination sites on MHC class I HC inhibits its retrotranslocation and degradation (18). To dissect the mechanism of US2-induced retrotranslocation of MHC class I HC, we establish an in vitro permeabilized cell assay that recapitulates key features of this reaction. In our in vitro assay, ubiquitin depletion completely abolishes US2-induced retrotranslocation, consistent with a previous report that US2-mediated retrotranslocation requires ubiquitination. In addition, the dislocation reaction requires ATP.
We show that in addition to ubiquitin, US2-dependent retrotranslocation also involves the p97 ATPase. At first glance, this appears to resemble US11-induced retrotranslocation. However, a thorough comparison of the two reactions indicates that the two HCMV proteins use distinct retrotranslocation complexes to down-regulate MHC class I HC. For ubiquitination, the US2 protein uses the TRC8 ubiquitin ligase to conjugate polyubiquitin chains on one or more lysine residues in HC and removal of ubiquitin acceptors in HC inhibits its retrotranslocation (23). By contrast, TRC8 is not involved in US11-induced retrotranslocation and lysine residues in HC molecules are dispensable for retrotranslocation (18, 23). Because MHC class I HC is polyubiquitinated in US11 cells and blocking this modification by either depletion of ubiquitin from the cytosol or inactivation of the ubiquitin activating enzyme abolishes US11-dependent retrotranslocation (15, 16, 31), we assumed that ubiquitin chains may be conjugated to one or more non-lysine residues in HC, as demonstrated recently (42). For the dislocation reaction, although both reactions appear to use the p97 ATPase, the mode of p97 action in these reactions is different. In US11-expressing cells, p97 interacts with both glycosylated retrotranslocation precursor as well as deglycosylated retrotranslocated product (38). In US2 cells, p97 primarily interacts with the deglycosylated species. This distinction suggests that US2 and US11 may employ different p97 complexes to dislocate MHC class I HC, a notion further supported by our finding that US2-mediated retrotranslocation does not require the cofactor complex Ufd1-Npl4. This model would explain why several p97 inhibitory proteins efficiently inhibit the dislocation of HC in permeabilized US11 cells but not in US2 cells. Perhaps, different p97 complexes may have differential accessibility to these inhibitory proteins.
The use of different p97 complexes by the two HCMV protein-induced retrotranslocation processes might be due to the different mechanisms by which these viral proteins recognize and target HC molecules for retrotranslocation. US11 uses its transmembrane domain to recognize both MHC class I HC and Derlin-1. The latter is a multispanning membrane protein implicated in retrotranslocation of some misfolded ER proteins (24, 25, 43). By serving as an adaptor that links MHC class I HC to Derlin-1, US11 targets HC to a retrotranslocation complex comprising Derlin-1, one or more membrane-bound ubiquitin ligases, and the AAA ATPase p97 (38, 44). In contrast, Derlin family members are dispensable for US2-mediated retrotranslocation. Instead, US2 uses its ER luminal domain to interact with HC and its cytosolic tail to bind the signal peptide peptidase (22). Signal peptide peptidase is in complex with the TRC8 E3 ubiquitin ligase, which appears to mediate the transfer of MHC class I HC across the ER membrane in US2 cells (23). Whether or not TRC8 also mediates retrotranslocation of misfolded ER proteins is currently unknown. Nonetheless, it becomes apparent that distinct retrotranslocation complexes exist in the ER membrane in both yeast and higher eukaryotes, which serve different cohorts of ERAD substrates. Because almost all retrotranslocation mechanisms studied to date use the p97 ATPase for substrate dislocation, the difference in the membrane machinery underscore the importance of tailoring p97 complex by affiliating it with different co-factors, which would allow the same enzyme to cooperate with different membrane-bound retrotranslocation complexes. In agreement with this idea, a recent study shows that the p97 adaptor UbxD8, a Ubx-domain-containing protein is only involved in retrotranslocation of HC molecules in US11 cells but not in US2 cells (27).
Acknowledgments
We thank Hemmo Meyer (University of Duisburg-Essen, Germany) for GST-p97 plasmid and αUfd1 antibody and Hidde Ploegh (MIT, MA) for US2-encoding plasmid.
This work was supported, in whole or in part, by a National Institutes of Health grant from the Intramural Research Program, NIDDK.
- ER
- endoplasmic reticulum
- HC
- heavy chain
- ERAD
- ER-associated protein degradation
- HCMV
- human cytomegalovirus
- ARS
- ATP regenerating system
- AAA
- ATPase associated with various cellular activities
- CLC
- cow liver cytosol.
REFERENCES
- 1.Tsai B., Ye Y., Rapoport T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 2.Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 3.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bays N. W., Hampton R. Y. (2002) Curr. Biol. 12, R366–371 [DOI] [PubMed] [Google Scholar]
- 5.Ye Y. (2005) Essays Biochem. 41, 99–112 [DOI] [PubMed] [Google Scholar]
- 6.Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 7.Ye Y., Meyer H. H., Rapoport T. A. (2003) J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flierman D., Ye Y., Dai M., Chau V., Rapoport T. A. (2003) J. Biol. Chem. 278, 34774–34782 [DOI] [PubMed] [Google Scholar]
- 9.Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. (2005) Cell 120, 73–84 [DOI] [PubMed] [Google Scholar]
- 10.Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. (2006) J. Cell Biol. 172, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q., Li L., Ye Y. (2006) J. Cell Biol. 174, 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Ye Y., Lencer W., Hansen T. H. (2006) J. Biol. Chem. 281, 8636–8644 [DOI] [PubMed] [Google Scholar]
- 13.Wiertz E. J., Tortorella D., Bogyo M., Yu J., Mothes W., Jones T. R., Rapoport T. A., Ploegh H. L. (1996) Nature 384, 432–438 [DOI] [PubMed] [Google Scholar]
- 14.Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 15.Kikkert M., Hassink G., Barel M., Hirsch C., van der Wal F. J., Wiertz E. (2001) Biochem. J. 358, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shamu C. E., Flierman D., Ploegh H. L., Rapoport T. A., Chau V. (2001) Mol. Biol. Cell 12, 2546–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman M. H., Loureiro J., Ploegh H. L., Tortorella D. (2003) J. Biol. Chem. 278, 34804–34811 [DOI] [PubMed] [Google Scholar]
- 18.Hassink G. C., Barel M. T., Van Voorden S. B., Kikkert M., Wiertz E. J. (2006) J. Biol. Chem. 281, 30063–30071 [DOI] [PubMed] [Google Scholar]
- 19.Oresic K., Ng C. L., Tortorella D. (2009) J. Biol. Chem. 284, 5905–5914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegde N. R., Chevalier M. S., Wisner T. W., Denton M. C., Shire K., Frappier L., Johnson D. C. (2006) J. Biol. Chem. 281, 20910–20919 [DOI] [PubMed] [Google Scholar]
- 21.Lee S. O., Cho K., Cho S., Kim I., Oh C., Ahn K.(XXXX)EMBO J. 29, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loureiro J., Lilley B. N., Spooner E., Noriega V., Tortorella D., Ploegh H. L. (2006) Nature 441, 894–897 [DOI] [PubMed] [Google Scholar]
- 23.Stagg H. R., Thomas M., van den Boomen D., Wiertz E. J., Drabkin H. A., Gemmill R. M., Lehner P. J. (2009) J. Cell Biol. 186, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 25.Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 26.Mueller B., Lilley B. N., Ploegh H. L. (2006) J. Cell Biol. 175, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 29.Wang Q., Li L., Ye Y. (2008) J. Biol. Chem. 283, 7445–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W., Tu D., Li L., Wollert T., Ghirlando R., Brunger A. T., Ye Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3722–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shamu C. E., Story C. M., Rapoport T. A., Ploegh H. L. (1999) J. Cell Biol. 147, 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones T. R., Hanson L. K., Sun L., Slater J. S., Stenberg R. M., Campbell A. E. (1995) J. Virol. 69, 4830–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blom D., Hirsch C., Stern P., Tortorella D., Ploegh H. L. (2004) EMBO J. 23, 650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ernst R., Mueller B., Ploegh H. L., Schlieker C. (2009) Mol. Cell 36, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flierman D., Coleman C. S., Pickart C. M., Rapoport T. A., Chau V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11589–11594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballar P., Zhong Y., Nagahama M., Tagaya M., Shen Y., Fang S. (2007) J. Biol. Chem. 282, 33908–33914 [DOI] [PubMed] [Google Scholar]
- 38.Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruderer R. M., Brasseur C., Meyer H. H. (2004) J. Biol. Chem. 279, 49609–49616 [DOI] [PubMed] [Google Scholar]
- 40.Hetzer M., Meyer H. H., Walther T. C., Bilbao-Cortes D., Warren G., Mattaj I. W. (2001) Nat. Cell Biol. 3, 1086–1091 [DOI] [PubMed] [Google Scholar]
- 41.Ramadan K., Bruderer R., Spiga F. M., Popp O., Baur T., Gotta M., Meyer H. H. (2007) Nature 450, 1258–1262 [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Herr R. A., Chua W. J., Lybarger L., Wiertz E. J., Hansen T. H. (2007) J. Cell Biol. 177, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lilley B. N., Tortorella D., Ploegh H. L. (2003) Mol. Biol. Cell 14, 3690–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lilley B. N., Ploegh H. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]