Abstract
Agrin released by motoneurons induces and/or maintains acetylcholine receptor (AChR) clustering and other aspects of postsynaptic differentiation at the vertebrate neuromuscular junction. Agrin acts by binding and activating a receptor complex containing LDL receptor protein 4 (Lrp4) and muscle-specific kinase (MuSK). Two critical downstream components of this signaling cascade, Dox-7 and rapsyn, have been identified. However, additional intracellular essential elements remain unknown. Prior observations by others and us suggested antagonistic interactions between agrin and neuregulin-1 (Nrg-1) signaling in cultured myotubes and developing muscle fibers in vivo. A hallmark of Nrg-1 signaling in skeletal muscle cells is the activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2). ERK1/2 are also activated in most cells by phorbol 12-myristate 13-acetate, a classical inhibitor of agrin-induced AChR clustering in myotubes. Here, it was investigated whether agrin activates ERK1/2 directly and whether such activation modulates agrin-induced AChR clustering. Agrin induced a rapid but transient activation of ERK1/2 in myotubes that was Lrp4/MuSK-dependent. However, blocking this ERK1/2 activation did not prevent but potentiated AChR clustering induced by agrin. ERK1/2 activation was dispensable for Nrg-1-mediated inhibition of the AChR clustering activity of agrin, but was indispensable for such activity by phorbol 12-myristate 13-acetate. Together, these results suggest agrin-induced activation of ERK1/2 is a negative modulator of agrin signaling in skeletal muscle cells.
Keywords: ERK, MAP Kinases (MAPKs), Nicotinic Acetylcholine Receptors, Signal Transduction, Skeletal Muscle, PMA, Agrin, Neuregulin, Neuromuscular Junction, synaptogenesis
Introduction
For more than a decade, three proteins have stood out as the essential signaling partners in vertebrate neuromuscular junction (NMJ)2 synaptogenesis. Agrin, a proteoglycan released by motoneurons that induces and/or maintains acetylcholine receptor (AChR) clustering and other aspects of postsynaptic differentiation (1). MuSK, a receptor tyrosine kinase activated by agrin (2, 3); and rapsyn, an intracellular peripheral membrane protein that binds to AChRs (4, 5). Recently, two additional key components of this signaling pathway have been discovered: Lpr4, a LDL-like receptor protein that binds agrin and associates with MuSK (6, 7); and Dok-7 (8), a phosphotyrosine-binding protein that binds to activated MuSK. In the absence of any of these proteins, neuromuscular synapses simply fail to form in vivo (8–12). Despite this impressive progress, how agrin-induced MuSK activation leads to AChR clustering remains elusive as additional critical intracellular components of the core pathway are yet to be identified.
Previously, Trinidad and Cohen (13) showed that in cultured myotubes agrin-induced AChR clustering was inhibited by co-treatment with Nrg-1. We showed that agrin failed to cluster AChRs in cultured myotubes expressing a constitutively active neuregulin receptor ErbB2 (14). Importantly, when expressed in developing muscle fibers in vivo, constitutively active ErbB2 led to synaptic loss resembling the neuromuscular phenotype in mice deficient for agrin (14). These results suggested antagonistic interactions between the neuregulin and agrin signaling pathways in developing skeletal muscle and raised the possibility that understanding the molecular mechanisms that mediate such interaction may lead to better understanding of normal agrin signaling. Sustained activation of ERK1/2 is a major signaling event induced by Nrg-1 on skeletal muscle cells (15, 16). Furthermore, phorbol 12-myristate 13-acetate (PMA), a protein kinase C (PKC) activator that was among the first identified inhibitors of agrin-induced AChR clustering on myotubes (17, 18), also induces activation of ERK1/2 in most cells. Here, it was investigated whether agrin can activate ERK1/2 directly and whether such activation can modulate agrin-induced AChR clustering. It was found that agrin indeed induces a rapid but transient activation of ERK1/2 in myotubes that is Lrp4/MuSK-dependent. However, blocking this ERK1/2 activation does not prevent but potentiates AChR clustering induced by agrin. ERK1/2 activation is not required for Nrg-1-mediated inhibition of the AChR clustering activity of agrin, but mediates such activity by PMA. These results implicate agrin-induced ERK1/2 activation in a feedback loop that negatively modulates agrin signaling to muscle cells.
EXPERIMENTAL PROCEDURES
Materials
Recombinant rat C-terminal agrin was from R&D Systems. Most experiments were done with the carrier-free product (catalogue number 550-AG/CF). The carrier (BSA)-containing product (catalogue number 550-AG) was also used in some experiments with similar results. Recombinant human Nrg-1-β1 EGF domain was from R&D Systems. PMA and U0126 were purchased from Sigma. PD0325901 was purchased from Selleck. PMA, U0126, and PD0325901 were dissolved in dimethyl sulfoxide (Sigma). DMEM, gentamicin, and phosphate-buffered saline (PBS) without CaCl2 and MgCl2 were from Invitrogen. FBS and horse serum were from Gemini Bio-Products. Matrigel was from BD Biosciences. Chick embryo extract was from Sera Laboratories International. Interferon-γ was from PeproTech. Anti-phospho-ERK1/2 (number 9101) and anti-total ERK1/2 (number 9102) primary antibodies were from Cell Signaling Technology. HRP-conjugated secondary antibodies were from Jackson ImmunoResearch. The Western Lighting Enhanced Chemiluminescence kit used for developing Western blots was from PerkinElmer Life Sciences. Rhodamine-α-bungarotoxin was from Molecular Probes (Invitrogen). Vectashield mounting medium was from Vector Labs.
Cell Growth and Differentiation
The C2 cells used in this study were initially derived from C2 cells in the Burden lab (Skirball Institute, New York University Medical Center). They have been used in the Rimer lab since 2001. Myoblasts were grown on Matrigel in DMEM, 15% FBS, 0.5% chick embryo extract, and 50 μg/ml of gentamicin. To slow their growth, myoblasts were often grown with the above medium but using 10% FBS. At confluence, myoblasts were switched into differentiation medium (C2DM) containing DMEM, 2.5% horse serum, and 50 μg/ml of gentamicin. C2 cells were grown and differentiated in a humidified incubator at 37 °C, 5% CO2. Myotubes were used for experiments after 2–3 days in C2DM.
Wild type, Lrp4−/−, and MuSK−/− immorto+ cells were a kind gift of Steven Burden and Ruth Herbst (Medical University of Vienna). Immorto+ cells were grown in a humidified incubator at 33 °C in 10% CO2 in the following medium: DMEM (with sodium pyruvate), 15% FBS, 2% chicken embryo extract, 50 μg/ml of gentamicin, and 100 μg/ml of interferon-γ. At confluence they were differentiated in a humidified incubator at 39 °C in 10% CO2 in the following medium: DMEM (with sodium pyruvate), 10% FBS, 10% horse serum, 0.5% chicken embryo extract, 50 μg/ml of gentamicin. Myotubes were used for experiments after 2–3 days in differentiation medium.
ERK1/2 Activation Assays
For these experiments C2 cells were grown and differentiated in 60-mm dishes. Myotubes were switched into DMEM + gentamicin for about 4 h before treatment. This medium was replaced with fresh medium previously warmed at 37 °C, with or without 0.1 nm agrin. Dishes were returned to the incubator and at different times thereafter, the dishes were taken out and the medium was aspirated and replaced with ice-cold PBS. Such dishes were kept on ice until lysates were prepared. 1 ml/dish of the following lysis buffer was used: 25 mm Tris-HCl, pH 7.4, 2% SDS, 95 mm NaCl, 10 mm EDTA, 5 mm EGTA, 5 mm NaFl, 2 mm Na+ orthovanadate, 2.5 mm Na+ pyrophosphate, 10% (v/v) protease inhibitor mixture (P8340, Sigma). Cell scrapers were used to detach the cells from the dish and the slurry was passed several times through a 21-gauge syringe. Lysates were cleared by centrifugation at 14,000 × g for 15 min at 4 °C and stored at −80 °C until use.
Western blotting was done as previously described (19). 75 μg of protein/sample were separated in 10% denaturing polyacrylamide gels. Membranes were probed first with anti-pERK1/2 antibodies diluted 1/1000 in Tris-buffered saline/Tween 20 (TBS-T), 1–5% BSA, and the above phosphatase inhibitors. Membranes were then stripped and reprobed with anti-tERK1/2 antibodies diluted 1/1000–1/3000 in TBS-T, 1–5% BSA. Bands were visualized by chemiluminescence following the manufacturer's instructions. During early experiments membranes were developed on BioMax x-ray film (Kodak). Later, a digital gel imaging system was used. All Western blot quantification was done using the digital system as described below.
For experiments with the immorto+ myotubes (Fig. 2) everything was the same as above except that cultures were incubated in C2DM for at least 4 h before adding 0.5 nm agrin. Myotubes were kept at 39 °C in 10% CO2 during this preincubation with C2DM and throughout the experiment.
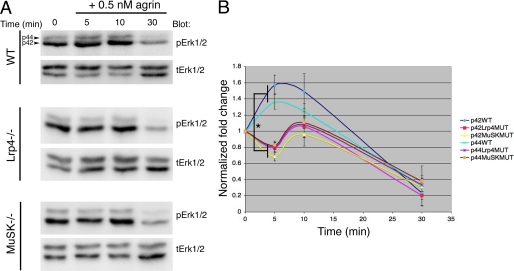
FIGURE 2.
The rapid agrin-induced ERK1/2 activation is Lrp4/MusK dependent. Time course of ERK1/2 activation following application of 0.5 nm recombinant neural agrin in wild-type (WT), Lrp4−/−, and MuSK−/− immorto+ myotubes. A, representative Western blots probed as in Fig. 1 for pERK1/2 and tERK1/2 for each cell line. These blots were developed using the gel digital imaging system described under “Experimental Procedures.” B, quantification. The rapid ERK1/2 activation seen in WT myotubes after agrin application was absent in mutant (MUT) myotubes. However, all 3 cell lines displayed the 3–4-fold reduction in active ERK1/2 at 30 min after agrin addition. Data are expressed as mean ± S.E. (error bars = +/− S.E.). All 4 pairwise statistical comparisons between active ERK1/2 levels at 5 min in WT versus either in Lrp4−/− or MuSK−/− cells using a t test were significantly different (*, p < 0.006).
For Western blot quantification, images of the developed membranes were captured with a CCD camera attached to a FluorChem Q MultiImage III system (Alpha Innotech). Exposure times (usually 45 s) were below saturation. ERK1 (p44) and ERK2 (p42) bands were quantified separately with AlphaView software (Alpha Innotech) following the manufacturer's instructions. Intensity values were corrected with the local background option of the program. For each band, values from the blot probed with anti-pERK1/2 were divided by values from the blot probed with anti-tERK1/2 to derive a phosphorylated/total ratio. These values were divided in turn by the ratio in the control (untreated) samples to derive normalized intensities for each band in each different experiment. Thus, the normalized intensity value for the control sample in each experiment was always 1.
The lysis buffer used to prepare samples in Fig. 4A was 30 mm triethanolamine, pH 7.5, 1% Nonidet P-40, 50 mm EGTA, 50 mm NaCl. Phosphatase and protease inhibitors were the same as for the previous lysis buffer except that 100 nm okadaic acid (Sigma) was also added.
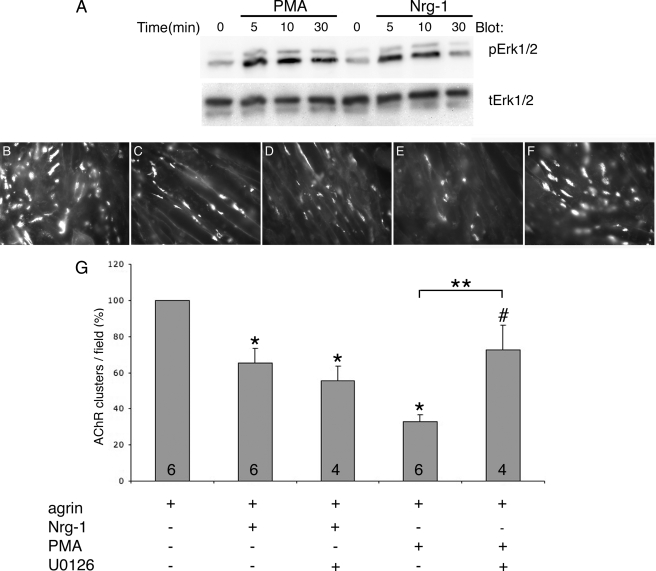
FIGURE 4.
PMA, but not Nrg-1, requires ERK1/2 activation to inhibit agrin-induced AChR clustering in C2 myotubes. A, both PMA (50 nm) and Nrg-1 (2 nm) induced ERK1/2 activation in C2 myotubes as detected by Western blotting. The lysis buffer used here was different from that used for Figs. 1–3 (see “Experimental Procedures”). B–F, representative 400× fields of C2 myotubes treated for 15–16 h with: 0.1 nm agrin (B); 0.1 nm agrin + 2 nm Nrg-1 (C); 0.1 nm agrin + 2 nm Nrg-1 + 10 μm U0126 (D); 0.1 nm agrin + 50 nm PMA (E); 0.1 nm agrin + 50 nm PMA + 10 μm U0126 (F). G, quantification. Nrg-1 produced a ∼35% inhibition, whereas PMA produced a ∼65% inhibition in agrin-induced AChR clustering. U0126 co-treatment rescued PMA-mediated inhibition to ∼75% control (agrin) levels, whereas it had no effect on Nrg-1-mediated inhibition. Data are presented as percentage of treatment with agrin alone. Data are expressed as mean + S.E. (error bars = + S.E.). n is given by numbers at the base of the bars. *, p versus control (agrin) < 0.01, one sample t test. #, p = 0.134 versus control, one sample t test. **, p = 0.009, agrin+PMA versus agrin + PMA + U0126, t test.
AChR Clustering Assays
For these experiments C2 cells were grown and differentiated in 35-mm dishes. Treatments were done in duplicate per experiment. Details of the specific treatments can be found under “Results” and in the figure legends. Following treatments, AChRs were stained live by incubation with 50 nm α-bungarotoxin for 30 min in the cell incubator. Following a rinse with PBS, dishes were fixed with 2% paraformaldehyde in PBS for 15 min at room temperature, washed 3 times in PBS for 10 min, and air-dried for ∼15 min inside a chemical hood. Coverslips were then mounted using Vectashield. Myotubes were visualized by epifluorescence with a Nikon Eclipse microscope equipped with rhodamine optics using a 40× lens (N.A. 1.30). Images from 10 fields per dish were captured using Methamorph software. The same software was used to count AChRs and determine the AChR area after manual thresholding of the images. Filters were set to exclude in the quantification AChR clusters ≤4 μm2. The number of clusters per dish was derived after averaging the number of clusters in each of the 10 fields. The average of clusters per treatment for a given experiment resulted from the average of the two duplicate dishes. For Fig. 3, results were expressed as AChR clusters/field. For Figs. 4 and 5 results were expressed as percentage of AChR clusters/field relative to positive control (i.e. agrin treatment) after subtracting clusters in dishes that received no treatment.
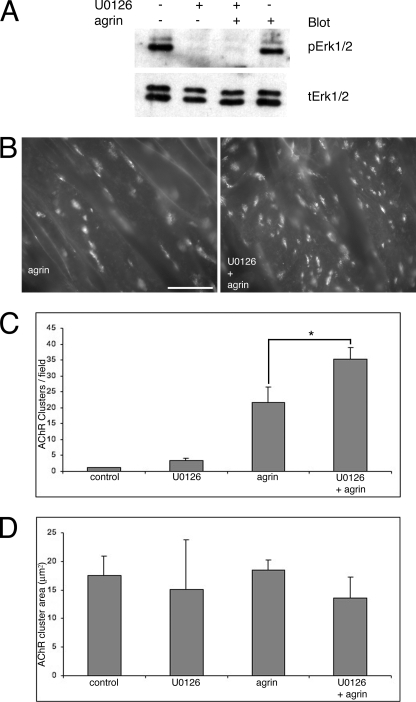
FIGURE 3.
Inhibition of ERK1/2 activation potentiates agrin-induced AChR clustering. A, Western blot for ERK1/2 at the end of a 1-h incubation with agrin, with or without U0126. The MEK inhibitor prevented ERK1/2 activation whether agrin was present or not. As shown in Fig. 1, levels of active ERK1/2 are not much different between untreated control and agrin-treated cells at 1 h after agrin addition. B, sample 400× fields of myotubes treated with agrin and agrin + U0126. There are many more AChR clusters (labeled with rhodamine-α-bungarotoxin) in the latter than the former. Scale bar, 50 μm. C, quantification of AChR clustering. There was ∼60% increase in AChR clusters in dishes treated with agrin + U0126 relative to dishes treated with agrin. Data are expressed as mean ± S.E. n = 4 for each treatment. *, p = 0.008, t test. D, quantification of AChR cluster area. No statistically significant differences were found in the AChR cluster area between any of the treatments. Data are expressed as mean + S.E. (error bars = + S.E.). n = 4 for each treatment.
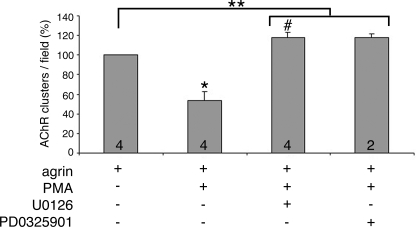
FIGURE 5.
Complete rescue of PMA-mediated inhibition of agrin-induced AChR clustering in myotubes treated for 5–6 h. PMA (50 nm) co-treatment for 5–6 h produced a ∼50% inhibition in agrin-induced AChR clustering. This effect was completely reversed to levels ∼20% higher than control by inhibition of ERK1/2 with either U0126 (10 μm) or PD0325901 (0.5 μm). n is given by numbers at the base of the bars. Data are expressed as mean + S.E. for agrin, agrin + PMA, and agrin + PMA + U0126 treatments. Data are expressed as mean + S.D. for agrin + PMA + PD0325901 treatment. *, p = 0.0152, agrin versus agrin + PMA. #, p = 0.0587, agrin versus agrin + PMA + U0126. **, p = 0.0055, agrin versus agrin + PMA + inhibitors (pooled data, n = 6).
Statistical Analysis
Two-tailed Student's t test was used to compare pairs of treatments. One sample t test was used to compare means in experimental and control samples in experiments where data were normalized relative to control. Statistical significance was set at p < 0.05.
RESULTS
Agrin Induces Rapid and Transient Activation of ERK1/2 in Cultured Myotubes
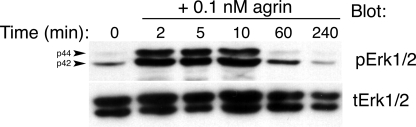
To begin investigating whether changes in ERK1/2 activation are involved in normal agrin signaling in muscle cells, C2 myotubes were treated with a saturating concentration of recombinant neural agrin (0.1 nm). Using Western blotting, a robust, rapid activation of ERK1/2 within 10 min was observed after agrin treatment (Fig. 1). Activated ERK1/2 returned to baseline around 1 h after agrin addition (Fig. 1). The transient ERK1/2 activation induced by agrin appeared specific in that agrin treatment failed to change the levels of PI 3-kinase signaling in the same cells as measured by phosphorylated Akt (data not shown). Agrin-induced ERK1/2 activation occurred as rapidly as agrin-induced MuSK activation observed before by others, however, the latter persists for much longer (3).
FIGURE 1.
Neural agrin induces a rapid but transient activation of ERK1/2 in C2 cells. Time course of ERK1/2 activation following application of 0.1 nm recombinant neural agrin to C2 myotubes. Top panel, Western blot probed with antibodies to active, phosphorylated ERK1/2 (pERK1/2). Bottom panel, same Western blot probed with antibodies to total ERK1/2 (tERK1/2). ERK1/2 were strongly activated within 2 min of neural agrin application. They peaked after 5–10 min and returned to control by 60 min. The arrowheads on the left side of the top panel point to ERK1 (p44) and ERK2 (p42). This blot was developed using film. This experiment was repeated once with similar results.
Agrin-induced ERK1/2 Activation Is Lrp4/MuSK-dependent
Recent work demonstrated that the LDL receptor-related protein Lrp4 binds to agrin and forms a complex with MuSK (6, 7). On its own, Lrp4 binds to neural, but not to aneural, agrin, however, its affinity for agrin is slightly increased by its association with MuSK. Like MuSK−/− mice (11), mice deficient in Lrp4 lack NMJs (12). Thus, the Lrp4-MuSK complex mediates agrin-induced signal transduction on the muscle surface that is essential for neuromuscular synaptogenesis. In addition to Lrp4/MuSK, agrin and specifically the C-terminal ∼95 kDa recombinant form of neural agrin used here also binds to other proteins on the muscle sarcolemma, such as α-dystroglycan (20) and β1-containing integrins (21), which might mediate agrin-induced ERK1/2 activation. Therefore, we sought to determine whether the agrin-induced ERK1/2 activation is Lrp4/MuSK-dependent. To this aim, agrin-induced ERK1/2 activation was measured in immortalized muscle cell lines derived from the Lrp4−/− (6) and MuSK−/− mice (22) and their wild-type control cells. Preliminary experiments determined that 0.5 nm agrin induced consistent, rapid and transient ERK1/2 activation in control immorto+ cells (Fig. 2A, top panels). The response in the immorto+ control cells was less robust than that in C2 cells. Thus, after 5 min normalized levels of activated ERK1 (p44) were 1.37 ± 0.09-fold over control, whereas activated ERK2 (p42) was 1.57 ± 0.12-fold over control (mean ± S.E., n = 6, Fig. 2, A and B). Interestingly, levels of activated ERK1/2 dropped to about 3–4-fold under control levels by 30 min after agrin addition in control immorto+ cells (Fig. 2, A and B, activated ERK1 = 0.33 ± 0.24-fold and activated ERK2 = 0.24 ± 0.16-fold relative to untreated control). Agrin failed to activate both ERK1 and ERK2 in Lrp4−/− and MuSK−/− cells within 10 min after its addition to the cultures (Fig. 2A). Thus, after 5 min activated ERK1 was 0.81 ± 0.07-fold and activated ERK2 was 0.80 ± 0.06-fold control levels in Lrp4-deficient cells (Fig. 2B, n = 3). In MuSK-deficient cells activated ERK1 was 0.83 ± 0.05-fold and activated ERK2 was 0.69 ± 0.05-fold control levels (Fig. 2B, n = 4). Table 1 presents the statistical analysis at 5 min for each cell line. Means were compared with the untreated control mean (i.e. 1) using a one-sample t test. The increase in both ERK1 and ERK2 activation was statistically significant in the wild-type cells, whereas the decrease in both ERK1 and ERK2 phosphorylation observed in the mutants at 5 min was statistically significant only for the MuSK-deficient cells. The data at 5 min for the Lrp4-deficient cells was not different from untreated control (Table 1). At 5 min the differences in activated ERK1/2 between wild-type and mutants were statistically significant (Fig. 2B, bracket). However, the drop in phosphorylated ERK1/2 observed at 30 min after agrin addition in control cells was also seen in Lrp4- and MuSK-deficient cells (Fig. 2, A and B). Thus, in immorto+ cells the agrin-induced, transient and rapid activation of ERK1/2 is Lrp4/MuSK-dependent, whereas the ERK1/2 inactivation that is evident later (30 min) is Lrp4/MuSK-independent. The biological significance of the decrease in ERK1/2 phosphorylation observed at 5 min in the mutant cell lines is unknown.
TABLE 1.
Statistical analysis of 5-min data for experiments with immorto+ cell lines (Fig. 2)
| ERK | Cell line | Mean ± S.E. | n | p versus controla |
|---|---|---|---|---|
| ERK2 (p42) | WT | 1.57 ± 0.12 | 6 | 0.0051 |
| Lrp4−/− | 0.80 ± 0.06 | 3 | 0.0794 | |
| MuSK−/− | 0.69 ± 0.05 | 4 | 0.0085 | |
| ERK1 (p44) | WT | 1.37 ± 0.09 | 6 | 0.0118 |
| Lrp4−/− | 0.81 ± 0.07 | 3 | 0.1132 | |
| MuSK−/− | 0.83 ± 0.05 | 4 | 0.0425 |
a One sample t test was used to compare means at 5 min versus means at 0 min, which were normalized at 1.
Agrin-induced ERK1/2 Transient Activation Is Dispensable for but Modulates Agrin-induced AChR Clustering in C2 Cells
Next, it was tested whether blocking agrin-induced ERK1/2 activation prevented formation of AChR clusters in cultured myotubes. C2 myotubes were pretreated with 10 μm U0126 for 1 h, and then 0.1 nm agrin was applied for another hour, keeping U0126 still present in the dishes. U0126 specifically blocks MEK 1 and 2 (23), the only upstream kinases that activate ERK1/2 directly. At the end of this treatment, dishes were extensively washed with medium without agrin or U0126, and returned to the incubator for another 7 h. Then, AChRs were labeled live with fluorescent-α-bungarotoxin and clusters were quantified after fixing and washing of the dishes. Fig. 3A shows that as expected the U0126 treatment was effective in lowering levels of activated ERK1/2 in the presence or absence of agrin. Fig. 3, B and C, show that in the presence of the MEK inhibitor agrin caused a 62% increase in the number of AChR clusters over agrin alone (agrin + U0126, 35.2 ± 3.8 clusters/field; agrin, 21.7 ± 4.7 clusters/field; n = 4 for each treatment). Statistical comparison between the agrin + U0126 and agrin treatments using a t test yielded a significant p = 0.008 (Fig. 3). The effect of U0126 pretreatment on agrin-induced AChR clustering was mainly on cluster number, rather than on cluster size, as there was no statistically significant difference in cluster area between the agrin and the agrin + U0126 treatments (Fig. 3D; p = 0.08, n = 4). Previously, Fuhrer and colleagues (24) showed that a 5-min pulse of 0.5 nm agrin was sufficient to induce AChR clustering at levels similar to those induced with standard protocols that use agrin at lower concentrations (e.g. 0.1 nm) for longer periods of times (e.g. at least 4 h, typically 8 h or overnight). These authors also demonstrated that the washing conditions that they used to remove agrin from the dishes, which were adopted here, clearly rid the cultures of agrin. One experiment using a 5-min pulse of 0.5 nm agrin in the presence or absence of U0126 pretreatment was performed. It was found that agrin + U0126 also induced more AChR clusters than agrin alone using this protocol (average of two dishes per treatment: agrin + U0126, 44.8 clusters/field; agrin, 34.2 clusters/field). Thus, blocking the agrin-induced transient ERK1/2 activation failed to prevent AChR clustering. Instead, it led to an increase in agrin-induced AChR clustering.
Nrg-1 and PMA, Two Known Inhibitors of Agrin-induced AChR Clustering, Display Differential Requirement for ERK1/2 Activation
The above results suggested that although dispensable for AChR clustering ERK1/2 activation does modulate the activity of agrin as blockage of ERK1/2 activation led to potentiation of agrin-induced clustering. One would predict that the converse, strong and persistent stimulation of ERK1/2 phosphorylation, would lead to inhibition of agrin-induced AChR clustering. Thus, we sought to determine whether ERK1/2 activation is required by Nrg-1 and PMA, two factors known to both induce ERK1/2 activation (Fig. 4A) (15, 25) and to inhibit the AChR clustering activity of agrin in myotubes (13, 17, 18). C2 myotubes were treated for 14–15 h with either 0.1 nm agrin or 0.1 nm agrin in the presence of 2 nm Nrg-1β EGF domain (13), or 50 nm PMA (17). In our hands, Nrg-1 co-treatment reduced agrin-induced AChR clustering to 65.54 ± 7.79% (mean ± S.E., n = 6) control levels, or about 35% inhibition (Fig. 4, B, C, and G). PMA treatment was more effective in reducing agrin-induced AChR clustering under our experimental conditions as it yielded 32.91 ± 3.95% control levels, or about 65% inhibition (Fig. 4, B, E, and G). Both agrin + Nrg-1 and agrin + PMA treatments were significantly different from the agrin alone treatment (p = 0.007 and p < 0.0001, respectively). To test whether ERK1/2 activation is required by Nrg-1 or PMA to inhibit agrin-induced AChR clustering, 10 μm U0126 was added to the agrin + Nrg-1 and agrin + PMA treatments. In the presence of U0126, the inhibition of AChR clustering mediated by Nrg-1 was essentially unchanged (55.62 ± 8.08% control levels, n = 4, Fig. 4, D and G; p = 0.41 versus agrin + Nrg-1), whereas PMA-mediated inhibition of agrin-induced AChR clustering was largely, albeit not completely, relieved (72.65 ± 13.43% control levels, Fig. 4, F and G; p = 0.009 versus agrin + PMA). Similar results were obtained if PD0325901, another highly specific MEK inhibitor (26), was used instead of U0126. Thus, in these experiments (n = 2) agrin + Nrg-1 treatment yielded 65.59 ± 0.52% control levels (mean ± S.D.) and agrin + Nrg-1 + PD0325901 yielded 75.35 ± 13.03% control levels. The agrin + PMA treatment yielded 28.54 ± 0.78% control levels, whereas the agrin + PMA + PD0325901 treatment yielded 72.76 ± 13.46% control levels.
Overnight treatment with agrin or other factors was tried here first because it was under such conditions that Trinidad and Cohen (13) first demonstrated the inhibitory effects of Nrg-1 on the activity of agrin. On the other hand, such effects mediated by PMA were first reported on chick primary myotubes treated for just 6 h (17). Early experiments established that as long as the incubation with PMA was for 6 h or less, its effect on AChR clustering was protein synthesis independent and did not alter the AChR number on the surface of the cell (17, 18). However, overnight incubation with PMA led to reduced AChR synthesis (27). To rule out that the inhibitory effects of PMA on the activity of agrin in C2 cells resulted from unspecific effects due to overnight treatment, experiments were performed in which agrin, PMA, and MEK inhibitor treatments were done for 5–6 h (Fig. 5). Under these conditions PMA led to about 50% inhibition of agrin-induced AChR clustering (53 ± 9.30, n = 4; p = 0.0152, agrin versus agrin + PMA). Co-incubation of agrin, PMA, and a MEK inhibitor completely rescued the inhibitory effects of PMA on agrin-induced AChR clustering (p = 0.0011, agrin + PMA versus agrin + PMA + U0126). Interestingly, both U0126 and PD0325901 restored agrin-induced AChR clustering to about 120% control levels. Thus, agrin + PMA + U0126 yielded 117.61 ± 5.91% control (i.e. agrin) levels (mean ± S.E., n = 4) and agrin + PMA + PD0325901 yielded 117.99 ± 3.70% control levels (mean ± S.D., n = 2) (Fig. 5). Although not quite statistically significant (p = 0.0587, agrin versus agrin + PMA + U0126) if the data from both MEK inhibitors are grouped, based on their similar effects on this experiment then p = 0.0055, agrin versus agrin + PMA + inhibitors (pooled data, n = 6). This 20% potentiation of agrin signaling is consistent with the 62% potentiation observed when agrin was applied to the cultures for just 1 h in the presence of U0126 (Fig. 3).
Thus, together these results indicate that activation of ERK1/2 is necessary for PMA-mediated, but not for Nrg-1-mediated, inhibition of agrin-induced AChR clustering. They further implicate active ERK1/2 as a negative modulator of agrin signaling in skeletal muscle cells.
DISCUSSION
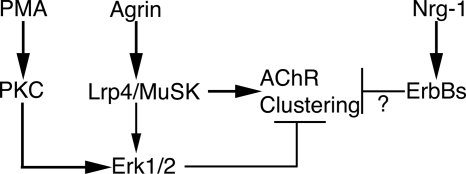
The most important and novel results from this work are summarized in Fig. 6. They are the following: (i) agrin induces a rapid but transient activation of ERK1/2 that is Lrp4/MuSK-dependent but unnecessary for AChR clustering (Figs. 1–3). (ii) ERK1/2 activation appears to be a negative modulator of agrin signaling in muscle cells because it mediates PMA inhibition of agrin-induced AChR clustering (Figs. 4 and 5) and because preventing early ERK1/2 activation leads to potentiation of agrin-induced AChR clustering (Figs. 3 and 5). (iii) ERK1/2 activation is dispensable for Nrg-1 inhibition of agrin-induced AChR clustering (Fig. 4).
FIGURE 6.
Summary of results. Agrin strongly activates Lrp4/MuSK to induce AChR clustering (represented by thick arrows). At the same time, agrin also activates ERK1/2 transiently (represented by thinner arrow) in an Lrp4/MuSK-dependent fashion. This, in turn, acts as a break on agrin-induced AChR clustering (represented as perpendicular (T), thin lines). PMA activates PKC, which in turn strongly activates ERK1/2 to inhibit agrin-induced AChR clustering. Thus, PMA inhibits AChR clustering by co-opting the feedback loop normally induced by agrin to modulate its own activity. Nrg-1 also inhibits AChR clustering but does so in an ERK1/2-independent manner (?).
A weak activation of ERK1/2 induced by agrin in C2 myotubes was previously reported (22). However, these investigators only examined the activation 30 and 60 min after agrin application. By then, most of the activation has passed, as it was the strongest within the first 10 min after agrin treatment (Figs. 1 and 2). In addition, it was not established in that work whether the ERK1/2 activation was Lrp4/MuSK-dependent as it has been done here. Herbst and Burden (22) failed to observe any effects on AChR clustering by the MEK inhibitor PD 09857. The difference with our results may lie in that we used different MEK inhibitors, U0126 and PD0325901, or that we inhibited MEK for short periods of time. In our hands, shorter periods of MEK inhibitor treatment led to bigger potentiation of agrin-induced AChR clustering. Long-term inhibition of MEK (e.g. overnight treatment) may engage a different set of homeostatic events that might compensate for the loss of early ERK1/2 activation. Alternatively, the levels of MEK inhibitor may simply decay below the effective concentration during overnight treatment with agrin.
Experiments with the immorto+ cell lines (Fig. 2) revealed a complex set of changes in ERK1/2 activation induced by neural agrin. Although it is clear that the rapid activation following agrin application was Lrp4/MuSK-dependent, Lrp4/MuSK-independent agrin-induced ERK1/2 inactivation, which was evident much later, was also observed. In C2 cells, levels of active ERK1/2 below those in untreated control were consistently seen 1 h or longer after the initial application (Fig. 1 and data not shown). Herbst and Burden (22) also reported a weak ERK1/2 activation induced by agrin after 30 min that was no longer detectable at 60 min. Although it remains to be tested, this protracted ERK1/2 inactivation could result from the interaction of agrin with either α-dystroglycan and/or β1-containing integrins, surface molecules that have been shown to bind to neural and non-neural forms of agrin (20, 28). It is tempting to speculate that this ERK1/2 inactivation might play some role in AChR clustering, not so much in the induction phase, which depends on Lrp4 and MuSK, but in the maintenance phase, for which a role for α-dystroglycan (29–31) and β1-containing integrins (21, 32) has been proposed previously.
More than 20 years ago, a comprehensive survey of drugs affecting agrin-induced AChR clustering on primary chick myotubes revealed that PMA is a potent inhibitor of this process (17). Since then, little has been learned about the mechanism used by PMA to inhibit AChR clustering, despite the appealing possibility that proteins targeted by PMA treatment may be critical components of the agrin signaling pathway. It is demonstrated here that in C2 myotubes activation of ERK1/2 is downstream of PKC activation by PMA and that it mediates the inhibitory effect of PMA on agrin-induced AChR clustering. As agrin activates ERK1/2 in an Lrp4/MuSK-dependent manner, it is likely that there are common downstream substrates between the ERK1/2 activated by PMA and ERK1/2 activated by agrin, especially because in both cases AChR clustering is inhibited. The former is just a more potent inhibitor than the latter because PMA induces sustained, higher levels of ERK1/2 activation, and because agrin also triggers other major signaling events that promote clustering. Thus, PMA may be co-opting pathways normally used by agrin to modulate its own activity (i.e. agrin-induced ERK1/2 activation) to drastically inhibit it. Rescue of agrin-induced AChR by MEK inhibitors was full when the incubation time with PMA was short (Fig. 5) and partial when the incubation time with PMA was long (Fig. 4). It is likely that long-term incubation with PMA triggers many cellular/molecular events that are unrelated to AChR clustering per se, such as changes in AChR gene expression (27), which are ERK1/2 independent and hence, cannot be restored with MEK inhibitors. On the other hand, short-term incubation may not produce these unspecific events, allowing full rescue of AChR clustering with MEK inhibitors.
Unlike PMA, Nrg-1 does not require ERK1/2 activation to inhibit agrin-induced AChR clustering in C2 myotubes. In our hands, levels of Nrg-1- and PMA-induced ERK1/2 activation were clearly comparable but more sustained than those induced by agrin (Figs. 1 and 4). Thus, differences in the amount of ERK1/2 activation are unlikely to account for the different requirements for ERK1/2 activation for PMA and Nrg-1 regarding inhibition of AChR clustering. Although persistence of ERK1/2 activation is an important issue, it may be as important where in the cell active ERK1/2 localizes. It is widely recognized that ERK1/2 signaling uses different scaffold proteins to localize the signaling to distinct parts of the cell and thus confer specificity to it (33). PMA activates PKC because it is analogous to diacylglycerol, which is generated at the inner leaflet of the plasma membrane by the action of phospholipase C on phosphoinositol biphosphate. Hence, most of the ERK1/2 activated downstream of PKC may be closely associated with the membrane via unknown scaffold proteins. Likewise, the ERK1/2 activated by agrin could also be closely associated with the sarcolemma, especially as this activation is rapid and transient. In this context, it has recently been reported that the juxtamembrane domain of β-dystroglycan can bind MEK and active ERK1 (but not ERK2) in non-muscle cells (34). The ERK1 binding site on β-dystroglycan (35) overlaps rapsyn's (36) suggesting potential competition between these β-dystroglycan ligands. Interestingly, overexpression of β-dystroglycan inhibits agrin-induced AChR clustering in cultured myotubes (37). On the other hand, most of the ERK1/2 activated by Nrg-1 presumably goes into the myonuclei, away from the sarcolemma where AChR clustering occurs, as it is necessary for inducing AChR transcription in cultured myotubes (15, 16).
The observation that Nrg-1 does not require ERK1/2 activation for inhibiting agrin-induced AChR clustering (this work), but does so for stimulating AChR transcription (38) provides a parsimonious explanation for these apparently paradoxical activities in Nrg-1. Thus, Nrg-1 can stimulate AChR transcription, whereas at the same time inhibit AChR clustering because it uses different signaling pathways to do so.
What is the biological significance of agrin-induced ERK1/2 activation? At least three possible roles could be considered here. (i) Close apposition between the pre- and post-synaptic apparatus during formation and maturation of the NMJ is of paramount importance. It is generated by several mechanisms such as local release of agrin at the nerve terminal and its deposition in the synaptic basal lamina as well as concentration of Lrp4/MuSK on the postsynaptic sarcolemma. Perhaps, the quenching of the agrin clustering signal produced by rapid agrin-induced ERK1/2 activation might contribute to further localize this signal at the synaptic site and to prevent its spreading. (ii) One of the earliest events following nerve-muscle contact, both in vitro and in vivo, is the formation of microprocesses on the muscle fiber surface called myopodia (39, 40). However, it is unclear what role myopodia play in NMJ synaptogenesis. Recombinant agrin can induce myopodia on rat or Xenopus muscle cells within 2–4 h after application, before AChR clusters appear (39, 40). Hence, it may be possible that the rapid ERK1/2 activation induced by agrin plays a role in the formation of myopodia. (iii) Mice deficient for agrin, Lrp4, MuSK, and Dok-7 have profound post- and pre-synaptic defects. Presynaptic defects are supposedly due to lack of proper agrin-induced retrograde signaling from muscle to nerve that accounts for the maturation of the growth cone into a nerve terminal. The intracellular pathways that mediate this signaling are unknown, however, it seems that β-catenin is a key player (41). It is tempting to suggest that agrin-induced ERK1/2 activation might be part of a retrograde signaling pathway that regulates pre-synaptic differentiation.
Acknowledgments
I thank Krunal Desai, Ximena Paez, Rami Weaver, and Liping Guo for technical assistance. I am grateful to Steve Burden and Ruth Herbst for providing wild-type and mutant immorto+ cell lines and Gregg Wells for statistical advice as well as for critical reading of the manuscript.
This work was supported by the Texas A&M Health Science Center College of Medicine.
- NMJ
- neuromuscular junction
- AChR
- acetylcholine receptor
- C2DM
- C2 differentiation medium
- ERK1/2
- extracellular signal-regulated kinases 1 and 2
- Lrp4
- LDL-related protein 4
- MuSK
- muscle specific kinase
- Nrg-1
- neuregulin 1
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1.McMahan U. J. (1990) Cold Spring Harbor Symp. Quant. Biol. 55, 407–418 [DOI] [PubMed] [Google Scholar]
- 2.Valenzuela D. M., Stitt T. N., DiStefano P. S., Rojas E., Mattsson K., Compton D. L., Nuñez L., Park J. S., Stark J. L., Gies D. R. (1995) Neuron 15, 573–584 [DOI] [PubMed] [Google Scholar]
- 3.Glass D. J., Bowen D. C., Stitt T. N., Radziejewski C., Bruno J., Ryan T. E., Gies D. R., Shah S., Mattsson K., Burden S. J., DiStefano P. S., Valenzuela D. M., DeChiara T. M., Yancopoulos G. D. (1996) Cell 85, 513–523 [DOI] [PubMed] [Google Scholar]
- 4.Froehner S. C. (1984) J. Cell Biol. 99, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frail D. E., McLaughlin L. L., Mudd J., Merlie J. P. (1988) J. Biol. Chem. 263, 15602–15607 [PubMed] [Google Scholar]
- 6.Kim N., Stiegler A. L., Cameron T. O., Hallock P. T., Gomez A. M., Huang J. H., Hubbard S. R., Dustin M. L., Burden S. J. (2008) Cell 135, 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B., Luo S., Wang Q., Suzuki T., Xiong W. C., Mei L. (2008) Neuron 60, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada K., Inoue A., Okada M., Murata Y., Kakuta S., Jigami T., Kubo S., Shiraishi H., Eguchi K., Motomura M., Akiyama T., Iwakura Y., Higuchi O., Yamanashi Y. (2006) Science 312, 1802–1805 [DOI] [PubMed] [Google Scholar]
- 9.Gautam M., Noakes P. G., Moscoso L., Rupp F., Scheller R. H., Merlie J. P., Sanes J. R. (1996) Cell 85, 525–535 [DOI] [PubMed] [Google Scholar]
- 10.Gautam M., Noakes P. G., Mudd J., Nichol M., Chu G. C., Sanes J. R., Merlie J. P. (1995) Nature 377, 232–236 [DOI] [PubMed] [Google Scholar]
- 11.DeChiara T. M., Bowen D. C., Valenzuela D. M., Simmons M. V., Poueymirou W. T., Thomas S., Kinetz E., Compton D. L., Rojas E., Park J. S., Smith C., DiStefano P. S., Glass D. J., Burden S. J., Yancopoulos G. D. (1996) Cell 85, 501–512 [DOI] [PubMed] [Google Scholar]
- 12.Weatherbee S. D., Anderson K. V., Niswander L. A. (2006) Development 133, 4993–5000 [DOI] [PubMed] [Google Scholar]
- 13.Trinidad J. C., Cohen J. B. (2004) J. Biol. Chem. 279, 31622–31628 [DOI] [PubMed] [Google Scholar]
- 14.Ponomareva O. N., Ma H., Vock V. M., Ellerton E. L., Moody S. E., Dakour R., Chodosh L. A., Rimer M. (2006) Mol. Cell. Neurosci. 31, 334–345 [DOI] [PubMed] [Google Scholar]
- 15.Si J., Luo Z., Mei L. (1996) J. Biol. Chem. 271, 19752–19759 [DOI] [PubMed] [Google Scholar]
- 16.Tansey M. G., Chu G. C., Merlie J. P. (1996) J. Cell Biol. 134, 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace B. G. (1988) J. Cell Biol. 107, 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross A., Rapuano M., Prives J. (1988) J. Cell Biol. 107, 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rimer M., Prieto A. L., Weber J. L., Colasante C., Ponomareva O., Fromm L., Schwab M. H., Lai C., Burden S. J. (2004) Mol. Cell. Neurosci. 26, 271–281 [DOI] [PubMed] [Google Scholar]
- 20.Bowe M. A., Deyst K. A., Leszyk J. D., Fallon J. R. (1994) Neuron 12, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 21.Martin P. T., Sanes J. R. (1997) Development 124, 3909–3917 [DOI] [PubMed] [Google Scholar]
- 22.Herbst R., Burden S. J. (2000) EMBO J. 19, 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., Copeland R. A., Magolda R. L., Scherle P. A., Trzaskos J. M. (1998) J. Biol. Chem. 273, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 24.Mittaud P., Camilleri A. A., Willmann R., Erb-Vögtli S., Burden S. J., Fuhrer C. (2004) Mol. Cell. Biol. 24, 7841–7854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Shanti N., Stewart C. E. (2008) J. Endocrinol. 198, 243–252 [DOI] [PubMed] [Google Scholar]
- 26.Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bursztajn S., Schneider L. W., Jong Y. J., Berman S. A. (1988) Biol. Cell 63, 57–65 [PubMed] [Google Scholar]
- 28.Burgess R. W., Dickman D. K., Nunez L., Glass D. J., Sanes J. R. (2002) J. Neurochem. 83, 271–284 [DOI] [PubMed] [Google Scholar]
- 29.Jacobson C., Côté P. D., Rossi S. G., Rotundo R. L., Carbonetto S. (2001) J. Cell Biol. 152, 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson C., Montanaro F., Lindenbaum M., Carbonetto S., Ferns M. (1998) J. Neurosci. 18, 6340–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heathcote R. D., Ekman J. M., Campbell K. P., Godfrey E. W. (2000) Dev. Biol. 227, 595–605 [DOI] [PubMed] [Google Scholar]
- 32.Schwander M., Shirasaki R., Pfaff S. L., Müller U. (2004) J. Neurosci. 24, 8181–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvo F., Agudo-Ibáñez L., Crespo P. (2010) Bioessays 32, 412–421 [DOI] [PubMed] [Google Scholar]
- 34.Spence H. J., Dhillon A. S., James M., Winder S. J. (2004) EMBO Rep. 5, 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore C. J., Winder S. J. (2010) Cell Commun. Signal 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cartaud A., Coutant S., Petrucci T. C., Cartaud J. (1998) J. Biol. Chem. 273, 11321–11326 [DOI] [PubMed] [Google Scholar]
- 37.Kahl J., Campanelli J. T. (2003) J. Neurosci. 23, 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Si J., Mei L. (1999) Brain Res. Mol. Brain. Res. 67, 18–27 [DOI] [PubMed] [Google Scholar]
- 39.Uhm C. S., Neuhuber B., Lowe B., Crocker V., Daniels M. P. (2001) J. Neurosci. 21, 9678–9689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madhavan R., Zhao X. T., Reynolds A. B., Peng H. B. (2006) J. Neurobiol. 66, 1511–1527 [DOI] [PubMed] [Google Scholar]
- 41.Li X. M., Dong X. P., Luo S. W., Zhang B., Lee D. H., Ting A. K., Neiswender H., Kim C. H., Carpenter-Hyland E., Gao T. M., Xiong W. C., Mei L. (2008) Nat. Neurosci. 11, 262–268 [DOI] [PubMed] [Google Scholar]