Abstract
Aggregation and fusion of lipoproteins trigger subendothelial retention of cholesterol, promoting atherosclerosis. The tendency of a lipoprotein to form fused particles is considered to be related to its atherogenic potential. We aimed to isolate and characterize aggregated and nonaggregated subfractions of LDL from human plasma, paying special attention to particle fusion mechanisms. Aggregated LDL was almost exclusively found in electronegative LDL (LDL(−)), a minor modified LDL subfraction, but not in native LDL (LDL(+)). The main difference between aggregated (agLDL(−)) and nonaggregated LDL(−) (nagLDL(−)) was a 6-fold increased phospholipase C-like activity in agLDL(−). agLDL(−) promoted the aggregation of LDL(+) and nagLDL(−). Lipoprotein fusion induced by α-chymotrypsin proteolysis was monitored by NMR and visualized by transmission electron microscopy. Particle fusion kinetics was much faster in agLDL(−) than in nagLDL(−) or LDL(+). NMR and chromatographic analysis revealed a rapid and massive phospholipid degradation in agLDL(−) but not in nagLDL(−) or LDL(+). Choline-containing phospholipids were extensively degraded, and ceramide, diacylglycerol, monoacylglycerol, and phosphorylcholine were the main products generated, suggesting the involvement of phospholipase C-like activity. The properties of agLDL(−) suggest that this subfraction plays a major role in atherogenesis by triggering lipoprotein fusion and cholesterol accumulation in the arterial wall.
Keywords: Atherosclerosis, Lipoprotein Structure, Low Density Lipoprotein (LDL), NMR, Phospholipase C, Electronegative LDL, Lipoprotein Aggregation
Introduction
Atherosclerosis is a consequence of the excessive deposition of cholesterol in the intimal vessel wall, coming mainly from plasma LDL. It is widely accepted that lipoprotein retention mediated by the binding of LDL to proteoglycans is a key event in atherogenesis (1). In this environment, chemical modification of trapped LDL triggers an inflammatory response that promotes localized leukocyte recruitment, cell proliferation, and apoptotic processes (2). Oxidation of LDL is the most widely studied mechanism of modification, and oxidized lipids play a central role in inflammation, proliferation, and apoptosis (3). However, other modifications, such as degradation by phospholipases, cholesteryl esterase, or proteases, also occur in the intima of the arterial wall and have a more relevant role than oxidation in lipoprotein trapping (4). These modifications favor LDL aggregation and fusion in the arterial wall, precluding its exit to the bloodstream and accelerating the development of the disease (5). Thus, the tendency of a lipoprotein to form fused particles is related to its atherogenic potential.
In addition to the modified forms of LDL that occur in the vessel wall, several minor forms have been detected in blood. A common characteristic is an increase of the electronegative charge of the particle. This feature has been used to isolate electronegative LDL (LDL(−)).7 It accounts for 5% of total LDL in healthy subjects (6). Growing evidence suggests a close relationship between LDL(−) and atherosclerosis. The proportion of LDL(−) is increased in pathologies with a high incidence of cardiovascular events, such as familial hypercholesterolemia (7), hypertriglyceridemia (8), diabetes (9), or severe renal disease (10). In addition, drugs known to decrease cardiovascular risk, such as statins or insulin, decrease LDL(−) proportion independently of changes in total LDL cholesterol (7, 9). Studies performed in endothelial and mononuclear cells have shown that LDL(−) induces inflammation and apoptosis and impairs angiogenesis (11–14). Regarding its physicochemical characteristics, apolipoprotein B-100 (apoB) in LDL(−) presents structural differences versus apoB in native LDL (15), binds poorly to the LDL receptor (16), is prone to aggregation (17), and presents amyloidogenic properties (18). Our group recently reported that LDL(−) has several populations with normal or high binding affinity to proteoglycans compared with native LDL (1). LDL(−) fractions with increased affinity to proteoglycans presented a higher content of aggregated particles. This finding suggests that an aggregated subfraction of LDL(−) could be retained more strongly in the arterial wall than the bulk of nonaggregated LDL(−). However, the specific characteristics of this aggregated subpopulation have not been studied to date.
The aim of the current study was to isolate aggregated and nonaggregated LDL(−) fractions from human plasma and study their physicochemical characteristics. Special attention was paid to the mechanism of particle fusion because this feature plays a key role in subendothelial lipoprotein retention (20). Our results show that aggregated LDL(−) has high intrinsic phospholipase (PLC)-like activity. This activity makes this subfraction prone to particle fusion and promotes the aggregation of nonaggregated LDL subfractions. These properties suggest that the aggregated LDL(−) subfraction has a major atherogenic role, favoring subendothelial cholesterol accumulation.
EXPERIMENTAL PROCEDURES
Materials
All of the reagents were purchased from Sigma unless otherwise stated.
Isolation of LDL Subfractions
The study was approved by the institutional ethics committee, and all volunteers gave informed consent. Total LDL (density range, 1.019–1.050 g/ml) was isolated from plasma of healthy volunteers by sequential ultracentrifugation using KBr gradients. VLDL and intermediate density lipoprotein (IDL) were isolated in 6 h at 200,000 × g (50,000 rpm), and LDL was then obtained after a further 14 h at the same centrifugal force. Total LDL was subfractionated in native LDL (LDL(+)) and LDL(−) by stepwise anion exchange chromatography in an AKTA-FPLC system with a Hi-Load 26/10 Q-Sepharose column (GE Healthcare) using buffer A (10 mmol/liter Tris, 1 mmol/liter EDTA, 2 μmol/liter butylated hydroxytoluene (BHT), pH 7.4) and buffer B (10 mmol/liter Tris, 1 mmol/liter EDTA, 2 μmol/liter BHT, 1 mol/liter NaCl, pH 7.4), as described (21). LDL(+) eluted at 0.24 mol/liter NaCl, and LDL(−) eluted at 0.5 mol/liter NaCl. Both subfractions were concentrated by centrifugation with Amicon microconcentrators (10,000 molecular weight cut-off; Amicon Ultra-4, Millipore, Cork, Ireland) and submitted to gel filtration chromatography to fractionate aggregated and nonaggregated fractions. Gel filtration chromatography was performed using two on-line connected Superose 6 columns in an AKTA-FPLC system (GE Healthcare). Two ml of LDL(+) or LDL(−) at 1 g of protein/liter were eluted with buffer 10 mmol/liter Tris, 150 mmol/liter NaCl, 1 mmol/liter EDTA, 2 μmol/liter BHT, pH 7.4, at a flow of 1 ml/min, and fractions of 1 ml were collected and concentrated with Amicon microconcentrators. All of the isolation steps were performed at 4 °C, and the buffers and density solutions contained 1 mmol/liter EDTA and 2 μmol/liter BHT. All of the analyses were performed within 3 days of isolation except HPLC and TLC studies, in which lipid extracts were made within 3 days and dry pellets were frozen at −80 °C under nitrogen and analyzed within 1 month.
Composition of LDL Subfractions
Major components of LDL subfractions, including apoB, total cholesterol, triglyceride, total phospholipids, nonesterified fatty acids (NEFA), and free glycerol were measured by commercial methods adapted to a Hitachi 917 autoanalyzer, as described (21, 22).
ApoE, apoC-III, apoA-I, and apoA-II (Kamiya, Seattle, WA) were quantified manually using commercial methods adapted to measure low concentrations (19). Briefly, 100 μl of reagent 1 (Tris buffer) were mixed with 100 μl of LDL at 0.4 g of protein/liter and incubated at 37 °C for 30 min. Afterward, 50 μl of reagent 2 (antiserum) were added and incubated at 37 °C for 30 min. Absorbance was measured in a microtiter plate reader at the wavelength recommended by the manufacturer. A standard curve was performed with an apolipoprotein multi-calibrator (Wako Chemicals, Richmond, VA) diluted 1/5 to 1/100. Total protein content was determined by the bicinchoninic acid method (Pierce).
Phosphatidylcholine (PC), sphingomyelin (SM), and lysophosphatidylcholine (LPC) were quantified by normal phase HPLC in a Gold System chromatograph equipped with a diode array detector (Beckman, Palo Alto, CA), as described (23). Dipalmitoyl-glycero-phosphodimethyl ethanolamine was used as an internal standard. Briefly, 25 μl of dipalmitoyl-glycero-phosphodimethyl ethanolamine at 20 g/liter in ethanol were added to 200 μl of LDL (0.5 g of protein/liter), and lipids were extracted by the Bligh and Dyer method. The pellet was resuspended with 80 μl of hexane/isopropanol/water (v/v/v 6:8:1), and 20 μl of this solution were injected into a normal phase column (Luna 5-μm silica; Phenomenex, Torrance, CA) at a flow rate of 1 ml/min. The mobile phase was acetonitrile:methanol:ammonium sulfate 5 mmol/liter (v/v/v 56:23:6). The peaks were detected at 205 nm using a photo diode array detector (model 168; Beckman). Lipoperoxidation was estimated by measuring the absorbance at 234 nm of the PC peak (that corresponds to oxidized PC) and expressed as the 205/234-nm ratio (17).
α-Tocopherol content in LDL subfractions was determined by reverse phase HPLC in a Gold System chromatograph (24). Tocopherol acetate was used as an internal standard. Briefly, 250 μl of tocopherol acetate at 10 mg/liter in ethanol were added to 250 μl of LDL (0.5 g of protein/liter), and lipids were extracted with 0.5 ml of heptane and intense agitation for 1 min. The organic phase was evaporated under nitrogen. The pellets were resuspended with 80 μl of ethanol, and 20 μl were injected into a reverse phase column (Ultrasphere ODS; Beckman, Palo Alto, CA) at a flow rate of 1 ml/min. The mobile phase was acetonitrile/isopropanol/water (v/v/v 85:13:2). The peaks were detected at 290 nm using a photo diode array detector. Susceptibility to oxidation was measured by continuous monitoring of conjugated diene formation at 234 nm at 37 °C, using 100 μl of LDLs (0.05 g of protein/liter) dialyzed in PBS and 5 μmol/liter CuSO4 in a UV microtiter plate.
TLC to identify major lipids was performed in silica gel plates (Partisil LK5DF; Whatmann, Middlestone, UK). The lipids were extracted according to Bligh and Dyer using 100 μl of LDL at 0.5 g of protein/liter. The pellets were resuspended with 20 μl of chloroform and applied to the plate. The plates were developed using three sequential phases: chloroform/methanol/water (v/v/v 65:40:5) to 5 cm, toluene/diethyl ether/ethanol (v/v/v 60:40:3) to 13 cm, and heptane to 17 cm. The lipids were stained by dipping the plates in phosphomolybdate solution (5% phosphomolybdate and 5% sulfuric acid in ethanol) at room temperature for 1 min and then incubated for 7 min at 100 °C.
Electrophoretic Studies
LDL particle size was analyzed by nondenaturing acrylamide gradient gel electrophoresis (GGE), in 2–16% gels, as described (8). Briefly, two solutions at 2 and 16% were prepared using a stock solution of acrylamide and bisacrylamide (30% total, 5% cross-linker) and mixed using two peristaltic pumps. Fifteen μl of each LDL subfraction at 0.3 g of protein/liter were preincubated for 15 min with 5 μl of Sudan Black (0.1% w/v in ethylene glycol), and 5 μl of sucrose (50% w/v). Fifteen μl of this mixture were electrophoresed at 4 °C for 30 min at 20 V, 30 min at 70 V, and 8 h at 100 V.
Phospholipolytic Activities
Platelet-activating factor acetylhydrolase (PAF-AH) activity was measured using 2-thio PAF (Cayman Chemicals, Tallin, Estonia) as substrate (25). LDL fractions (10 μl of LDL at 0.25 g of protein/liter, in Tris 10 mm, EGTA 1 mm, pH 7.2) were incubated with 2-thio-PAF and 5,5′-dithiobis(2-nitrobenzoic) acid as indicated by the manufacturer. Absorbance was measured at 414 nm at increasing times in a microtiter plate, and the slope was used to calculate PAF-AH activity (expressed as μmol/min·mg of protein).
PLC-like activity was quantified by the Amplex Red method (Cayman Chemicals) using LPC (lysophospholipase C activity) or SM (sphingomyelinase (SMase) activity) as substrate (17). LDL subfractions (100 μl at 0.3 g of protein/liter) were mixed with 10 μl of substrate (LPC or SM at 5 mmol/liter in reaction buffer containing ethanol 5% and Triton X-100 2%; final concentration, 0.25 mmol/liter) and with the appropriate amount of Amplex Red, alkaline phosphatase, choline oxidase, peroxidase, and reaction buffer (100 mmol/liter Tris, 10 mmol/liter MgCl2, pH 7.4), following the manufacturer's instructions. Staphylococcus sp. SMase (100 μl at 0, 0.1, 1, 10, 50, 100, and 200 milliunits/ml, final concentration) was used as a standard curve. Fluorescence production (λ excitation, 530 nm; λ emission, 590 nm) was monitored for 3 h, and the results were calculated from the maximum curve slope.
SMase activity was also determined by measuring the degradation of fluorescently labeled SM (BODIPY-SM) and separation of substrate (BODIPY-SM) and product (BODIPY-ceramide) by TLC (17). Activity of LDL subfractions (0.3 g of protein/liter) was evaluated by incubation with BODIPY-FL-C12-SM at 0.025 mmol/liter, (Molecular Probes, Leiden, The Netherlands) as substrate for 3 h at 37 °C in buffer 10 mmol/liter Tris, 10 mmol/liter Cl2Mg, pH 7.4, and separation by TLC, as described (17). The TLC plate was developed with 1,2-dichloroethane/methanol/water (v/v/v 90:20:0.5) to separate BODIPY-ceramide from BODIPY-SM.
α-Chymotrypsin-induced Fusion of LDL Subfractions
LDL subfractions at 1.33 g of protein/liter in PBS, containing 10 μmol/liter EDTA, 1 μmol/liter BHT, and 1 μmol/liter sodium azide were mixed with α-chymotrypsin at 0.4 g/liter in the same buffer to render a final concentration of 1.0 g of protein/liter of LDL protein and 0.1 g/liter of protease, as previously described (26). Aggregation and fusion of LDL particles were analyzed by GGE, 1H NMR, and transmission electron microscopy (TEM), as follows.
1H NMR Spectroscopy of α-Chymotrypsin-treated Subfractions
A sealed coaxial insert (outer diameter, 1.7 mm; supported by a Teflon adapter) containing 10 mmol/liter 2,2-dimethyl-2-silapentane-5-sulfonic acid in H2O was used with one of the samples as an external reference for the 1H chemical shift. This calibration was transferred to the other samples. 1H NMR spectra were acquired every 30 min for 48 h at 37 °C on a Bruker 800 MHz spectrometer. A period of 25 min was necessary for sample insertion, experimental set-up, and thermal stabilization before the acquisition of the first NMR spectrum. For this reason, the spectra at time 0 could not be acquired. The content of choline-containing phospholipids (3.170–3.280 ppm) (27, 28), ceramide (5.250–5.350 ppm) (28), and bisallylic compounds (2.650–5.850 ppm) (29) was measured by area integration of the corresponding resonance signal.
TEM of α-Chymotrypsin-treated Subfractions
LDL subfractions treated with α-chymotrypsin for 0, 3, and 24 h were absorbed and processed for negative staining with 2% potassium phosphotungstate, pH 7.0, over carbon-coated grids (30). Micrographs were obtained using a Jeol 120-kV JEM-1400 transmission electron microscope, with an Erlangshen ES1000W CCD camera (Gatan, Abingdon, UK).
Statistical Analysis
The results are expressed as the means ± SD. The SPSS 17 statistical package was used. Differences between LDL subfractions were tested with Wilcoxon's t test for paired data.
RESULTS
Separation of LDL Subfractions
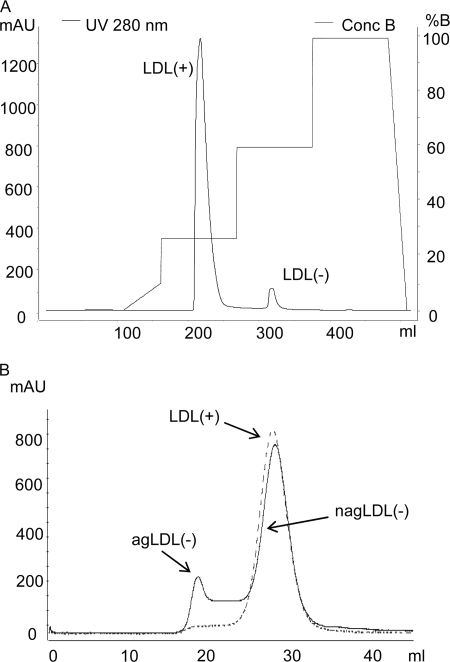
Fig. 1 shows representative chromatograms performed to isolate, in a first step, LDL(+) and LDL(−) fractions by anion exchange chromatography (Fig. 1A) and, in a second step, aggregated (agLDL(−)) and nonaggregated (nagLDL(−)) subfractions by gel filtration chromatography (Fig. 1B). The total LDL(−) proportion ranged from 3.5 to 7.0% (5.3 ± 1.2%, n = 15). Gel filtration chromatography showed that LDL(+) presented a single peak corresponding to nonaggregated LDL, although traces of aggregated particles were detectable. In contrast, LDL(−) had two well defined peaks of agLDL(−) and nagLDL(−) particles. In all of the plasma pools analyzed, agLDL(−) ranged from 5 to 20% of total LDL(−) (mean ± S.D. 14 ± 8%, n = 15), measured by its cholesterol content. Thus, agLDL(−) represents ∼0.25–1% (mean ± S.D. 0.74 ± 0.61%) of total LDL in human normolipemic plasma.
FIGURE 1.
Representative chromatograms of anion exchange (A) and gel filtration (B) chromatography. A, 50 ml of total LDL were injected into a HiLoad Q-Sepharose 26/10 column using an AKTA-FPLC system and eluted at 8 ml/min using a stepwise gradient method. After concentration of subfractions, 2 ml of LDL(+) (dashed line) or LDL(−) (solid line) were injected in two on-line connected Superose 6 columns in an AKTA-FPLC system and eluted at 1 ml/min.
The presence of aggregated and/or fused particles in LDL(−) subfractions was confirmed by measuring the turbidity of samples and by performing GGE. Supplemental Fig. 1S (panel A) shows the turbidity of LDL subfractions measured at 450 nm, as an index of aggregation. Total LDL(−) had a higher turbidity than LDL(+), whereas agLDL(−) presented higher turbidity than nagLDL(−). Supplemental Fig. 1S (panel B) shows a representative GGE of LDL(+) and LDL(−) fractions, where lipoproteins run according to their size.
To demonstrate that aggregated LDL was not formed during isolation, two strategies were used. First, a simplified protocol that uses a smaller salt concentration and skips the anion exchange chromatography (see supplemental material for details) allowed the separation of a fraction with similar characteristics to agLDL(−) including high PLC-like activity (supplemental Fig. 2S). Second, LDL(+) isolated by the regular method and depleted of aggregated particles by gel filtration (nagLDL(+)) was reisolated by ultracentrifugation (see supplemental material for details). No evidence of aggregated particle formation was observed in reisolated nagLDL(+) (supplemental Fig. 3S). These observations indicate that agLDL(−) is not an artifact of the isolation procedure and that it is present in blood.
Lipid and Protein Composition
Differences between LDL(+) and total LDL(−) in the relative content of major components (Table 1) concurred with previous data (21). The composition of LDL subfractions showed dramatic differences between agLDL(−) and nagLDL(−), with total LDL(−) presenting an intermediate composition between the two subfractions. agLDL(−) had less cholesterol and phospholipid and more triglyceride than nagLDL(−). These features are compatible with VLDL or IDL, the metabolic precursors of LDL. Both lipoproteins have lower density, higher size, and higher content of apoE and apoC-III than LDL. However, the content of total protein was similar in all LDL subfractions, indicating similar density (Table 1). To investigate the possibility that VLDL or IDL could contaminate agLDL(−), we measured the content of minor apolipoproteins. ApoC-III, apoE, apoA-I, and apoA-II were increased in LDL(−) subfractions compared with LDL(+), in agreement with previous reports (21, 31). agLDL(−) had a lower content of apoE and apoC-III and higher content of apoA-I and apoA-II than nagLDL(−). The amount of apoE and apoC-III in agLDL(−) was much lower than usually reported (32) in VLDL or IDL. Indeed, apoA-I and apoA-II also were increased in both LDL(−) subfractions, and their content was higher in agLDL(−) compared with nagLDL(−). Taken together, these data make it unlikely that agLDL(−) could be remnant VLDL or IDL.
TABLE 1.
Composition of LDL subfractions (n = 9; except for apoE, apoC-III, apoA-I, and apoA-II, for which n = 4)
| LDL(+) | Total LDL(−) | nagLDL(−) | agLDL(−) | |
|---|---|---|---|---|
| Major componentsa | ||||
| Cholesterol (%) | 42.5 ± 1.9 | 41.4 ± 2.2 | 42.2 ± 2.0 | 38.3 ± 3.3c,d,e |
| Triglyceride (%) | 6.5 ± 1.0 | 10.0 ± 2.5c | 7.8 ± 1.5c,d | 15.0 ± 3.8c,d,e |
| Phospholipid (%) | 29.6 ± 1.2 | 27.4 ± 1.9c | 28.4 ± 1.2 | 24.9 ± 2.6c,d,e |
| Total protein (%) | 21.4 ± 1.8 | 21.2 ± 2.1 | 21.6 ± 1.7 | 21.8 ± 1.8 |
| Minor componentsb | ||||
| NEFA (mol/mol protein) | 25.0 ± 5.5 | 49.7 ± 15,4c | 43.9 ± 13.4c | 57.9 ± 16.9c,d,e |
| ApoE (mmol/mol protein) | 21.2 ± 15.6 | 90.5 ± 21.4 | 87.8 ± 17.3 | 61.7 ± 23.7 |
| ApoC-III (mmol/mol protein) | 37.2 ± 20.2 | 160.0 ± 108.9 | 166.5 ± 116.2 | 80.2 ± 68.1 |
| ApoA-I (mmol/mol protein) | 54.1 ± 6.6 | 259.9 ± 45.2 | 151.7 ± 6.7 | 328.6 ± 111.8 |
| ApoA-II (mmol/mol protein) | 14.2 ± 8.3 | 49.0 ± 6.4 | 39.5 ± 6.1 | 61.3 ± 11.0 |
a Cholesterol, triglyceride, phospholipid, and protein are expressed as percentages of total mass.
b NEFA, apoE, apoC-III, apoA-I, and apoA-II are expressed as mol/mol protein or as mmol/mol protein, considering the molecular mass of apoB (550 kDa).
c p < 0.05 vs. LDL(+).
d p < 0.05 vs. total LDL(−).
e p < 0.05 vs. nagLDL(−).
Oxidative Characteristics
We estimated the oxidative level of LDL subfractions by measuring the ratio of the PC peak areas at 234 nm (corresponding to conjugated diene) and 205 nm (corresponding to the maximum of absorbance of PC) and by quantifying the α-tocopherol content. In contrast with in vitro oxidized LDL, which presented a much lower 205/234 PC ratio and α-tocopherol content, no difference was observed among LDL subfractions (supplemental Table 1S). The evaluation of LDL susceptibility to CuSO4-induced oxidation showed that LDL(+) was slightly more susceptible to oxidation than nagLDL(−), whereas agLDL(−) was much more resistant than LDL(+) and nagLDL(−). Representative conjugated diene kinetics is shown in supplemental Fig. 4S. The percentage of increase in lag phase time is shown in supplemental Table 1S.
Phospholipolytic Activities
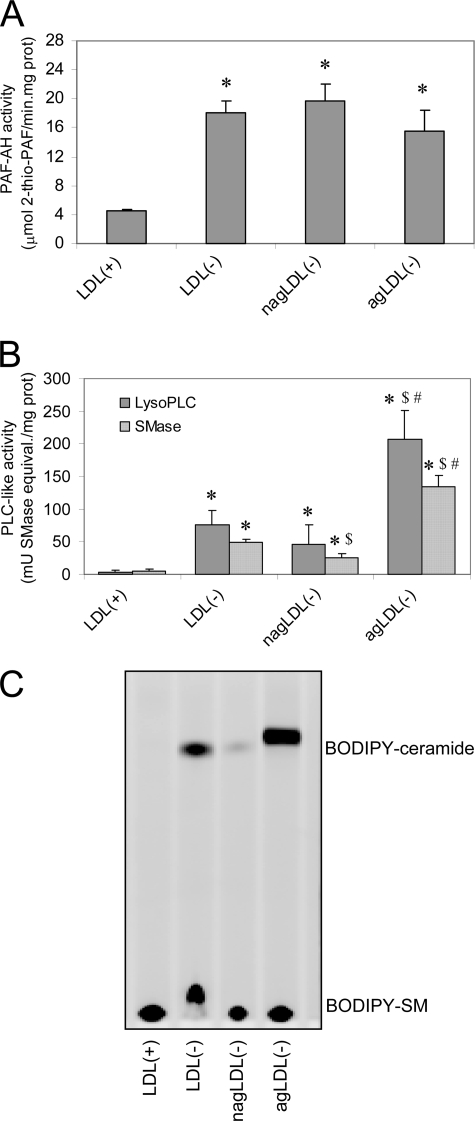
PAF-AH activity was increased in all LDL(−) subfractions compared with LDL(+), but no difference was observed between agLDL(−) and nagLDL(−) subfractions (Fig. 2A). In contrast, dramatic differences were observed in SMase and lysophospholipase C activities between agLDL(−) and nagLDL(−) (Fig. 2B). Both LDL(−) subfractions had higher PLC-like activities than LDL(+). However, agLDL(−) presented an activity 4–6-fold higher than nagLDL(−). To confirm differences among LDL subfractions in PLC-like activity, SMase activity was tested by evaluating the degradation of exogenous SM labeled with the fluorescent probe BODIPY (BODIPY-SM); TLC was used to separate the product of the reaction (BODIPY-ceramide) from the substrate (BODIPY-SM). Fig. 2C shows the increased SMase activity in nagLDL(−) compared with LDL(+) and that of agLDL(−) compared with nagLDL(−). PLC-like activity in VLDL, IDL, and HDL, measured by the Amplex Red method, was similar to LDL(+) (data not shown).
FIGURE 2.
Phospholipolytic activities associated to LDL subfractions. A, PAF-AH activity was measured by monitoring the degradation of 2-thio-PAF. The data are the means ± S.D. of four independent experiments. *, p = 0.064 versus LDL(+). B, PLC-like activities were measured by the Amplex Red method that quantifies the formation of choline, using SM (SMase activity) or LPC (lysophospholipase C (LysoPLC) activity) as a substrate. The data are the means ± S.D. of five independent experiments. *, p < 0.05 versus LDL(+); $, p < 0.05 versus LDL(−); #, p < 0.05 versus nagLDL(−). C, representative TLC of LDL subfractions after incubation with BODIPY-SM. Fluorescence of BODIPY was detected in a Chemi-Doc densitometer. BODIPY-SM stands at the bottom of the plate, whereas BODIPY-ceramide runs to the top of the plate.
Aggregation of Nonaggregated LDL Subfractions Induced by agLDL(−)
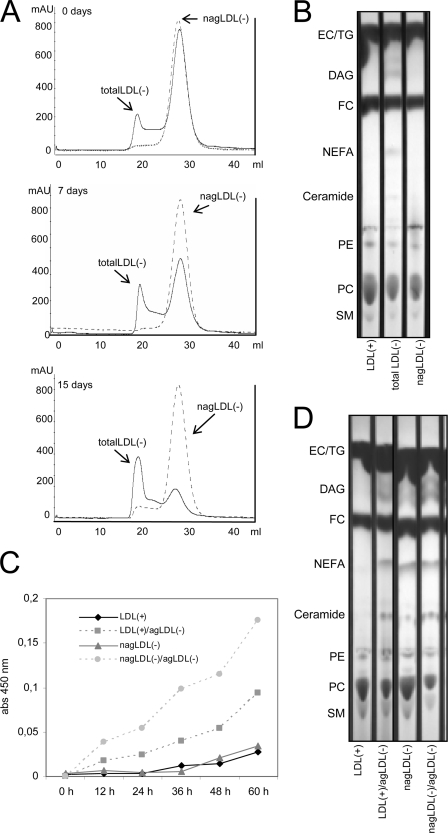
agLDL(−) promoted the aggregation of LDL(+) and nagLDL(−) at 4 °C and 37 °C. The formation of agLDL(−) in total LDL(−) and nagLDL(−) was determined by gel filtration chromatography after 15 days of storage at 4 °C. The proportion of agLDL(−) increased progressively in total LDL(−) but not in nagLDL(−) (Fig. 3A). In the experiments performed at 37 °C, LDL(+) and nagLDL(−) (at 0.5 g/liter) were incubated alone or with agLDL(−) (at 0.1 g/liter) up to 60 h, and aggregation was monitored at 450 nm. LDL(+) and nagLDL(−) progressively increased their absorbance when coincubated with agLDL(−) (Fig. 3C). At both 4 and 37 °C, evidence of increased phospholipid degradation (decrease of PC and SM and increase of diacylglycerol (DAG), NEFA, and ceramide spots in TLC) was observed in samples containing agLDL(−) (total LDL(−) stored at 4 °C for 15 days (Fig. 3B) and LDL(+) and nagLDL(−) incubated with agLDL(−) at 37 °C for 60 h (Fig. 3D)). The fact that DAG and ceramide are the main products generated by the PLC-like activity present in agLDL(−) supports the involvement of such activity in the degradation of phospholipids.
FIGURE 3.
agLDL(−)-induced aggregation of LDL subfractions. A and B, LDL(+), total LDL(−), and nagLDL(−) were stored at 4 °C for 15 days in presence of 1 mmol/liter EDTA and 2 μmol/liter BHT. A, gel filtration chromatograms of total LDL(−) (continuous line) and nagLDL(−) (dotted line) at 0, 7, and 15 days of storage at 4 °C. B, TLC of LDL(+), total LDL(−), and nagLDL(−) after 15 days of storage at 4 °C. C and D, LDL(+) and nagLDL(−) (at 0.5 g/liter) were incubated for 60 h at 37 °C in the presence or absence of agLDL(−) (at 0.1 g/liter) (final volume, 200 μl). C, absorbance monitored at 450 nm. D, TLC of samples in C after 60 h of incubation. SM, sphingomyelin; PE, phosphatidylethanolamine; FC, free cholesterol; EC, esterified cholesterol; TG, triglyceride.
Susceptibility to Aggregation or Fusion Induced by Proteolysis of LDL Subfractions
This behavior is difficult to reproduce because the aggregation of total LDL(−), LDL(+), and nagLDL(−) induced by agLDL(−) was highly variable. To standardize the experimental conditions and accelerate the process, LDL subfractions were proteolysed with α-chymotrypsin, one of the most commonly used methods to induce lipoprotein aggregation and fusion (20).
GGE
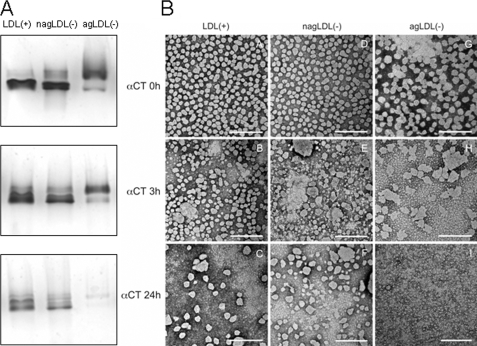
GGE showed that proteolytic degradation of apoB promoted the formation of aggregated and/or fused particles of greater size in both LDL(+) and nagLDL(−) (Fig. 4A). agLDL(−) bands corresponding to aggregated/fused particles decreased in size at 3 h of α-chymotrypsin and almost disappeared at 24 h.
FIGURE 4.
Effect of α-chymotrypsin treatment on the formation of aggregated or fused particles in LDL subfractions. A, GGE. Five μg of LDL (expressed as total protein) were run in each lane for 8 h at 100 V. LDLs were prestained with Sudan Black (lipid staining). B, electron micrographs of LDL subfractions. LDL(+) (panels A–C), nagLDL(−) (panels D–F), and agLDL(−) (panels G–I) subfractions were proteolysed by incubation with α-chymotrypsin (αCT) for 0 (panels A, D, and G), 3 (panels B, E, and H), and 24 h (panels C, F, and I). Transmission electron microscopy was performed in a JEM-1400 microscope, using 2% potassium phosphotungstate, pH 7.0, for negative staining. Bars, 200 nm.
TEM
GGE does not discriminate between aggregated or fused particles. This difference is relevant because aggregation is a reversible process, whereas fusion is irreversible (20). Hence, to study whether LDL subfractions were fused or aggregated after proteolytic treatment, TEM analysis was performed. Fig. 4B shows TEM micrographs of LDL subfractions at 0 (native samples), 3, and 24 h of α-chymotrypsin treatment. The particle size distribution is shown in supplemental Fig. 5S. At 0 h, LDL(+) and nagLDL(−) presented mainly monomeric particles, whereas agLDL(−) had evidence of aggregates, and some fused particles (>30 nm) were detected. At 3 h of proteolysis, fusion of particles was evident in all LDL subfractions, although this process was more intense in both LDL(−) subfractions than in LDL(+). Indeed, small 5–10-nm particles were observed in agLDL(−). After 24 h of proteolysis, LDL(+) and nagLDL(−) presented numerous fused particles and also small 5–10-nm particles. These small 5–10-nm particles were the only structures observed in agLDL(−) at 24 h. The appearance of particles smaller than 5–10 nm suggests LDL decomposition.
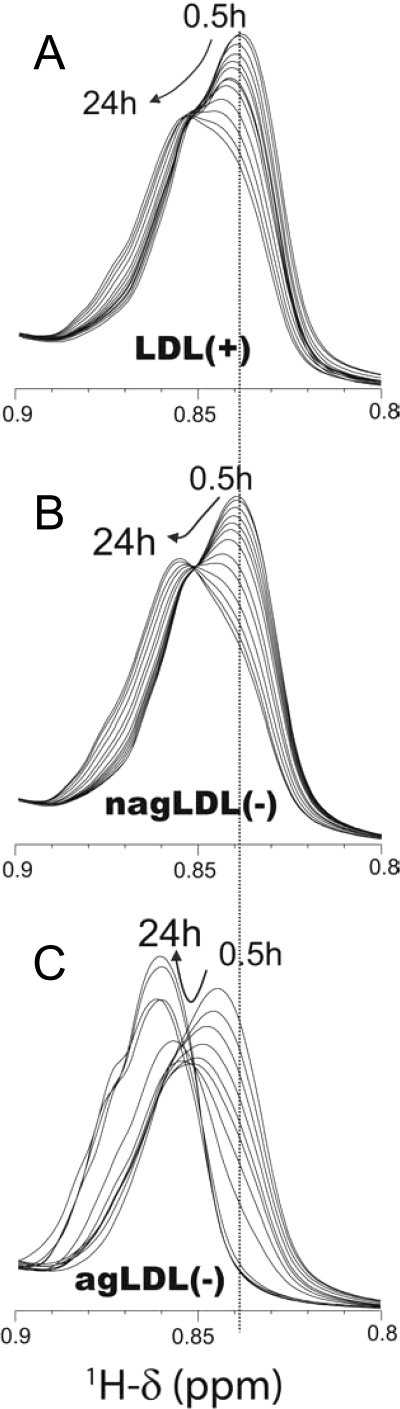
NMR
1H NMR has been used to evaluate the fusion of LDL particles by measuring the chemical shifts of fatty acid-CH3 resonances (33). Resonance shifting to larger ppm values indicates an increase in particle size. Fig. 5 shows 1H NMR spectra of LDL subfractions proteolysed with α-chymotrypsin from 30 min up to 24 h. At 30 min, LDL(+) and nagLDL(−) showed a resonance at 0.839 ppm, with a similar signal shape (Fig. 5, A and B). After 24 h of proteolysis, the resonance of LDL(+) and nagLDL(−) shifted up to 0.856 ppm. The only difference was slightly slower kinetics for the LDL(+) subfraction than for the nagLDL(−) (Fig. 5, A and B). In contrast, the behavior of agLDL(−) was dramatically different (Fig. 5C). The spectrum of agLDL(−) was already shifted up to 0.845 ppm at 30 min, and further shifts were progressively observed, reaching 0.861 ppm at 24 h. Thus, because agLDL(−) is aggregated at time 0 (as shown with GGE and TEM), what we observed at 30 min in the NMR spectra was the rapid fusion of the particles. These results are consistent with TEM data showing that agLDL(−) has a higher susceptibility to fusion than LDL(+) and nagLDL(−). The spectra of the three subfractions remained almost unchanged after 24 h (monitored for a total time of 48 h; data not shown), except for the signal of the fatty acid-CH3 in agLDL(−), which increased in intensity, probably because of particle decomposition, as was observed by TEM.
FIGURE 5.
Overlay of the regions with the fatty acid-CH3 resonances in 1H NMR spectra of LDL at different times of particle fusion induced by α-chymotrypsin proteolysis. A, LDL(+). B, nagLDL(−). C, agLDL(−). LDLs were mixed with α-chymotrypsin inside the NMR tube, and spectra were recorded at 37 °C at intervals of 30 min. The spectra shown correspond to 0.5, 1, 1.5, 2, 2.5, 3, 4, 6, 8, 12, 16, 20, and 24 h after α-chymotrypsin addition.
1H NMR spectra also provides information on degradation of phospholipids by monitoring the intensity of the resonance signals corresponding to choline-containing phospholipids (3.170–3.280 ppm) and ceramide (5.250–5.350 ppm) (28). In addition, the progression of oxidation can be detected by monitoring the decrease of the signal corresponding to bisallylic groups of fatty acids (CH-CH2-CH, 2.650–2.850 ppm) (27). These results are explained in detail in supplemental Figs. 6S–8S.
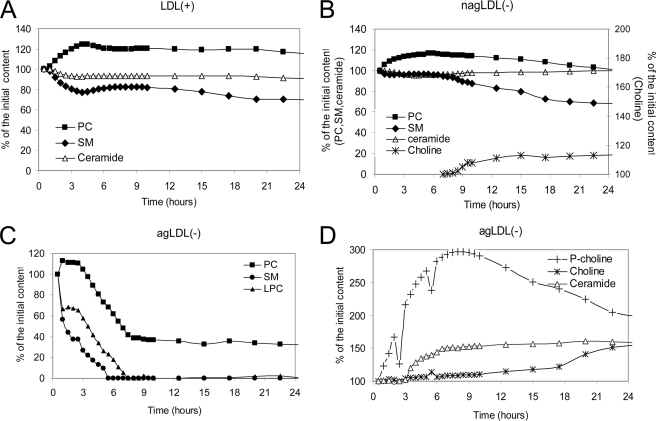
Changes in phospholipid composition induced by proteolysis, calculated from the corresponding NMR signal intensity, are summarized in Fig. 6. The phospholipid content of LDL(+) remained relatively constant, and the initial increase of PC and decrease of SM could be better explained by fusion-induced microenvironment changes than by degradation (26). nagLDL(−) presented low levels of degradation of SM and PC simultaneously with the appearance of free choline (NMR signal observed after 8 h of proteolysis). In contrast, agLDL(−) initially suffered a massive and rapid degradation of PC, SM, and LPC. Signals corresponding to SM and LPC disappeared at 8 h of proteolysis. This promoted an immediate increase of phosphorylcholine, an increase of ceramide after 3 h, and an increase of free choline after 4 h.
FIGURE 6.
Changes in phospholipid composition during α-chymotrypsin proteolysis of LDL particles quantified from NMR data. A, LDL(+). B, nagLDL(−). C, agLDL(−) (PC, SM, and LPC). D, agLDL(−) (phosphorylcholine (P-choline), free choline, and ceramide). LDLs were proteolysed by α-chymotrypsin up to 24 h. The 1H NMR spectra were acquired at 37 °C on a Bruker 800 MHz spectrometer. The concentration was calculated by integration of the area of the corresponding signals.
Phospholipid Degradation Quantified by HPLC
Analysis of phospholipid signals monitored by NMR is a first approach to quantitative changes promoted by proteolysis. However, the intensity and position of these signals can be modified by changes in the phospholipid environment (26). To confirm the changes in phospholipid composition during proteolysis, PC, SM, and LPC levels were also measured by normal phase HPLC. HPLC analysis (Fig. 7A) was consistent with NMR data. Phospholipids in agLDL(−) were much more labile to degradation than those in LDL(+) and nagLDL(−). LPC was only detected in nagLDL(−) and agLDL(−) and only at 3 h of proteolysis. We investigated the possibility that lipoperoxidative processes could occur during proteolysis by measuring the 205/234 ratio of the PC peak (indicative of the formation of conjugated dienes) and by quantifying the α-tocopherol content. No evidence of extensive oxidation was observed because the α-tocopherol content and the ratio 205/234 nm of the PC peak were not modified during proteolysis in the analyzed subfractions (Fig. 7, B and C). The ratio 205/234 could not be calculated in agLDL(−) at 24 h because the area of the PC peak at 234 nm was too low and could not be reliably integrated.
FIGURE 7.
HPLC analysis of LDL subfractions during α-chymotrypsin proteolysis. A, PC, SM, and LPC content in LDL subfractions measured by normal phase HPLC. B, ratio 205/234 nm of the PC peak. ND, not determined. C, α-tocopherol content in LDL subfractions measured by reverse phase HPLC. LDLs were proteolyzed by α-chymotrypsin up to 24 h. The data are the means ± S.D. of three independent experiments.
Changes in Major Lipids during Proteolysis
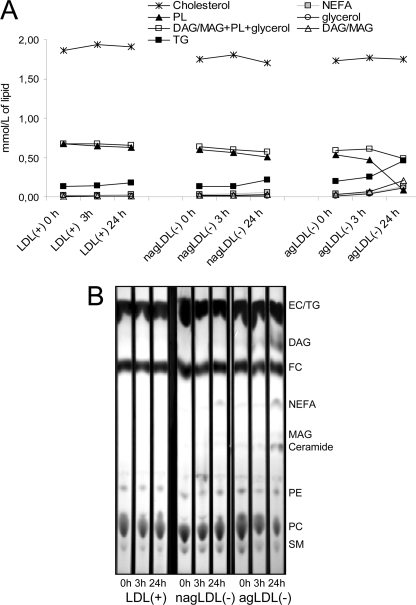
Changes in major lipids were measured by commercial methods in an autoanalyzer (Fig. 8A). Regarding neutral lipids, cholesterol and triglyceride were not degraded during proteolysis. The triglyceride content appeared to increase, but this measurement was erroneous because the commercial enzymatic method measures glycerol after the degradation of acylglycerols by triglyceride lipase. This increase was thus the consequence of DAG and/or monoacylglycerol (MAG) generated from the degradation of PC and LPC, respectively. This can be deduced from the observation that phospholipids decreased at the same rate as the sum of DAG/MAG + triglyceride + glycerol increased. Hence, the sum of DAG/MAG + phospholipids + glycerol after proteolysis was similar to that of initial phospholipids.
FIGURE 8.
Changes in major lipids during proteolysis. A, total cholesterol, triglyceride (TG), total phospholipids (PL), glycerol, and NEFA were quantified by commercial methods in an autoanalyzer. DAG and MAG were estimated from the difference between the initial content of triglyceride and the concentration obtained at every time point. The data are the means of three independent experiments. B, representative TLC of lipids from LDL subfractions. EC, esterified cholesterol; FC, free cholesterol; PE, phosphatidylethanolamine.
TLC supported data obtained by NMR, HPLC, and autoanalyzer (Fig. 8B). Briefly, massive degradation of phospholipids was observed in agLDL(−) after 24 h of proteolysis, and as a consequence, bands corresponding to ceramide, MAG, NEFA, and DAG, which were apparent at 0 and 3 h of proteolysis, increased their intensity at 24 h. nagLDL(−) also presented light bands of ceramide, DAG, MAG, and NEFA after 24 h of proteolysis.
DISCUSSION
The ability of LDL(−) to modulate the expression of molecules involved in inflammation (11, 12, 21), apoptosis (13), or angiogenesis (14) in endothelial cells and monocytes suggests a strong atherogenic role. However, there is some controversy regarding several properties of LDL(−), and a detailed knowledge of its physicochemical characteristics remains elusive. These discrepancies could be due to the fact that several independent mechanisms, such as oxidation (34), nonenzymatic glycosylation (9), hemoglobin derivatization (35), phospholipase A2 degradation (15, 23), or NEFA loading (36), could be involved in its generation. These observations strongly suggest that LDL(−) is a heterogeneous group of distinct modified LDL particles that share increased electronegativity as a common feature. Based on this assumption, the current study focused on the aggregated subfraction of LDL(−).
We observed that PLC-like activity differed in the three LDL subfractions. This activity, which degrades with increasing efficiency, PC < SM < LPC (17), was absent in LDL(+) and 5-fold higher in agLDL(−) than in nagLDL(−). The origin of such phospholipolytic activity is unknown. Studies using proteomic approaches have reported the presence of more than 20 minor proteins in total LDL (37, 38), but none of these proteins have a known PLC activity. The only proteins in LDL with a well established phospholipolytic activity are PAF-AH and lecithin:cholesterol acyl transferase. However, we have previously demonstrated that SM and LPC are not hydrolyzed by purified PAF-AH (17). Rather, LPC is the product of the reaction degrading PAF-like phospholipids. Indeed, no difference was observed between the two LDL(−) subfractions in PAF-AH activity. Regarding lecithin:cholesterol acyl transferase, this activity is not increased in LDL(−) versus LDL(+) (25). Indeed, 5,5′-dithiobis(2-nitrobenzoic) acid, a known inhibitor of lecithin:cholesterol acyl transferase, had no effect on LDL(−) PLC-like activity (data not shown). It has been suggested that PLC-like activity of LDL(−) could arise from conformation differences of apoB between LDL(+) and LDL(−) (17, 39).
The most striking property observed in agLDL(−) in this study was its ability to promote the aggregation of nonaggregated LDL subfractions. This process was paralleled by phospholipid degradation. These results concur with previous studies reporting that in the absence of physical or chemical stimuli, total LDL(−) aggregates to form amyloid-like fibrils in a period of one to several days and that it can promote the spontaneous aggregation of native LDL (18). Our results suggest that amyloidogenesis promoted by LDL(−) could be specifically due to agLDL(−) in a process involving PLC-like activity.
In addition to its aggregation induction ability, agLDL(−) showed high lability to protease digestion, leading first to fusion and later to particle degradation. α-Chymotrypsin proteolysis is known to promote LDL fusion and destabilize particle integrity in advanced stages (20). However, we found that this process was much faster in agLDL(−) than in LDL(+) or nagLDL(−) because of the accelerated degradation of phospholipids during the early phases of proteolysis, mainly SM and LPC. In this context, it is well established that SM degradation by SMase also promotes lipoprotein fusion (40). The exact mechanism by which proteolysis of apoB leads to phospholipid degradation is unknown; however, the observation that SM and LPC are rapid and almost totally degraded suggests the involvement of the PLC-like activity associated to LDL(−), at least in the early phases of the process. This concurs with the observation that products of such activity, such as phosphorylcholine, MAG, DAG, and ceramide, increased throughout the proteolytic digestion. Interestingly, cholesterol and triglyceride content remained unchanged during proteolysis, indicating that this process had a strong effect on the molecules present on the surface of the lipoprotein but little effect on the core components.
It could be argued that extensive lipoperoxidation could promote the changes observed in agLDL(−) after proteolysis, because oxidation induces aggregation and fusion of lipoproteins. The appearance of LPC in nagLDL(−) and agLDL(−) at 3 h of proteolysis is likely a consequence of PAF-AH activity, which is high in both LDL(−) subfractions. Because PAF-AH does not degrade intact PC but oxidation-fractionated PC (41), the increase of LPC could indicate mild oxidation. However, other observations rule out a major role for extensive oxidative processes in the high lability to proteolysis of agLDL(−). First, proteolysis was performed in presence of EDTA and BHT, which respectively act as metal and free radical chelants. Moreover, the relation between aggregation and oxidation is not reciprocal, because although oxidation promotes aggregation, the current study and previous (42) reports show that aggregated lipoproteins are more resistant to oxidation. In addition, although PC is a much better substrate for lipoperoxidation than SM, we observed that SM was degraded faster than PC in agLDL(−). In addition, α-tocopherol was not consumed during proteolysis, as occurs with oxidized LDL, and finally, the ratio 205/234 nm of the PC peak decreased only slightly in all of the LDL subfractions during proteolysis. Taken together, these results indicate that although minor oxidation cannot be ruled out to occur in all subfractions, its specific involvement in the massive degradation of phospholipids in agLDL(−) is unlikely.
It is well known that several proteases are up-regulated in the atherosclerotic lesion (2). Thus, degradation of the apoB protein in agLDL(−) would trigger its fusion and subsequently promote LDL retention. In agreement with this observation, our group previously reported that the subfraction of LDL(−) with the highest affinity to proteoglycans also had the highest aggregation level (19). A very recent report has shown that proteolysis sensitizes LDL particles to phospholipolysis mediated by phospholipases (43). Our data indicate that phospholipid degradation and subsequent fusion of agLDL(−) after proteolysis occurs in the absence of external phospholipases. The fact that LDL(−) has amyloidogenic properties caused by α-helix to β-sheet transition in the secondary structure of apoB (18) would also contribute to lipoprotein aggregation and subsequent cholesterol accumulation. Taken together, these observations suggest a picture in which circulating agLDL(−), aggregated but not fused, could enter the arterial wall, and once in this microenvironment, it could fuse itself and trigger the fusion of nonaggregated LDL, favoring the binding of LDL to proteoglycans and, subsequently, promoting subendothelial cholesterol accumulation and atherosclerosis progression.
Supplementary Material
Acknowledgments
We thank Carolyn Newey for editorial assistance and Sara González for technical assistance.
This work was supported by Instituto de Salud Carlos III Grants PI060500 and PI070148 and by an ETORTEK-2008 grant from Sociedad para la Promoción y Reconversión Industrial.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table 1S, and Figs. 1S–8S.
- LDL(−)
- electronegative LDL
- agLDL(−)
- aggregated electronegative LDL
- apo
- apolipoprotein
- BHT
- butylated hydroxytoluene
- DAG
- diacylglycerol
- GGE
- nondenaturing acrylamide gradient gel electrophoresis
- IDL
- intermediate density lipoprotein
- LDL(+)
- native electropositive LDL
- LPC
- lysophosphatidylcholine
- MAG
- monoacylglycerol
- nagLDL(−)
- nonaggregated electronegative LDL
- NEFA
- nonesterified fatty acid
- oxLDL
- oxidized LDL
- PAF-AH
- platelet-activating factor-acetylhydrolase
- PC
- phosphatidylcholine
- PLC
- phospholipase C
- SM
- sphingomyelin
- SMase
- sphingomyelinase
- TEM
- transmission electron microscopy.
REFERENCES
- 1.Camejo G., Hurt-Camejo E., Wiklund O., Bondjers G. (1998) Atherosclerosis 139, 205–222 [DOI] [PubMed] [Google Scholar]
- 2.Ross R. (1999) N. Engl. J. Med. 340, 115–126 [DOI] [PubMed] [Google Scholar]
- 3.Navab M., Berliner J. A., Watson A. D., Hama S. Y., Territo M. C., Lusis A. J., Shih D. M., Van Lenten B. J., Frank J. S., Demer L. L., Edwards P. A., Fogelman A. M. (1996) Arterioscler. Thromb. Vasc. Biol. 16, 831–842 [DOI] [PubMed] [Google Scholar]
- 4.Bhakdi S., Lackner K. J., Han S. R., Torzewski M., Husmann M. (2004) Thromb. Haemost. 91, 639–645 [DOI] [PubMed] [Google Scholar]
- 5.Williams K. J., Tabas I. (1998) Curr. Opin. Lipidol. 9, 471–474 [DOI] [PubMed] [Google Scholar]
- 6.Sánchez-Quesada J. L., Benítez S., Ordóñez-Llanos J. (2004) Curr. Opin. Lipidol. 15, 329–335 [DOI] [PubMed] [Google Scholar]
- 7.Benítez S., Ordóñez-Llanos J., Franco M., Marín C., Paz E., López-Miranda J., Otal C., Pérez-Jiménez F., Sánchez-Quesada J. L. (2004) Am. J. Cardiol. 93, 414–420 [DOI] [PubMed] [Google Scholar]
- 8.Sánchez-Quesada J. L., Benítez S., Otal C., Franco M., Blanco-Vaca F., Ordóñez-Llanos J. (2002) J. Lipid. Res. 43, 699–705 [PubMed] [Google Scholar]
- 9.Sánchez-Quesada J. L., Pérez A., Caixàs A., Rigla M., Payés A., Benítez S., Ordóñez-Llanos J. (2001) J. Clin. Endocrinol. Metab. 86, 3243–3249 [DOI] [PubMed] [Google Scholar]
- 10.Ziouzenkova O., Sevanian A. (2000) Blood Purif. 18, 169–176 [DOI] [PubMed] [Google Scholar]
- 11.Benítez S., Camacho M., Bancells C., Vila L., Sánchez-Quesada J. L., Ordóñez-Llanos J. (2006) Biochim. Biophys. Acta 1761, 1014–1021 [DOI] [PubMed] [Google Scholar]
- 12.Benítez S., Bancells C., Ordóñez-Llanos J., Sánchez-Quesada J. L. (2007) Biochim. Biophys. Acta 1771, 613–622 [DOI] [PubMed] [Google Scholar]
- 13.Chen H. H., Hosken B. D., Huang M., Gaubatz J. W., Myers C. L., Macfarlane R. D., Pownall H. J., Yang C. Y. (2007) J. Lipid Res. 48, 177–184 [DOI] [PubMed] [Google Scholar]
- 14.Tai M. H., Kuo S. M., Liang H. T., Chiou K. R., Lam H. C., Hsu C. M., Pownall H. J., Chen H. H., Huang M. T., Yang C. Y. (2006) Atherosclerosis 186, 448–457 [DOI] [PubMed] [Google Scholar]
- 15.Asatryan L., Hamilton R. T., Isas J. M., Hwang J., Kayed R., Sevanian A. (2005) J. Lipid Res. 46, 115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benítez S., Villegas V., Bancells C., Jorba O., González-Sastre F., Ordóñez-Llanos J., Sánchez-Quesada J. L. (2004) Biochemistry 43, 15863–15872 [DOI] [PubMed] [Google Scholar]
- 17.Bancells C., Benítez S., Villegas S., Jorba O., Ordóñez-Llanos J., Sánchez-Quesada J. L. (2008) Biochemistry 47, 8186–8194 [DOI] [PubMed] [Google Scholar]
- 18.Parasassi T., De Spirito M., Mei G., Brunelli R., Greco G., Lenzi L., Maulucci G., Nicolai E., Papi M., Arcovito G., Tosatto S. C., Ursini F. (2008) FASEB J. 22, 2350–2356 [DOI] [PubMed] [Google Scholar]
- 19.Bancells C., Benítez S., Jauhiainen M., Ordóñez-Llanos J., Kovanen P. T., Villegas S., Sánchez-Quesada J. L., Oörni K. (2009) J. Lipid Res. 50, 446–455 [DOI] [PubMed] [Google Scholar]
- 20.Oörni K., Pentikäinen M. O., Ala-Korpela M., Kovanen P. T. (2000) J. Lipid Res. 41, 1703–1714 [PubMed] [Google Scholar]
- 21.Sánchez-Quesada J. L., Camacho M., Antón R., Benítez S., Vila L., Ordóñez-Llanos J. (2003) Atherosclerosis 166, 261–270 [DOI] [PubMed] [Google Scholar]
- 22.De Castellarnau C., Sánchez-Quesada J. L., Benítez S., Rosa R., Caveda L., Vila L., Ordóñez-Llanos J. (2000) Arterioscler. Thromb. Vasc. Biol. 20, 2281–2287 [DOI] [PubMed] [Google Scholar]
- 23.Benítez S., Camacho M., Arcelus R., Vila L., Bancells C., Ordóñez-Llanos J., Sánchez-Quesada J. L. (2004) Atherosclerosis 177, 299–305 [DOI] [PubMed] [Google Scholar]
- 24.Sánchez-Quesada J. L., Ortega H., Payés-Romero A., Serrat-Serrat J., González-Sastre F., Lasunción M. A., Ordóñez-Llanos J. (1997) Atherosclerosis 132, 207–213 [DOI] [PubMed] [Google Scholar]
- 25.Benítez S., Sánchez-Quesada J. L., Ribas V., Jorba O., Blanco-Vaca F., González-Sastre F., Ordóñez-Llanos J. (2003) Circulation 108, 92–96 [DOI] [PubMed] [Google Scholar]
- 26.Pentikäinen M. O., Hyvönen M. T., Oörni K., Hevonoja T., Korhonen A., Lehtonen-Smeds E. M., Ala-Korpela M., Kovanen P. T. (2001) J. Lipid Res. 42, 916–922 [PubMed] [Google Scholar]
- 27.Soininen P., Oörni K., Maaheimo H., Laatikainen R., Kovanen P. T., Kaski K., Ala-Korpela M. (2007) Biochem. Biophys. Res. Commun. 360, 290–294 [DOI] [PubMed] [Google Scholar]
- 28.Duarte I. F., Marques J., Ladeirinha A. F., Rocha C., Lamego I., Calheiros R., Silva T. M., Marques M. P., Melo J. B., Carreira I. M., Gil A. M. (2009) Anal Chem 81, 5023–5032 [DOI] [PubMed] [Google Scholar]
- 29.Lodge J. K., Sadler P. J., Kus M. L., Winyard P. G. (1995) Biochim. Biophys. Acta 1256, 130–140 [DOI] [PubMed] [Google Scholar]
- 30.Forte T., Nichols A. V. (1972) Adv. Lipid Res. 10, 1–41 [PubMed] [Google Scholar]
- 31.Gaubatz J. W., Gillard B. K., Massey J. B., Hoogeveen R. C., Huang M., Lloyd E. E., Raya J. L., Yang C. Y., Pownall H. J. (2007) J. Lipid Res. 48, 348–357 [DOI] [PubMed] [Google Scholar]
- 32.Campos H., Perlov D., Khoo C., Sacks F. M. (2001) J. Lipid Res. 42, 1239–1249 [PubMed] [Google Scholar]
- 33.Ala-Korpela M., Pentikäinen M. O., Korhonen A., Hevonoja T., Lounila J., Kovanen P. T. (1998) J. Lipid Res. 39, 1705–1712 [PubMed] [Google Scholar]
- 34.Sevanian A., Bittolo-Bon G., Cazzolato G., Hodis H., Hwang J., Zamburlini A., Maiorino M., Ursini F. (1997) J. Lipid Res. 38, 419–428 [PubMed] [Google Scholar]
- 35.Ziouzenkova O., Asatryan L., Akmal M., Tetta C., Wratten M. L., Loseto-Wich G., Jürgens G., Heinecke J., Sevanian A. (1999) J. Biol. Chem. 274, 18916–18924 [DOI] [PubMed] [Google Scholar]
- 36.Ursini F., Sevanian A. (2002) Biol. Chem. 383, 599–605 [DOI] [PubMed] [Google Scholar]
- 37.Banfi C., Brioschi M., Barcella S., Wait R., Begum S., Galli S., Rizzi A., Tremoli E. (2009) Proteomics 9, 1344–1352 [DOI] [PubMed] [Google Scholar]
- 38.Karlsson H., Leanderson P., Tagesson C., Lindahl M. (2005) Proteomics 5, 551–565 [DOI] [PubMed] [Google Scholar]
- 39.Parasassi T., Bittolo-Bon G., Brunelli R., Cazzolato G., Krasnowska E. K., Mei G., Sevanian A., Ursini F. (2001) Free Radic. Biol. Med. 31, 82–89 [DOI] [PubMed] [Google Scholar]
- 40.Oörni K., Hakala J. K., Annila A., Ala-Korpela M., Kovanen P. T. (1998) J. Biol. Chem. 273, 29127–29134 [DOI] [PubMed] [Google Scholar]
- 41.Stafforini D. M. (2009) Cardiovasc. Drugs Ther. 23, 73–83 [DOI] [PubMed] [Google Scholar]
- 42.Hermann M., Gmeiner B. (1992) Arterioscler. Thromb. 12, 1503–1506 [DOI] [PubMed] [Google Scholar]
- 43.Plihtari R., Hurt-Camejo E., Oörni K., Kovanen P. T. (2010) J. Lipid Res. 51, 1801–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.