Abstract
ERM proteins are regulated by phosphorylation of the most C-terminal threonine residue, switching them from an activated to an inactivated form. However, little is known about the control of this regulation. Previous work in our group demonstrated that secretion of acid sphingomyelinase acts upstream of ERM dephosphorylation, suggesting the involvement of sphingomyelin (SM) hydrolysis in ERM regulation. To define the role of specific lipids, we employed recombinant bacterial sphingomyelinase (bSMase) as a direct probe of SM metabolism at the plasma membrane. bSMase induced a rapid dose- and time-dependent decrease in ERM dephosphorylation. ERM dephosphorylation was driven by ceramide generation and not by sphingomyelin depletion, as shown using recombinant sphingomyelinase D. The generation of ceramide at the plasma membrane was sufficient for ERM regulation, and no intracellular SM hydrolysis was required, as was visualized using Venus-tagged lysenin probe, which specifically binds SM. Interestingly, hydrolysis of plasma membrane bSMase-induced ceramide using bacterial ceramidase caused ERM hyperphosphorylation and formation of cell surface protrusions. The effects of plasma membrane ceramide hydrolysis were due to sphingosine 1-phosphate formation, as ERM phosphorylation was blocked by an inhibitor of sphingosine kinase and induced by sphingosine 1-phosphate. Taken together, these results demonstrate a new regulatory mechanism of ERM phosphorylation by sphingolipids with opposing actions of ceramide and sphingosine 1-phosphate. The approach also defines a tool kit to probe sphingolipid signaling at the plasma membrane.
Keywords: Cellular Regulation, Cytoskeleton, Lipid, Plasma Membrane, Sphingolipid, Ceramidase, Ceramide, Ezrin Radixin Moesin (ERM), Sphingosine-1-Phosphate, Sphingomyelinase
Introduction
Ezrin (82 kDa), radixin (80 kDa), and moesin (75 kDa), known as the ERM proteins, link the plasma membrane to the cortical cytoskeleton. These proteins have been found to be enriched in specialized plasma membrane domains such as microvilli, lamellipodia, membrane ruffles, and other membrane protrusions (1). ERM proteins have been implicated in regulation of cell shape (2), cell polarization (3, 4), membrane enzyme localization, membrane transport, cell adhesion/migration (5–7), and signal transduction (8). The function of ERM proteins is regulated by a two-step process based on an open (active) and closed (inactive) conformation. In the closed conformation, the N terminus domain (FERM) and the C terminus domain (C-ERMAD) interact with each other in a self-folded, dormant state, and the proteins rest in the cytosol. This folding is regulated by phosphorylation on the very C-terminal threonine residue (ezrin Thr567, radixin Thr564, and moesin Thr558), which leads to the open, active conformation. In the active conformation, the FERM domain interacts with the plasma membrane, and with several membrane-associated proteins (CD95, CD44, intercellular adhesion molecule (I-CAM), CD43, cystic fibrosis transmembrane conductance regulator (CFTCR), and NHE1), and the C-ERMAD domain interacts with actin filaments in the submembrane cortex (9–13). Several kinases have been shown to phosphorylate the C terminus of ERM, including classical protein kinase C (14), atypical protein kinase C (15, 16), Rho kinase (17), myotonic dystrophy kinase-related Cdc42-binding kinase (18), and G-protein-coupled receptor kinase 2 (19). However, only myosin phosphatase has been clearly implicated in ERM dephosphorylation, although other phosphatases have been suggested, including PP2A (20) or PP2C in vitro studies (21).
As shown previously for ERM, sphingolipids (SLs)2 have also been implicated in regulating the actin cytoskeleton (22, 23) and cell migration (24, 25). Thus, sphingomyelin (SM) hydrolysis to ceramide mediates apoptosis, cell senescence, and other cell stress responses (26) and cytoskeleton remodeling (26–28). Previous work in our group implicated acid sphingomyelinase as an upstream regulator of ERM dephosphorylation and that activity could be mimicked by bacterial sphingomyelinase (bSMase) (20, 29). Given these connections, it became important to identify the specific bioactive SLs required for this dephosphorylation as well as their topological implications and how other SLs in the SL network would affect ERM phosphorylation.
In this study, we demonstrate that ERM dephosphorylation is acutely regulated by hydrolysis of SM at the plasma membrane. We developed tools (Fig. 1) to show that the effect of sphingomyelinases on ERM dephosphorylation is caused by generation of ceramide and not by a decrease in sphingomyelin per se, or by generation of more complex SLs. We also show that dephosphorylation of ERM can be strongly overcome by hydrolysis of ceramide. Using for the first time bacterial ceramidase (bCDase) in cells, we found that hydrolysis of ceramide at the plasma membrane rapidly induces hyperphosphorylated ERM proteins. We further implicate S1P as a novel inducer of ERM phosphorylation. We propose a new model of regulation of ERM proteins by competing roles for ceramide and S1P generated at the plasma membrane.
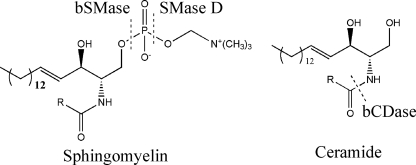
FIGURE 1.
Hydrolysis of sphingomyelin and ceramide with the exogenous enzymes used in this work.
EXPERIMENTAL PROCEDURES
Materials
High glucose Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum, and rhodamine phalloidin were from Invitrogen. Sphingomyelinase from Bacillus cereus, bovine serum albumin (BSA), essentially fatty acid free, and protamine sulfate were from Sigma. Bovine sphingomyelin, N-hexadecanoyl-d-erythro-sphingosine (C16 ceramide), N-nervonoyl-d-erythro-sphingosine (C24:1 ceramide), d-erythro-sphingosine, and d-erythro-sphingosine 1-phosphate (S1P) were from Avanti Polar Lipids (Alabaster, AL). Antiphospho-ERM proteins, anti-ezrin, anti-moesin, and anti-radixin, were from Cell Signaling Technology (Danvers, MA). Draq5 was from Alexis (Carlsbad, CA).
Cell Culture
HeLa cells were originally purchased from American Type Culture Collection (ATCC, Manassas, VA). Cells were grown in DMEM supplemented with 10% FBS and incubated at 37 °C, 5% CO2. When FBS-free medium was used, medium was supplemented with 0.1% of BSA.
Protein Expression and Purification of Bacterial Sphingomyelinase, SMase D, Bacterial Ceramidase, and Lysenin
All recombinant protocols were carried out following standard protocols. The plasmid encoding sphingomyelinase D (SMase d-pET30) from Loxosceles reclusa containing a N-terminal His6 tag, was a kind gift from Dr. Kevin R. Lynch (University of Virginia, School of Medicine, Charlottesville, VA). The plasmid and recombinant bacteria for Pseudomonas aeruginosa ceramidase and Bacillus cereus sphingomyelinase were obtained previously by our group (29); the plasmid pRSET-His-Venus-lysenin (30), which encodes six residues of histidine followed by Venus and a nontoxic mutant of lysenin, was kindly provided by Dr. Toshihide Kobayashi from Supra-Biomolecular System Research Group, RIKEN. In all cases, the plasmid was introduced by transformation into an Escherichia coli BL21(DE3) pLysS competent cell strain (Stratagene) according to the manufacturer's instructions. For optimization of expression, the E. coli SMase d-BL21 was grown in Luria-Bertani broth (LB) containing selective antibiotic. The protein induction was tested at different concentrations of inducer isopropyl-β-thiogalactopyranoside (IPTG; 0.1–1 mm), various times of induction (3, 8, and 24 h), and different temperatures (25, 30, and 37 °C), following standard procedures. After optimization, the induction condition for bCDase expression was carried out using 0.1 mm IPTG in M9 minimum medium for 24 h at 30 °C before harvest. For bSMase and SMase D, the conditions were 0.5 mm IPTG in LB medium for 24 h at 25 °C. For lysenin, the optimum conditions were 1 mm IPTG in LB medium for 24 h at 37 °C.
Immunofluorescence and Confocal Microscopy
For immunofluorescence analysis of cells with/without bSMase, SMase D, and/or bCDase treatment or exogenous SL treatments, HeLa cells were plated onto uncoated glass-bottomed dishes (MatTek, Ashland, MA) at 100,000 cells/dish and incubated overnight; medium was changed to FBS-free DMEM, 0.1% BSA for 2 h, and cells were treated with the exogenous enzymes/exogenous SLs for the times indicated under “Results.” Cells were fixed by addition of warmed (37 °C) 3.7% paraformaldehyde solution for 15 min, and samples were washed with PBS, if required, permeabilized with 0.1% Triton X-100 for 10 min and blocked for 20 min at room temperature in PBS containing 2% of human serum. Primary antibodies were added for 18 h at 4 °C diluted in PBS with 2% of human serum. Samples were examined, and images were collected using a confocal microscope (LSM510 META; Carl Zeiss, Inc.) using a 64× objective, and data were imported into Volocity software (Improvision).
Immunoblotting
Medium was removed from cells, and the lysis buffer containing 1% (w/v) SDS was added immediately, sonicated briefly (10 s), and boiled for 5 min. Samples were normalized to protein using BCA protein assay, and 10–20 μg were subjected to SDS-PAGE (10% or 4–20% gradient Tris-HCl) using the Bio-Rad Criterion system. Proteins were transferred to nitrocellulose membranes, blocked with PBS/0.1% Tween 20 (PBS-T) containing 5% nonfat milk, and incubated overnight at 4 °C with primary antibody following the manufacturer's protocol.
Mass Spectrometry to Measure Endogenous SL Levels
After treatment, cells were washed two times with cold PBS and collected by centrifugation, and the cell pellets were used for lipid extraction. Two ml of 2:3 70% isopropanol/ethyl acetate were added directly to the cells. Internal standards for SLs (50 pmol each of analyzed species) were added to each sample. The samples were centrifuged at 2000 × g, and the upper phase was transferred to a new glass tube, and the lower phase was re-extracted adding 2 ml of the same extraction solution (4 ml). The two upper phases were combined in the same tube and used to measure SLs and inorganic phosphate. The identification of SL species was performed on a Thermo Finnigan TSQ 7000 triple quadrupole MS operating in a multiple reaction monitoring positive ionization mode as described previously (31). Results from MS analysis were normalized by total inorganic phosphate (32) present in the sample after a Bligh & Dyer extraction (33).
Kinetics Measurements
Km was measured using detergent/lipid mixed micelles with d-erythro-C16 or C24:1 ceramide as a substrate at different concentrations (from 0.1 to 20 μm) in a 50 mm (pH 7.1) Tris buffer containing 1 mm CaCl2, 0.1% (w/v) Triton X-100 (final concentration), with a total volume of 100 μl. The enzymatic reaction was started by adding 1 ng of purified enzyme. The enzymatic reaction was continued for 30 min at 37 °C, and it was stopped by addition of chloroform/methanol (1:1). The organic phase was analyzed by mass spectrometry. The Km was calculated using Prism software.
Statistical Analysis
Mean values ± S.E. of five different experiments are reported (**, p < 0.05 and ***, p < 0.01 as determined by one-way analysis of variance, Dunnett's post-test).
RESULTS
Bacterial Sphingomyelinase (bSMase) Hydrolyzes SM in Serum and Induces Ceramide Formation
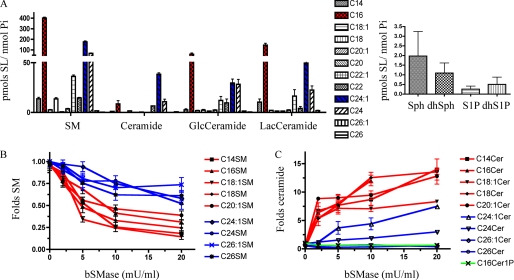
SM is a structural constituent of mammalian cell membranes, mainly found at the outer leaflet of the plasma membrane but is also found extracellularly in serum lipoproteins (34). The effects of bSMase on ERM proteins reported previously by our group (20) could be exerted by the effect of bSMase on cellular SM or on the SM associated with lipoproteins found in FBS. We observed that FBS used in cell culture is rich in SM, consistent with previous studies (35). Thus, bSMase treatment of FBS medium caused hydrolysis of SM not only in the cells but also in the medium. Mass spectrometric analysis of FBS demonstrated high levels of SM (Fig. 2A), although ceramide, Sph, and sphingosine 1-phosphate were not detected. Bacterial SMase rapidly hydrolyzed SM from FBS medium generating ceramide (Fig. 2B). Thus, FBS contains SM that can be converted into ceramide, as an extra source of SL to the cells, and makes the system more complex. Additionally, bSMase-mediated hydrolysis of lipoprotein SM to ceramide has been shown to induce LDL aggregation. LDL aggregates can act as potent signaling molecules on their own (36). Because bSMase in FBS-free medium was sufficient to induce ERM dephosphorylation (Fig. 2C) and to avoid this confounding source of extracellular signaling, all subsequent experiments were conducted in FBS-free medium.
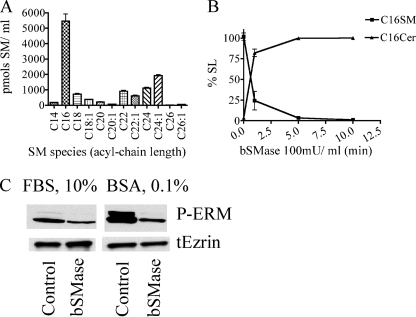
FIGURE 2.
Effects of bSMase on sphingomyelin in FBS medium. A, the SL profile was measured in FBS medium, and different species of SM were detected. B, after bSMase treatment (25 milliunits (mU)/ml), ceramide formation was measured. Only the C16 acyl chain is shown for clarity, but all sphingomyelin species had similar kinetics. C, HeLa cells were cultured in 10% FBS-containing DMEM medium or FBS-free medium and treated with bSMase (25 milliunits/ml for 10 min), and phospho-medium and total ezrin were measured by Western blotting.
Effects of bSMase on Dephosphorylation of ERM Proteins and Their Association with Plasma Membrane
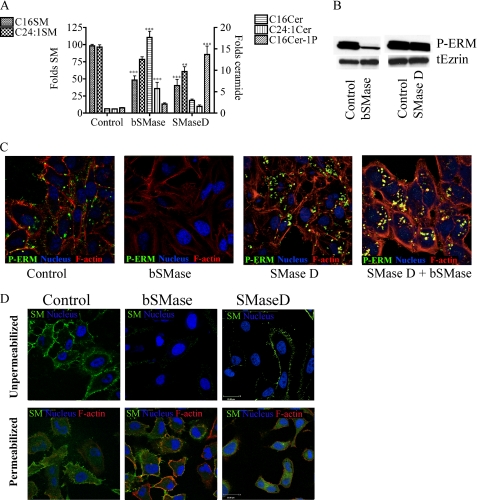
Previous results from our group implicated acid sphingomyelinase in ezrin dephosphorylation (20). To determine the specific bioactive SLs involved and their roles, HeLa cells were treated with bSMase using different doses and different times. After bSMase treatment, ERM proteins underwent dephosphorylation as early as 2 min and started recovering after 20–30 min with maximal effects seen at 50 milliunits/ml (Fig. 3A). Because phosphorylation of ERM proteins is closely related to their association with the plasma membrane and cortical actin, the localization of ERM proteins after bSMase treatment was studied. HeLa cells were treated with bSMase (25 milliunits/ml) for 10 min and examined by confocal microscopy (Fig. 3B). In resting cells, phospho-ERM proteins were localized in protrusions of the plasma membrane colocalizing with actin filaments, and total ERM protein was localized to both the cytosol and plasma membrane, particularly enriched in plasma membrane protrusions. After bSMase treatment, the phospho-ERM signal disappeared; thus demonstrating the cellular dephosphorylation of ERM and ruling out artifacts resulting from post-lysis of cells. Importantly, the total ERM signal became more cytosolic and disappeared from the plasma membrane protrusions. Therefore, bSMase was sufficient to induce ERM dephosphorylation and its relocalization from plasma membrane to cytosol.
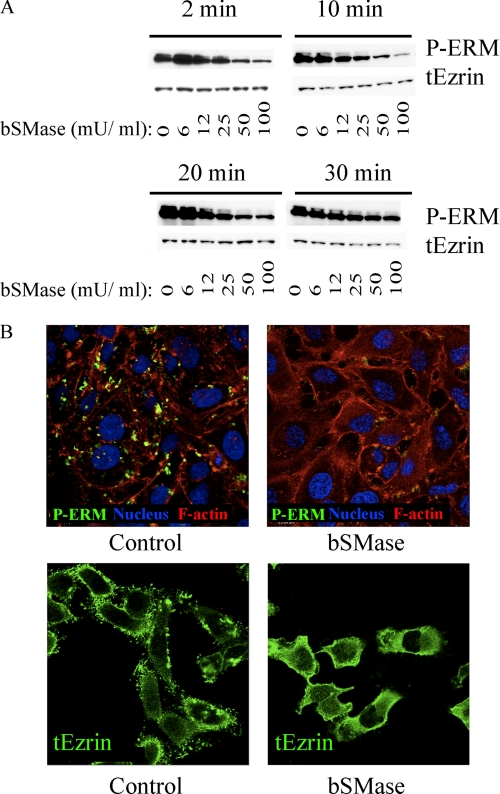
FIGURE 3.
BSMase effects on ERM dephosphorylation. A, HeLa cells were treated with vehicle or with different doses of bSMase: 6, 12, 25, 50, and 100 milliunits (mU)/ml for 2, 10, 20, and 30 min. Phospho-ERM and total ezrin were analyzed by Western blotting. B, HeLa cells were treated with vehicle bSMase (25 milliunits/ml, 10 min) and stained with phospho-ERM specific antibody (P-ERM, green). Nuclei and F-actin were visualized with Draq5 (blue) and rhodamine phalloidin (red) staining or stained with total ezrin antibody (tEzrin, green).
Effects of bSMase on Plasma Membrane and Intracellular Sphingomyelin
Although SM is enriched in the outer leaflet of the plasma membrane, it also exists in other intracellular membranes. Using cell fractionation on HL-60 cells, Linardic and Hannun (37) showed that bSMase acted only at the plasma membrane but not on intracellular SM. To corroborate and extend these data, we purified a Venus-tagged, nontoxic mutant of lysenin protein, which specifically binds SM (38) and has been used as an SM-staining probe (39). Thus, HeLa cells were treated with bSMase for different times, and nonpermeabilized cells were stained with lysenin-Venus protein and analyzed by confocal microscopy (Fig. 4, upper panels). Afterward, the cells were permeabilized and restained to allow lysenin-Venus to bind the intracellular SM (Fig. 4, lower panels). Thus, before bSMase treatment, SM was distributed evenly across the cell surface, including cell protrusions. After bSMase treatment, as early as 1 min, SM at the plasma membrane was decreased significantly, and it disappeared completely after 10 min of treatment. However, the intracellular SM was not hydrolyzed by bSMase. Furthermore, to support these data, HeLa cells were treated with tagged bSMase-His6 and analyzed by Western blotting, bSMase-His6 was not detected in the cell lysates in up to 1-h treatments (results not shown). Considering that bSMase caused ERM dephosphorylation as early as 1–2 min, and bSMase only hydrolyzed SM from the outer leaflet of the plasma membrane but could not hydrolyze intracellular SM, at least at short times, we could conclude that hydrolysis of SM from the outer leaflet was sufficient to induce ERM dephosphorylation.
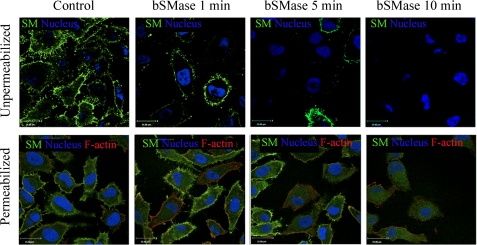
FIGURE 4.
Effects of bSMase on plasma membrane and intracellular SM. Presence of SM was monitored by lysenin-Venus (green channel) binding to SM after bSMase treatment (25 milliunits/ml). Cells were fixed, and the nonpermeabilized (upper panels) or permeabilized (lower panels) cells were analyzed by confocal microscope. Nuclei was visualized with Draq5 (blue), and in permeabilized cells, F-actin was visualized with rhodamine phalloidin staining (red).
Selective Hydrolysis of Long Chain Sphingomyelin at Plasma Membrane Was Sufficient to Induce ERM Dephosphorylation
In mammalian cells, the composition of sphingomyelin analyzed by mass spectrometry has been shown to be cell- and organism-specific. Furthermore, it is not known which species of SM are enriched in specific subcellular membranes. Moreover, different acyl chain length SLs as well as different subcellular localizations have been proposed to drive diverse actions in cells. To study the effect of bSMase on the SM profile of the external leaflet of the plasma membrane, various SL species were analyzed before and after bSMase treatment. Before bSMase treatment, the main SM species in HeLa cells were the C16 and C24:1 acyl chain, which also were the main acyl chains for ceramide, hexosyl ceramide, and lactosyl ceramide (Fig. 5A). After bSMase treatment, no major changes were measured for hexosyl ceramides, lactosyl ceramides, Sph, or S1P (data not shown). On the other hand, almost all the sphingomyelin species were decreased. Importantly, bSMase action showed preference for the long acyl chain species (e.g. C16) at early times (2 and 5 min), with additional hydrolysis of C24 and C24:1 SM at later times, and little changes were detected for C26 and C26:1 (Fig. 5B). Simultaneously, the long acyl chain ceramide species increased at early points (2 min) and faster than the very long acyl chain species that increased only after 5 min. As seen with SM, no changes were detected for the C26 and C26:1 ceramide species (Fig. 5C). Thus, the above results showed a correlation between bSMase-induced dephosphorylated ERM at 2 min and the lipidomic data, showing a preferential hydrolysis of long chain species (C14 to C20) versus very long species (C22 to C26) at 2 min. Thus, the decrease of long chain SM or the generation of long chain ceramide was sufficient to dephosphorylate ERM proteins, although a role for very long chain ceramide in regulation of ERM phosphorylation at later time points cannot be excluded.
FIGURE 5.
Changes in SLs after bSMase treatment in HeLa cells. A, the SL profile of HeLa cells was measured in serum-free conditions. C16 (red), C24:1 (blue), and C24 were the main acyl chains among the different acyl chain species analyzed. Sph and S1P and their dihydro (dh) forms were also measured. B, levels of SM and ceramide species. After bSMase treatment (25 milliunits (mU)/ml, 10 min), the SL profile was monitored, and only SL species that underwent significant changes are presented (red, LC species; blue, VLC species): Sphingomyelin (B) and ceramide (C) species. GlcCeramide, glucosyl ceramide; LacCeramide, lactosyl ceramide; Cer, ceramide.
Decrease in SM Levels at Plasma Membrane Was Insufficient to Dephosphorylate ERM Proteins
Normally, when bSMase is used in cells, the effects of bSMase have been attributed to ceramide generation or sphingomyelin removal from the plasma membrane, but distinguishing the effects of SM loss versus ceramide formation is challenging. In this context, SMase D hydrolyzes sphingomyelin to generate ceramide 1-phosphate (C1P) instead of ceramide. To distinguish the effects on ERM phosphorylation by decreasing sphingomyelin from generating ceramide at the plasma membrane, His-tagged SMase D from the spider Loxosceles sp. was expressed in E. coli and purified (supplemental Table 2). Next, HeLa cells were treated with bSMase and SMase D at concentrations producing a similar decrease in sphingomyelin (Fig. 6A). As expected, whereas bSMase produced ceramide, SMase D produced mainly C1P although a small increase in ceramide levels were detected at short times, presumably by dephosphorylation of C1P (Fig. 6A). Next, the effects of the two sphingomyelinases on ERM phosphorylation was evaluated by Western blot. Only bSMase treatment but not SMase D treatment led to ERM dephosphorylation, showing that ceramide generation and not the decrease of SM induced ERM dephosphorylation (Fig. 6B). Analysis by confocal microscopy also confirmed that although bSMase was able to induce ERM dephosphorylation, SMase D was not (Fig. 6C). Moreover, when HeLa cells were first treated with SMase D and then with bSMase, the latter lost its ability to induce ERM dephosphorylation (Fig. 6C, last panel), consistent with the loss of the SM substrate for bSMase after SMase D treatment. As seen with bSMase, SMase D hydrolyzed plasma membrane SM but had no effect on intracellular SM (Fig. 6D). These data strongly suggest that SM is not the active compound required to keep ERM phosphorylated at the plasma membrane but that ceramide at the plasma membrane is required to induce the dephosphorylation.
FIGURE 6.
Effects of depletion of SM versus ceramide generation on ERM proteins. A, HeLa cells were cultured in FBS-free medium for 2 h and treated with bSMase (25 milliunits/ml) or SMase D (300 milliunits/ml) to cause the same fold decrease in SM species for 10 min, and ceramide (Cer) and ceramide 1-phosphate were monitored. B, HeLa cells were treated with bSMase or SMase D as before and phospho-ERM, and total ezrin was measured by Western blotting. C, HeLa cells were treated with bSMase (10 min), SMase D (10 min), or with SMase D (10 min), and then with bSMase (10 min) and stained against phospho-ERM (green), actin filaments (stained with rhodamine phalloidin, red) and nuclei (stained with Draq5, blue). D, HeLa cells were treated with bSMase or SMase D as in A, and the presence of SM was monitored by lysenin-Venus (green channel). Cells were fixed, and the nonpermeabilized (upper panels) or permeabilized (lower panels) cells were analyzed by confocal microscope. Nuclei were visualized with Draq5 (blue) and in permeabilized cells F-actin was visualized with rhodamine phalloidin staining (red). tEzrin, total ezrin.
Effects of Hydrolysis of Ceramide at Plasma Membrane on Phosphorylation of ERM Proteins
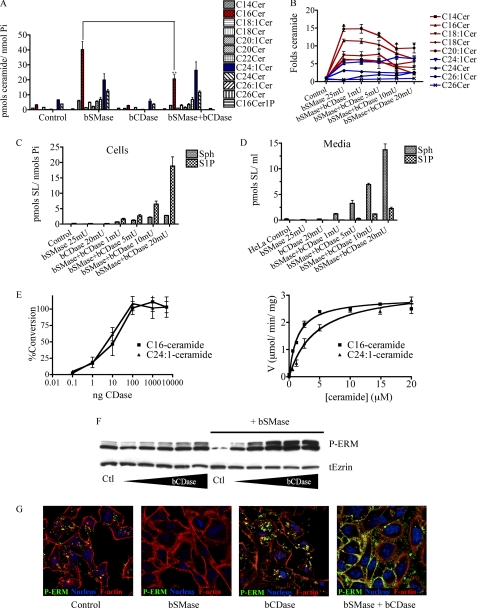
To further implicate ceramide at the plasma membrane and distinguish its effects from those of potential subsequent metabolites, we purified bCDase from Pseudomonas aeruginosa (supplemental Table 1) to decrease the ceramide levels at the plasma membrane and study how that would affect ERM phosphorylation. Initially, to study how the basal levels of ceramide could affect ERM proteins, HeLa cells were treated with bCDase and harvested at different times. Surprisingly, no changes in ceramide levels were detected after bCDase treatment, suggesting that ceramide levels in the plasma membrane of resting cells are very low. However, when HeLa cells were pretreated with bSMase to generate ceramide and then treated with bCDase, the ceramide generated after bSMase treatment started to decrease (Fig. 7A), generating Sph, and, somewhat unexpectedly, very high levels of S1P in cells (Fig. 7C) as well as in medium (Fig. 7D). Notably, bCDase preferentially decreased long acyl chain ceramides but had no effect on very long acyl chain ceramides (Fig. 7B). Functionally, ceramide hydrolysis generated by bCDase had a profound effect on ERM phosphorylation. As shown for ceramide production, when HeLa cells were pretreated with bSMase, bCDase caused significant ERM hyperphosphorylation in a dose-dependent manner (Fig. 7F). The hyperphosphorylation of ERM appeared in cell protrusions surrounding the cells completely (Fig. 7G). Thus, ceramide hydrolysis produced not only a recovery in ERM phosphorylation levels but also a hyperphosphorylation of ERM, suggesting that formation of Sph and/or S1P might promote ERM phosphorylation.
FIGURE 7.
Effect of bCDase on SL profile and ERM proteins. A, HeLa cells were treated with bSMase (25 milliunits (mU)/ml), bCDase (20 milliunits/ml), or with bSMase and bCDase together, and the levels of the different species of ceramide were analyzed by mass spectrometry (red, C16 ceramide; blue, C24:1 ceramide). B, HeLa cells were treated with bSMase (25 milliunits/ml) and different amounts of bCDase, and the fold change in different ceramide species are presented; long chain species are displayed in red, and very long chain species are shown in blue. C and D, after bSMase and bCDase treatment, Sph and S1P levels were measured in cells (C) and in medium (D). E, the amount of protein required for in vitro assays were ∼5000 times less (1 ng) than those required for in vivo assays (left panel). In vitro kinetics of bCDase on pure C16 (Km 1.4 ± 0.1) and C24:1 (Km 4.6 ± 0.8) ceramide species are shown (right panel). F, HeLa cells were treated with different doses of bCDase (1, 2, 5, 10, and 20 milliunits/ml) and treated with both bSMase (25 milliunits/ml) and different doses of bCDase, and the effect on phospho-ERM and total ezrin (tEzrin) were studied by Western blotting. G, HeLa cells were treated with bSMase (25 milliunits/ml), bCDase (20 milliunits/ml), or co-treated with both, and phospho-ERM (green) was analyzed by confocal microscopy. Nuclei (stained with Draq5, blue) and F-actin (stained with rhodamine phalloidin, red) were also visualized.
Bacterial Ceramidase Showed Same Vmax and Similar although Preferential Km toward Long Chain Ceramide Species
To study whether the selective loss of long chain ceramide in cells treated with bCDase was due to accessibility to different pools of ceramide or to a different substrate preference, the in vitro kinetics of bCDase toward a long chain ceramide (C16 ceramide) and very long chain ceramide (C24:1) were determined (Fig. 7E). BCDase showed similar Vmax values toward C16 and C24:1 ceramide species, and moderately different in Km values from 1.4 μm in C16 ceramide to 4.6 μm in C24:1 ceramide (Fig. 7E, right panel), indicating the enzymes do exhibit a slight in vitro preference for long chain ceramide over very long chain ceramide. However, the amount required for the in vitro assays was 1/5000 the amount used in the studies in cell culture (Fig. 7E, left panel) to avoid excess of enzyme and maintain Michaelis-Menten conditions of modest substrate consumption. The higher concentrations used in cell studies were designed to achieve total conversion of ceramide in the membrane and, as such, should be much less susceptible to relatively small differences in Km. Thus, the selective loss of C16 ceramide but not C24:1 ceramide in cells treated with bSMase and bCDase suggests that bCDase is able to access C16 ceramide pools more readily.
S1P-induced Hyperphosphorylation of ERM Proteins
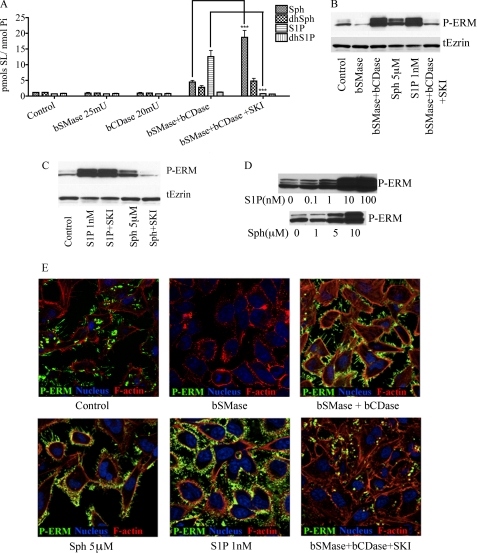
The above results suggested that ERM hyperphosphorylation caused by the combined action of bSMase and bCDase treatment could be explained by a decrease in long chain ceramide species or production of Sph or S1P. To distinguish the effects of decreasing ceramide levels and generating Sph from those caused by generating S1P, HeLa cells were pretreated with a Sph kinase inhibitor (SKI) (40) before bSMase and bCDase treatment. Pretreatment (6 h) with SKI effectively inhibited the S1P accumulation following combined bSMase/bCDase treatment, and Sph and dhSph levels were increased (Fig. 8A). Interestingly, SKI also blocked ERM hyperphosphorylation following bSMase/bCDase co-treatment, suggesting that formation of S1P by sphingosine kinase was responsible for enhanced ERM phosphorylation (Fig. 8B) and arguing against either the drop in ceramide or the rise in Sph. To confirm these data, HeLa cells were also treated with exogenous Sph and S1P, both of which induced a dose-dependent increase in ERM phosphorylation (Fig. 8, B and D); however, at least 5 μm Sph was required for the phosphorylation, whereas only 1 nm S1P was sufficient (Fig. 8C). Taken together, these results demonstrate that hydrolysis of ceramide by bCDase drives ERM hyperphosphorylation by formation of endogenous S1P.
FIGURE 8.
Differential effects of S1P, Sph, and long-chain ceramides on ERM. A, HeLa cells were treated with bSMase (25 milliunits/ml), bCDase (20 milliunits/ml), or both of them, and pretreated with either vehicle or sphingosine kinase inhibitor (SKI, 10 mm for 6 h), and the levels of Sph and S1P, as well as their dihydro forms were measured. B, phospho-ERM and total ezrin were measured by Western blotting in HeLa cells after being treat with vehicle, bSMase, bSMase, and bCDase together, exogenous Sph (5 μm), exogenous S1P (1 nm), and with bSMase (25 milliunits/ml) and bCDase (20 milliunits/ml) in cells pretreated 6 h with SKI. C, phospho-ERM levels after being treated with S1P (1 nm) or Sph (5 μm) in presence or absence of sphingosine kinase inhibitor (SKI, 10 mm, 6-h pretreatment). D, phospho-ERM levels after being treated with different doses of S1P or Sph. E, HeLa cells were treated in the same way that in B and stained for phospho-ERM (green). Nuclei (stained with Draq5, blue) and F-actin (stained with rhodamine phalloidin, red) also were visualized.
Competing Effects of Ceramide and S1P in Regulation of Phospho-ERM Levels
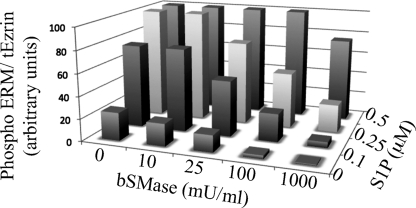
As shown above, whereas generation of ceramide at the plasma membrane rapidly dephosphorylates ERM, very low concentrations of S1P recover and hyperphosphorylate ERM levels. To study this relationship, HeLa cells were treated with different amounts of bSMase for 5 min, and for each amount of bSMase, the cells were also co-treated with different amounts of S1P. As shown in Fig. 9, very small amounts of S1P (100 nm) were able to phosphorylate ERM proteins beyond the basal levels, except when 1 unit/ml of bSMase was used, which required significantly higher concentrations of S1P. Using 0.5 μm of S1P, phosphorylation of ERM reached a plateau, and up to 1 unit/ml of bSMase was necessary to detect some dephosphorylation. Thus, these results demonstrate dynamic and reciprocal effects of ceramide and S1P on ERM phosphorylation.
FIGURE 9.
Balance between ceramide and S1P. HeLa cells were treated with different concentrations of bSMase, and for each bSMase treatment were also treated with different amounts of S1P. After 5 min of treatment, cells were harvested, and the phospho-ERM protein was detected by Western blotting, quantified, and normalized by total ezrin signal.
DISCUSSION
In this manuscript, we present a new mechanism of ERM (ezrin, radixin, and moesin) regulation by interconversion of SLs in the plasma membrane. Thus, hydrolysis of SM at the plasma membrane producing long chain ceramide species induced a rapid ERM dephosphorylation, which can be overcome by hydrolysis of a specific fraction of plasma membrane ceramide to Sph. The Sph generated at the plasma membrane is rapidly transformed into S1P, which, at nanomolar levels (suggesting receptor mediated effects), induced hyperphosphorylation of ERM. These results have implications for the regulation of ERM, the functions of ceramide at the plasma membrane, and the interconversion of bioactive SLs.
Previous studies showed that ERM are regulated by phosphorylation at the C terminus, switching them from an inactive, closed conformation, where the N and C terminus interact with each other, to an active, open conformation. Thus, in the open conformation, the N terminus can interact with the plasma membrane, and the C terminus interacts with F-actin fibers at the cell cortex. However, little is known about regulation of ERM phosphorylation, although a few kinases and phosphatases have been involved. Not much is known about the signaling that triggers ERM regulation. A previous study in our group invoked acid sphingomyelinase as a possible regulator of ERM phosphorylation, opening a whole new pathway for ERM regulation. Continuing that work, in the present study, we demonstrate that generation of ceramide at the plasma membrane regulates ERM dephosphorylation.
Ceramide is a metabolic hub that can be metabolized toward different structural and bioactive derivatives through the action of various ceramide-acting enzymes. This metabolic interconversion has emerged as a common and recalcitrant problem in defining lipid-mediated signaling. For example, although our previous studies implicated acid sphingomyelinase, one may not directly conclude that it is ceramide per se that mediates the effects. It could be the loss of SM or the generation of subsequent metabolites of ceramide that mediate the dephosphorylation of ERM proteins. To this end, we employed several specific tools to probe the roles of SM and ceramide metabolites. First, the effects on ERM dephosphorylation by bSMase were observed using low doses and short treatments. In those conditions, no changes in hexosyl or lactosyl ceramide were observed, and no Sph or S1P generation was observed. Second, ceramide 1-phosphate was excluded by both, SMase D treatment, which generates C1P at the plasma membrane, and by applying exogenous C1P (data not shown). Third, the results with SMase D also rule out a role for the drop in SM as the main trigger of dephosphorylation. Fourth, the addition of bCDase to bSMase resulted in a drop in ceramide and extensive formation of both Sph and S1P. This not only prevented the dephosphorylation effect of bSMase but converted it to a stimulatory one. Finally, pretreatment with SMase D prevented the action of bSMase, demonstrating that the effects of bSMase on ERM are due to its action in producing ceramide from SM (and not some other fortuitous effect of bSMase). Thus, taken together, the results point directly to ceramide as the main mediator. However, minor as-yet-unknown ceramide metabolites cannot be excluded by the current approaches.
As shown by other studies (41, 42), phosphorylation of ERM proteins is an important regulatory mechanism that has been related to cell motility, migration, and cell invasion. We found that SL are not just involved in ERM inactivation but are also involved in a rapid activation driven by S1P. We observed that exogenous S1P led to ERM phosphorylation. Furthermore, endogenous production of S1P from hydrolysis of plasma membrane ceramide by the action of bCDase, also resulted in a dramatic phosphorylation of ERM proteins. This was inhibited by inhibitors of SK, strongly suggesting it was S1P and not Sph. Moreover, the potent effects of S1P, with addition of very low concentrations of S1P (100 nm) induced a dramatic phosphorylation of ERM proteins. This low concentration of S1P strongly suggests an S1P receptor-mediated event. Thus, S1P is emerging as a novel, potent, and robust inducer of ERM phosphorylation.
These observations have relevance to a possible interaction of S1P and ERM in cancer progression as several cancers have been shown to have high levels of SK and S1P and also have been related to ERM activation. The activation of ERM by S1P suggests a strong link between these two pathways regulating cell adhesion and motility. This is currently under investigation.
The opposing effects between ceramide and S1P have been reported in several important biologic responses, although such a duality has not been much described for acute signaling effects. Ceramide has been associated with cell death, cell cycle arrest, and other antitumoral functions, whereas S1P has been described to promote cell growth, stimulate cell motility, and enhance tumor progression. In the current study, we provide a quantitative analysis of the reciprocal effects of ceramide at the plasma membrane and S1P on ERM phosphorylation, showing conclusively that the status of ERM phosphorylation is regulated in opposite directions by ceramide and S1P. It should be noted that the relative levels of SM, ceramide, and S1P in the cell are in a nearly logarithmical scale; thus, a small hydrolysis of SM may result in a dramatic change in ceramide, and a small hydrolysis in ceramide can also result in a huge change in Sph and S1P folds. We show here that hydrolysis of moderate amounts of SM generated large changes in ceramide that caused a profound dephosphorylation of ERM. Likewise, a modest hydrolysis of the generated ceramide was sufficient to overcome ERM dephosphorylation by ceramide, hyperphosphorylating ERM beyond basal levels following bSMase treatment. These results support a model of dual and reciprocal regulation of an acute signaling function, ERM phosphorylation, by ceramide and S1P.
The results from this study raise an intriguing possibility on the relative paucity of “basal” ceramide in the outer leaflet of the plasma membrane. The subcellular localization of bioactive SLs has been poorly studied, primarily because of the lack of tools to investigate these processes at the subcellular level. Ceramide can be generated in the cell by different pathways through the action of various enzymes that themselves show distinct subcellular localization. An important finding in this study was the near absence of any increase of Sph after bCDase treatment alone. Sph was detected only if bSMase was added to the cell prior to bCDase, thus generating the ceramide substrate for the bCDase. These results demonstrate that bCDase is capable of hydrolyzing the bSMase-generated ceramide; and thus, we are led to suggest that there is little ceramide substrate for bCDase in HeLa cells not treated with bSMase.
It is not clear whether these results apply only to the outer leaflet or to both leaflets of the plasma membrane. Some biophysical studies (43–45) suggested that ceramide is able to flip-flop freely between the two layers of the plasma membrane. If these data are applicable to biological membranes, we also could assume that ceramide is not just poor in the outer leaflet but also in the entire plasma membrane in nonstimulated conditions. Furthermore, not only is the topology of ceramide important but the species of ceramide involved in cell signaling is also important. Thus, under certain conditions, long acyl chain species (LC; from C14 to C18), mainly C16 ceramide, have shown distinct effects from very long species (VLC; from C20 to C26) (46). In this work, in HeLa cells, limiting concentrations of bSMase caused an early increase of long chain ceramide species, followed by a later increase of some very long chain ceramides (C24, C24:1) but not others (C26, C26:1). On the other hand, in vitro studies with bSMase did not show preferential substrate hydrolysis between LC and VLC SM. These results suggest that the plasma membrane is enriched in LC species, although other more complex possibilities could explain these results (e.g. LC and VLC chains could be structured differently in the plasma membrane, showing different accessibility to bSMase in biologic membranes). Furthermore, in HeLa cells, bCDase also acted preferentially on LC ceramides but had no effect on VLC species. These results further support the hypothesis that mainly LC SM species are present at the outer leaflet of the plasma membrane, or at least they present different organization than VLC, because bCDase showed the same Vmax between C16 and C24:1 ceramide, and bCDase was applied in excess, minimizing the effects of the small differences in Km. Regardless of the mechanisms for the differential generation of LC ceramides, the results clearly suggest that ERM dephosphorylation is much more tied to the formation of LC ceramides than VLC ceramides. Thus, these studies are beginning to point to very specific metabolic and functional differences for LC versus VLC SMs and ceramides.
Different tools have been widely used to study SL signaling in cells. Exogenous SLs have been added directly into the cell medium. Also, the enzymes involved in SL metabolism have been overexpressed, knocked down, or pharmacologically inhibited. In this work, we present a tool kit of different exogenous enzymes involved in SL metabolism that can be used to study specifically SL signaling at the plasma membrane. Unlike other methods used, we show that exogenous enzymes can be used for very short treatment, affecting only the SL profile at the plasma membrane, at least over short time treatments. Using Venus-tagged lysenin, which specifically binds SM, we showed that bSMase hydrolyzed plasma membrane SM but not intracellular SM, consistent with previous cell fractionation studies (37). We observed that at short time treatments, neither bSMase, SMase D nor bCDase was detected to be internalized into the cell, and, therefore, the effects of those enzymes appear to be caused by effects on the plasma membrane. Using bSMase versus SMase D in parallel to produce the same degree of SM hydrolysis allowed to distinguish the effects caused by SM depletion versus those caused by formation of ceramide or ceramide-1phosphate. Furthermore, bCDase was used here for the first time in cell culture, demonstrating its ability to hydrolyze plasma membrane ceramide. Thus, this combination of tools can be used to study SL signaling at the plasma membrane.
In conclusion, the results show that ERM proteins are regulated by SLs. Thus, ceramide and S1P regulate the phosphorylation state of ERM, with the long chain ceramides at the plasma membrane sufficient to dephosphorylate ERM proteins with S1P countering these effects and producing robust phosphorylation of ERMs. In addition, these studies have implications for the subcellular and topological organization of SL signaling.
Supplementary Material
Acknowledgments
We thank all members in the Hannun and Obeid laboratories, LC-MS/MS analysis of sphingolipids was performed by Lipidomics Shared Resource, Medical University of South Carolina (MUSC) and conducted in the facility constructed with support from the National Institutes of Health, Grant Number C06 RR018823 from the Extramural Research Facilities Program of the National Center for Research Resources. We also thank the MUSC Cell and Molecular Imaging Core, and the MUSC Synthetic Lipidomics Core. We give special thanks to Dr. Kevin R Lynch (University of Virginia, School of Medicine, Charlottesville, VA) and Dr. Toshihide Kobayashi (Supra-Biomolecular System Research Group, RIKEN, Japan) for sharing His-SMase D and His-Venus-lysenin plasmid constructs, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grants CA87584 and CA97132.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- SL
- sphingolipid
- bSMase
- bacterial sphingomyelinase
- bCDase
- bacterial ceramidase
- SMase D
- sphingomyelinase D
- SKI
- sphingosine kinase inhibitor (p-hydroxyanilino)-4-(p-chlorophenyl)
- SM
- sphingomyelin
- Sph
- sphingosine
- S1P
- sphingosine 1-phosphate
- C1P
- ceramide 1-phosphate
- LC
- long acyl chain
- VLC
- very long acyl chain.
REFERENCES
- 1.Bretscher A., Edwards K., Fehon R. G. (2002) Nat. Rev. Mol. Cell Biol. 3, 586–599 [DOI] [PubMed] [Google Scholar]
- 2.Lamb R. F., Ozanne B. W., Roy C., McGarry L., Stipp C., Mangeat P., Jay D. G. (1997) Curr. Biol. 7, 682–688 [DOI] [PubMed] [Google Scholar]
- 3.Serrador J. M., Alonso-Lebrero J. L., del Pozo M. A., Furthmayr H., Schwartz-Albiez R., Calvo J., Lozano F., Sánchez-Madrid F. (1997) J. Cell Biol. 138, 1409–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Harada T., Juang Y. T., Kyttaris V. C., Wang Y., Zidanic M., Tung K., Tsokos G. C. (2007) J. Immunol. 178, 1938–1947 [DOI] [PubMed] [Google Scholar]
- 5.Zohar R., Suzuki N., Suzuki K., Arora P., Glogauer M., McCulloch C. A., Sodek J. (2000) J. Cell. Physiol. 184, 118–130 [DOI] [PubMed] [Google Scholar]
- 6.Endo K., Kondo S., Shackleford J., Horikawa T., Kitagawa N., Yoshizaki T., Furukawa M., Zen Y., Pagano J. S. (2009) Oncogene 28, 1725–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prag S., Parsons M., Keppler M. D., Ameer-Beg S. M., Barber P., Hunt J., Beavil A. J., Calvert R., Arpin M., Vojnovic B., Ng T. (2007) Mol. Biol. Cell 18, 2935–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukita S., Yonemura S. (1997) Curr. Opin. Cell Biol. 9, 70–75 [DOI] [PubMed] [Google Scholar]
- 9.Hirao M., Sato N., Kondo T., Yonemura S., Monden M., Sasaki T., Takai Y., Tsukita S., Tsukita S. (1996) J. Cell Biol. 135, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone F., Lugini L., Matarrese P., Luciani F., Federici C., Iessi E., Margutti P., Stassi G., Malorni W., Fais S. (2004) J. Biol. Chem. 279, 9199–9207 [DOI] [PubMed] [Google Scholar]
- 11.Fais S., Malorni W. (2003) J. Leukoc. Biol. 73, 556–563 [DOI] [PubMed] [Google Scholar]
- 12.Helander T. S., Carpén O., Turunen O., Kovanen P. E., Vaheri A., Timonen T. (1996) Nature 382, 265–268 [DOI] [PubMed] [Google Scholar]
- 13.Hao J. J., Liu Y., Kruhlak M., Debell K. E., Rellahan B. L., Shaw S. (2009) J. Cell Biol. 184, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuan Y. C., Pang S. T., Cedazo-Minguez A., Norstedt G., Pousette A., Flores-Morales A. (2006) J. Biol. Chem. 281, 29938–29948 [DOI] [PubMed] [Google Scholar]
- 15.Pietromonaco S. F., Simons P. C., Altman A., Elias L. (1998) J. Biol. Chem. 273, 7594–7603 [DOI] [PubMed] [Google Scholar]
- 16.Wald F. A., Oriolo A. S., Mashukova A., Fregien N. L., Langshaw A. H., Salas P. J. (2008) J. Cell Sci. 121, 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S., Tsukita S. (1998) J. Cell Biol. 140, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura N., Oshiro N., Fukata Y., Amano M., Fukata M., Kuroda S., Matsuura Y., Leung T., Lim L., Kaibuchi K. (2000) Genes Cells 5, 571–581 [DOI] [PubMed] [Google Scholar]
- 19.Cant S. H., Pitcher J. A. (2005) Mol. Biol. Cell 16, 3088–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeidan Y. H., Jenkins R. W., Hannun Y. A. (2008) J. Cell Biol. 181, 335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hishiya A., Ohnishi M., Tamura S., Nakamura F. (1999) J. Biol. Chem. 274, 26705–26712 [DOI] [PubMed] [Google Scholar]
- 22.Smith E. R., Merrill A. H., Obeid L. M., Hannun Y. A. (2000) Methods Enzymol. 312, 361–373 [DOI] [PubMed] [Google Scholar]
- 23.Meivar-Levy I., Sabanay H., Bershadsky A. D., Futerman A. H. (1997) J. Biol. Chem. 272, 1558–1564 [DOI] [PubMed] [Google Scholar]
- 24.Hla T. (2004) Prostaglandins Other Lipid Mediat. 73, 1–2 [DOI] [PubMed] [Google Scholar]
- 25.Yu H., Okada T., Kobayashi M., Abo-Elmatty D. M., Jahangeer S., Nakamura S. (2009) Genes Cells 14, 597–605 [DOI] [PubMed] [Google Scholar]
- 26.Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 27.Takenouchi H., Kiyokawa N., Taguchi T., Matsui J., Katagiri Y. U., Okita H., Okuda K., Fujimoto J. (2004) J. Cell Sci. 117, 3911–3922 [DOI] [PubMed] [Google Scholar]
- 28.Gassert E., Avota E., Harms H., Krohne G., Gulbins E., Schneider-Schaulies S. (2009) PLoS Pathog. 5, e1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu B. X., Snook C. F., Tani M., Büllesbach E. E., Hannun Y. A. (2007) J. Lipid Res. 48, 600–608 [DOI] [PubMed] [Google Scholar]
- 30.Ishitsuka R., Yamaji-Hasegawa A., Makino A., Hirabayashi Y., Kobayashi T. (2004) Biophys. J. 86, 296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. (2006) Methods 39, 82–91 [DOI] [PubMed] [Google Scholar]
- 32.Van Veldhoven P. P., Bell R. M. (1988) Biochim. Biophys. Acta 959, 185–196 [DOI] [PubMed] [Google Scholar]
- 33.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 34.Jenkins R. W., Canals D., Hannun Y. A. (2009) Cell Signal 21, 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chigorno V., Giannotta C., Ottico E., Sciannamblo M., Mikulak J., Prinetti A., Sonnino S. (2005) J. Biol. Chem. 280, 2668–2675 [DOI] [PubMed] [Google Scholar]
- 36.Yan Z. Q., Hansson G. K. (2007) Immunol. Rev. 219, 187–203 [DOI] [PubMed] [Google Scholar]
- 37.Linardic C. M., Hannun Y. A. (1994) J. Biol. Chem. 269, 23530–23537 [PubMed] [Google Scholar]
- 38.Kiyokawa E., Makino A., Ishii K., Otsuka N., Yamaji-Hasegawa A., Kobayashi T. (2004) Biochemistry 43, 9766–9773 [DOI] [PubMed] [Google Scholar]
- 39.Gupta V. R., Patel H. K., Kostolansky S. S., Ballivian R. A., Eichberg J., Blanke S. R. (2008) PLoS Pathog. 4, e1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.French K. J., Schrecengost R. S., Lee B. D., Zhuang Y., Smith S. N., Eberly J. L., Yun J. K., Smith C. D. (2003) Cancer Res. 63, 5962–5969 [PubMed] [Google Scholar]
- 41.Elliott B. E., Meens J. A., SenGupta S. K., Louvard D., Arpin M. (2005) Breast Cancer Res. 7, R365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abiatari I., Esposito I., De Oliveira T., Felix K., Xin H., Penzel R., Giese T., Friess H., Kleeff J. (2010) J. Cell Mol. Med. 14, 1166–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Contreras F. X., Sanchez-Magraner L., Alonso A., Goni F. M. (2010) FEBS Lett. 584, 1779–1786 [DOI] [PubMed] [Google Scholar]
- 44.Contreras F. X., Villar A. V., Alonso A., Goñi F. M. (2009) Methods Mol. Biol. 462, 155–165 [DOI] [PubMed] [Google Scholar]
- 45.Pohl A., López-Montero I., Rouvière F., Giusti F., Devaux P. F. (2009) Mol. Membr. Biol. 26, 194–204 [DOI] [PubMed] [Google Scholar]
- 46.Liu X., Elojeimy S., Turner L. S., Mahdy A. E., Zeidan Y. H., Bielawska A., Bielawski J., Dong J. Y., El-Zawahry A. M., Guo G. W., Hannun Y. A., Holman D. H., Rubinchik S., Szulc Z., Keane T. E., Tavassoli M., Norris J. S. (2008) Front Biosci 13, 2293–2298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.