Abstract
Synucleins and apolipoproteins have been implicated in a number of membrane and lipid trafficking events. Lipid interaction for both types of proteins is mediated by 11 amino acid repeats that form amphipathic helices. This similarity suggests that synucleins and apolipoproteins might have comparable effects on lipid membranes, but this has not been shown directly. Here, we find that α-synuclein, β-synuclein, and apolipoprotein A-1 have the conserved functional ability to induce membrane curvature and to convert large vesicles into highly curved membrane tubules and vesicles. The resulting structures are morphologically similar to those generated by amphiphysin, a curvature-inducing protein involved in endocytosis. Unlike amphiphysin, however, synucleins and apolipoproteins do not require any scaffolding domains and curvature induction is mediated by the membrane insertion and wedging of amphipathic helices alone. Moreover, we frequently observed that α-synuclein caused membrane structures that had the appearance of nascent budding vesicles. The ability to function as a minimal machinery for vesicle budding agrees well with recent findings that α-synuclein plays a role in vesicle trafficking and enhances endocytosis. Induction of membrane curvature must be under strict regulation in vivo; however, as we find it can also cause disruption of membrane integrity. Because the degree of membrane curvature induction depends on the concerted action of multiple proteins, controlling the local protein density of tubulating proteins may be important. How cellular safeguarding mechanisms prevent such potentially toxic events and whether they go awry in disease remains to be determined.
Keywords: Apolipoproteins, Electron Microscopy (EM), Membrane, Parkinson Disease, Synuclein, 11 Amino Acid Repeats, Amphipathic Helix, Membrane Remodeling
Introduction
Several familial forms of Parkinson disease (PD)2 have been linked to mutations in α-synuclein and animal studies further support a causative role of this protein in neurodegeneration (1, 2). α-Synuclein is a predominantly presynaptic protein, but in PD it forms fibrils and deposits in intracellular inclusions called Lewy bodies (3).
The membrane interaction of α-synuclein has become of significant interest for the pathological as well as the physiological functions of this protein (4, 5). Although its physiological roles are not fully understood, it is thought that α-synuclein binds to synaptic vesicles, plays a role in neuronal plasticity, modulates the release of neurotransmitters, plays a role in vesicular trafficking and affects brain lipid metabolism (6–14). A role in endocytosis has recently been supported by the finding that overexpression of α-synuclein increases basal and evoked synaptic vesicle endocytosis in hippocampal neurons (15). However, α-synuclein has also been shown to be disruptive to the integrity of cellular membranes. Overexpression of α-synuclein in cell lines as well as animal models of PD have provided evidence for Golgi fragmentation (16, 17), mitochondrial degeneration (18, 19), damage to lysosomes (20), as well as damage to endoplasmic reticulum and nuclear membranes (21). Moreover, lipids represent a significant component of Lewy bodies and have been proposed to be derived from degraded membrane organelles (22). Although oligomerization and aggregation of α-synuclein could be an important factor in destabilizing membrane integrity (23–26), the molecular mechanisms by which α-synuclein promotes disruption of cellular membranes remain largely unknown. Thus, a better understanding of α-synuclein-membrane interaction might help to shed light on its physiological as well as its pathological roles.
A number of biophysical studies have concluded that the membrane interaction of α-synuclein is curvature-sensitive (27–29) and that curvature-sensitive membrane binding is mediated by an extended helical structure (27, 30–34). Recent work revealed the existence of other proteins, mainly involved in membrane remodeling and vesicle trafficking events, with curvature-sensing abilities akin to those of α-synuclein (35–37). Interestingly, some of these proteins have the additional ability to convert moderately curved bilayers into small and highly curved vesicles or tubules (35, 36, 38). Endophilin and amphiphysin are examples of such curvature-inducing proteins involved in endocytosis. Both proteins have a highly curved BAR domain whose concave surface is rich in basic residues (39–41). These structures suggest a scaffolding mechanism wherein the concave surface directly interacts with membranes and molds their shapes (35, 36, 39–42). However, both proteins also have an N-terminal amphipathic helix which could promote curvature by wedging into the bilayer (36, 38, 39, 43, 44). In addition endophilin contains a central insert region, which also forms a membrane-inserting amphipathic helix (39, 45, 46).
Considering that the wedging of amphipathic helices alone might be sufficient to induce membrane curvature, we set out to test whether α-synuclein might be capable of this effect. The possibility that α-synuclein can induce morphological changes in membranes has been suggested (31, 47, 48), but it has not been investigated in detail. For example, it is not clear whether such interactions predominantly promote increased or decreased membrane curvature, as α-synuclein has been reported to increase as well as decrease vesicle size (31, 47, 48). Several studies have reported that membrane interaction of α-synuclein, as well as that of other amyloid proteins, can cause projections to emanate from vesicles. While it has been suggested that these projections are membrane tubules induced by the α-helical form (47), they have often been considered to be derived from β-sheet-rich fibrils or other misfolded forms that were mixed with lipids (49–52).
To further test whether the ability to induce membrane curvature might be a shared property of α-synuclein and related 11 amino acid repeat-containing proteins, we also included β-synuclein and apolipoprotein A-1 (apoA-1) in the present study. As with α-synuclein, the 11 amino acid repeat regions of both of these proteins can also form amphipathic helices that interact with membranes (53–56). β-Synuclein has a high sequence similarity to α-synuclein, but lacks one 11 amino acid repeat and does not readily form fibrils (57). ApoA-1 interacts with membranes and lipid particles in vivo and is known to play an important role in lipid metabolism (53, 58). In vitro apolipoproteins are known to form lipid-containing disks but the mechanisms by which they form are not well understood (59). Whereas it has long been suspected that synucleins and apolipoproteins might have similar effects on lipid membranes, this has not been shown directly.
EXPERIMENTAL PROCEDURES
Preparation of α-Synuclein, β-Synuclein, Amphiphysin, and ApoA-I
The human α-synuclein and β-synuclein were expressed in Escherichia coli BL21 (DE3) pLysS cells and generated as reported earlier (60). Briefly, cells were lysed by boiling, followed by acid precipitation. Supernatant was passed through anion exchange columns, and eluted with a 0–1.0 m NaCl gradient. The human β-synuclein was further subjected to gel filtration using Superdex 200 column.
Human apoA-I containing a hexa-His affinity tag at the N terminus was expressed in E. coli strain BL21 Star (DE3) cells (Invitrogen) using a pET-20b vector as described previously (61). ApoA-I was purified on a His-Trap-Nickel-chelating (GE) column using phosphate-buffered saline (PBS), pH 7.4 with 3 m guanidine. The protein was then washed in PBS (pH 7.4) containing 100 mm imidazole, and then eluted with PBS containing 500 mm imidazole. Imidazole was removed from the protein sample by using Bio-spin columns (Bio-Rad) equilibrated with PBS, pH 7.4.
The plasmid containing His6-tagged N-BAR domain (amino acids 1–244) of Drosophila amphiphysin was kindly provided by Dr. Harvey McMahon (Medical Research Council). The protein was expressed in E. coli BL21(DE3) pLysS cells and purified using nickel-nitrilotriacetic acid-agarose, followed by Superdex 200 gel filtration, and finally monoS cation exchange chromatography was performed using buffer gradient composed of the following two buffers: buffer A (20 mm Hepes pH 7.4, 1 mm dithiothreitol (DTT), and buffer B (20 mm Hepes pH 7.4 2 m NaCl and 1 mm DTT) with the protein eluting around 600 mm NaCl.
Preparation of Phospholipid Vesicles
The following synthetic lipids were used to prepare different membrane compositions: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-RAC-(1-glycerol)] (POPG), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), cholesterol, sphingomyelin. All lipids were purchased from Avanti Polar Lipids Inc. (Alabaster, AL). Large non-extruded vesicles used for electron microscopy studies were prepared by vortexing the dried lipid film in the required buffer. Addition of 2 mm EGTA to the buffer showed that the observed tubulation does not depend on metal ions such as Ca2+ (supplemental Fig. S5C).
Phospholipid Vesicle Clearance Assay
The ability of α-synuclein to clear large lipid vesicles was monitored by measuring change in light scattering as a function of time using a Jasco V-550 UV/Visible spectrophotometer. The monitoring wavelength was set at 500 nm with a slit width of 2 nm and medium response time. Briefly, lipid vesicles were suspended in 20 mm Hepes pH 7.4 with 100 mm NaCl at a final volume of 500 μl in a quartz cuvette.
Circular Dichroism (CD)
All CD spectra were obtained using a Jasco J-810 spectropolarimeter with 1 mm quartz cell at room temperature. A scan rate of 50 nm/minute, bandwidth of 1 nm, 0.1 nm time response and step resolution of 0.5 nm was used for all experiments. Protein concentration was determined using the extinction coefficient of protein at 280 nm based on the number of tryptophan and tyrosine molecules in the protein. Appropriate blanks were collected under similar conditions and were subtracted to obtain the final spectra. A 10 mm sodium phosphate pH 7.4 buffer was used for all CD studies.
Fluorescence Microscopy
Giant vesicles were prepared as follows: Lipid mixtures were prepared with POPG, POPG/POPE (1:1), POPG/POPC (1:1), and supplemented with 0.5% 1,2 dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)-Atto633 (Attotec) and 0.5% DOPE-biotin (Avanti). A thin lipid film was formed in a teflon cup and rehydrated overnight at 37 °C. Liposomes were prepared with 2 days of rehydration at 37 °C and no extrusion. The giant liposomes were immobilized on an assay surface and incubated with protein for 30 min at room temperature.
Alexa488-labeled α-synuclein was obtained by reacting α-synuclein Y136C derivative with 10× molar excess of Alexa-488 (Invitrogen) for 4 h. Unreacted dye was removed by gel filtration using PD-10 column (GE). Glass surfaces were passivated with a BSA:BSA-Biotin mixture and subsequently coated with streptavidin onto which the biotinylated vesicles tethered, as described previously (62). Microscopy was performed on a Leica TCS SP5 Confocal Fluorescence Microscope, with an AOBS/AOTF system allowing tunable wavelength detection intervals. The objective used was an oil immersion HCX PL APO with ×100 magnification and numerical aperture 1.4. Alexa488-labeled α-synuclein was excited at 488 nm, using an argon laser, while detecting from 495 nm to 555 nm. Vesicles containing DOPE-Atto633 dye were excited at 633 nm, detecting from 640 nm to 790 nm. Both channels were recorded simultaneously for the duration of the experiment, with a time resolution of ∼1.5 s. The microscope was kept at a constant temperature of 22 °C.
Electron Microscopy
Samples were negatively stained for transmission electron microscopy studies. Carbon-coated formvar films mounted on copper grids (EMS) were floated on a 10-μl droplet of sample for 5 min and the excess liquid was removed from the grids with a filter paper. The grids were then stained with 2% uranyl acetate. A JEOL 1400 transmission electron microscope accelerated to 100 kV was used for specimen observation.
Dye Leakage Assay
Leakage assay was modified from a previous method (63). Large unilamellar vesicles (LUVs) composed of either 66%POPG/33%POPE, 100% POPG or POPG/POPC (1:1 molar ratio) were prepared by resuspending dried lipid in 9 mm ANTS (8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt) and 25 mm DPX [p-xylene-bis(pyridinium bromide)] (Invitrogen). This lipid solution was treated to 10 cycles of freeze/thaw, and large 1 μm diameter vesicles were formed by passing lipid mixture through mini-extruder (20×) with 1 μm cutoff polycarbonate membrane (Avanti Polar Lipids Inc.) Unencapsulated dye was removed by gel filtration using PD-10 column (GE). 100% leakage was attained using a final concentration of 0.04% Triton-X 100. All data were normalized to 100% leakage. Fluorescence measurements were recorded using a JASCO fluorometer (FP-6500), setting excitation and emission at 380 nm and 520 nm with slits of 5 nm and 20 nm, respectively.
RESULTS
α-Synuclein Transforms Negatively Charged Vesicles into Smaller Structural Entities
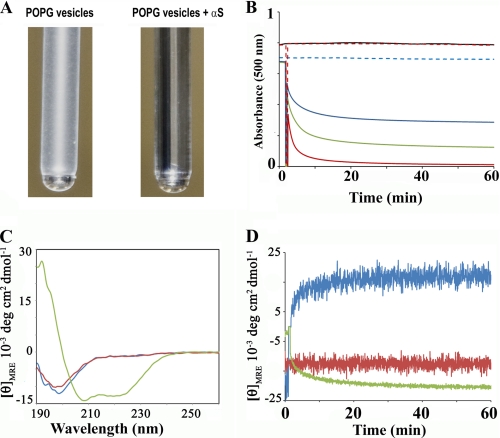
To test whether α-synuclein can induce changes in the shape and size of phospholipid vesicles, we incubated α-synuclein with large, POPG-containing vesicles and visually inspected the vesicle suspension. This assay is analogous to those previously established for investigating effects of apolipoproteins and detergents on vesicle structure (64–66). Because of their large size, non-extruded vesicles exhibit strong scattering and have a milky appearance (Fig. 1A). Remarkably, after only 1 h of incubation with α-synuclein, the vesicle suspension became completely clear. In a more quantitative approach, we monitored the light scattering by recording the absorption of the suspension at 500 nm. As shown in Fig. 1B, the decrease in scatter exhibited very fast kinetics for the first 5 min and then began to level off. This effect was dose-dependent, as increasing amounts of α-synuclein progressively reduced scattering of the vesicle suspension. These results showed that α-synuclein can remodel large POPG-containing vesicles into smaller structural entities. In contrast, no change was observed when analogous experiments were performed using large vesicles containing the zwitterionic POPC.
FIGURE 1.
Phospholipid vesicle clearance by α-synuclein. A, photograph of test tubes containing 600 μm POPG vesicles before and 60 min after addition of 60 μm α-synuclein. Because of their large size, the vesicles scatter light, but the addition of α-synuclein causes the suspension to become clear. B, clearance of phospholipid vesicles in the presence of varying amounts of α-synuclein was continually monitored by recording the apparent absorbance at 500 nm. Control traces for POPG vesicles (400 μm) in the absence of α-synuclein are indicated by the broken blue trace. POPG vesicles (400 μm) incubated with 10 μm, 20 μm and 40 μm of α-synuclein are shown with the blue, green, and red traces, respectively. Control POPC vesicles (400 μm) in the absence and presence of 40 μm α-synuclein are given by the black and broken red lines, respectively. Large non-extruded vesicles were used. C, circular dichroism was used to distinguish whether the observed vesicle clearing effect was mediated by α-helical or misfolded, β-sheet containing α-synuclein. α-synuclein (20 μm) was incubated for 5 min with large non-extruded vesicles. The spectrum remained essentially unchanged in the presence of POPC-containing vesicles (1:20 protein to lipid molar ratio, red line) in agreement with previous findings that α-synuclein does not significantly interact with such lipids (27). α-Synuclein alone (blue); α-synuclein with POPC vesicles (red); α-synuclein with POPG vesicles (green). D, change in the mean residue ellipticity value is plotted as a function of time. A protein to lipid molar ratio of 1:20 was used for the experiment. 197.5 nm (blue); 203 nm (red); 222 nm (green). As expected for a simple two-state transition from random coil to α-helical structure, the ellipticity at the isosbestic point (203 nm) did not show any temporal changes. In contrast, the traces obtained at 197.5 nm and 222 nm exhibited spectral changes indicative of rapid α-helix formation within the first 5 min.
Measurements using circular dichroism and ThT fluorescence enhancement revealed that the membrane-clearing effect of α-synuclein was mediated by its α-helical rather than a misfolded β-sheet conformation (Fig. 1C). Interestingly, α-synuclein converted from a random coil in solution into a membrane-bound helical structure at a rate that closely matched that of vesicle clearing (Fig. 1D). Thereafter, the sample retained its helical structure for days (supplemental Fig. S1A).
Interaction of α-Synuclein with Giant Vesicles
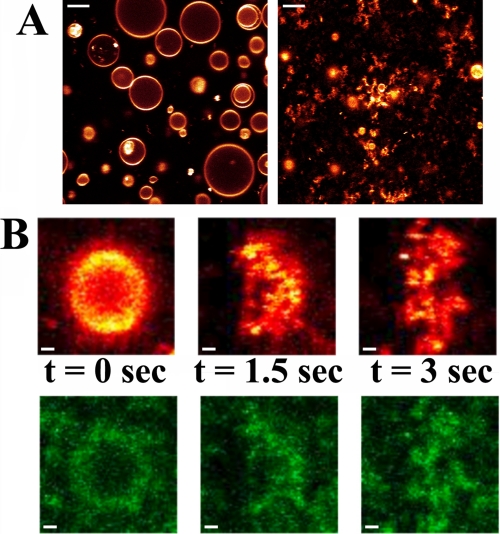
In an effort to directly visualize the interaction of α-synuclein with POPG-containing vesicles, we performed optical imaging experiments with immobilized uni- and multilamellar giant vesicles in an aqueous environment (62, 67, 68). Before addition of protein, the vesicles appeared round (Fig. 2A). After addition of α-synuclein (∼1:50 protein/lipid [P/L] molar ratio); however, they lost their spherical integrity (Fig. 2A). This process was completed within ∼15 min. At lower P/L ratios, only a fraction of the giant vesicles was disrupted within the same timeframe in good agreement with the light scattering data (Fig. 1B). Above a certain threshold concentration of added protein, vesicles disrupted very quickly, on a time scale faster than the experimental time resolution of ∼1.5 s. The deformation of a single giant vesicle is depicted in Fig. 2B, which shows corresponding images (t = 0, 1.5 s, 3 s) of a vesicle (red) and bound α-synuclein (green).
FIGURE 2.
Interaction of α-synuclein with POPG vesicles. A, giant vesicles in buffer, tethered to a glass surface, were imaged by fluorescence microscopy before (left) and after (right) incubation with α-synuclein at 1:50 protein to lipid molar ratio for 15 min. Nearly all vesicles were dispersed into smaller lipidic structures. Scale bar is 5 μm. B, disruption of individual POPG vesicles occurs on a very rapid time scale. A single vesicle (top, red) was imaged over a period of 3 s, together with corresponding images of bound α-synuclein (bottom, green). Vesicle disruption occurs faster than the 1.5 s time resolution. Scale bar is 500 nm.
Addition of α-synuclein to vesicles consisting of a 1:1 molar ratio of POPG/POPC or POPG/POPE resulted in less aggressive deformation than for POPG vesicles; while some vesicles were completely disrupted, the most common observation was blebbing, that is formation of protrusions from the vesicles as illustrated in video S1. This blebbing was accompanied by a local increase in the protein fluorescence/concentration indicating that this process is cooperative and requires multiple proteins to act in concert.
α-Synuclein Tubulates Large Vesicles Consisting of Negatively Charged Phospholipids
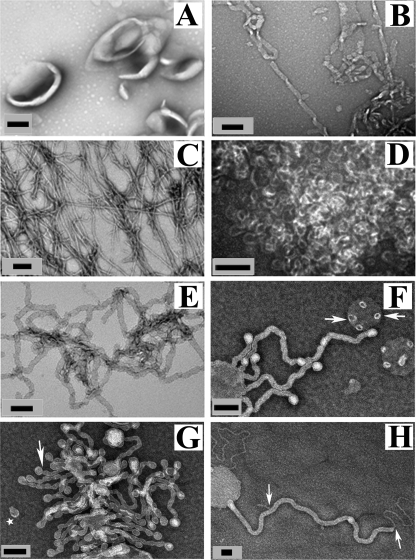
In view of the resolution limit of optical microscopy, we used transmission electron microscopy to obtain more detailed insights to this process (Fig. 3 and supplemental Fig. S2, A–C). In the absence of α-synuclein, vesicles had spherical morphologies (Fig. 3A). After 5 min of incubation with α-synuclein at a P/L molar ratio of 1:40, we mainly observed thicker tubules, ∼30–40 nm in diameter (Fig. 3B). Smaller tubules (10–15 nm in diameter) were mostly detected at a 1:20 molar ratio (Fig. 3C). Often, the tubules had a width modulated appearance (Fig. 3, B and C). The tubules as well as the α-helical structure appeared to be stable for days and no change in tubule morphology was seen after 24 h, consistent with the stable secondary structure of α-synuclein (supplemental Fig. S1B). At a 1:10 molar ratio, we often observed small circular structures that were mostly about 25 nm across. These structures are consistent with being highly curved vesicles although it cannot be excluded that smaller, non-vesicular structures (31, 69) might have also formed (Fig. 3D). The progressive increase in membrane curvature with increasing amounts of α-synuclein is reminiscent of a previous study, which investigated the morphological effects of detergent on vesicle structure (64). Moderate amounts of detergent caused tubulation without significant changes in light scattering. Increasing amounts of detergent, however, caused formation of much smaller spherical structures, which had pronounced effects on light scattering. In agreement with this study we find that conditions which give rise to the most pronounced formation of smaller circular structures (1:10 P/L molar ratio; Fig. 3D) also lead to the most pronounced vesicle clearance.
FIGURE 3.
Electron microscopy reveals α-synuclein-dependent tubulation and curvature induction of phospholipid vesicles. A, negative stain electron micrograph of POPG vesicles in the absence of α-synuclein; B–D, negative stain electron micrographs of POPG vesicles incubated with α-synuclein at a protein-to-lipid (P/L) ratio of 1:40 (B), 1:20 (C), and 1:10 (D). E, β-synuclein with POPG vesicles at 1:20 P/L molar ratio (F and G) I, POPG/POPC (1:1 molar ratio) vesicles incubated with α-synuclein at 1:20 P/L molar ratio. Arrow indicates “budding vesicle,” and star indicates “budded vesicle”. H, POPG/POPC (1:4 molar ratio) vesicles incubated with α-synuclein at 1:10 P/L molar ratio. Arrows show a smaller tube coming out from a larger tube. Large non-extruded vesicles were used. Black scale bar is 100 nm.
Analogous experiments with β-synuclein, which does not readily form fibrils, indicated that this protein is also capable of generating tubular structures with morphology similar to those observed for α-synuclein (Fig. 3E). As in the case of α-synuclein increasing amounts of β-synuclein caused the formation of more curved membrane structures (supplemental Fig. S2, D–F). However, β-synuclein was less efficient in inducing tubulation, a higher protein concentration being required to observe tubulation (Fig. 3E and supplemental Fig. S2). Again, membrane remodeling was accompanied by an induction of α-helical structure (supplemental Fig. S3). These results further supported the finding that it is the helical structure rather than a misfolded β-sheet structure that is responsible for the membrane remodeling.
The ability of α-synuclein to induce tubulation was not limited to multilamellar POPG-containing vesicles; tubulation was also observed for giant vesicles and large extruded vesicles (supplemental Fig. S4, A–B, but not for small unilamellar vesicles). Moreover, vesicles with less negative charge density also gave rise to tubulation (supplemental Figs. S4, D–G, S4, I–K, S3, F–H, and S5, C—D). POPG is not a mammalian phospholipid and hence we also included vesicles with phosphatidylserine-containing phospholipids which more closely resemble the intracellular surfaces of mammalian membranes (supplemental Fig. S4H). The ability of α-synuclein to tubulate these vesicles showed that POPG is not required and that α-synuclein is able to remodel vesicles with physiologically relevant phospholipid compositions. Overall, the morphology of the tubules was similar, but it appeared that tubulation of less charged vesicles was not as complete as with POPG vesicles. A distinctive feature often observed for POPG/POPC- containing vesicles was the presence of rounded structures at the ends of tubules, giving the impression of nascent vesicles that are “budding off” (Fig. 3, F and G and supplemental Fig. S4G). The inference of budding events is further supported by the presence of small vesicular structures of comparable size that could be detected in solution (Fig. 3G). In some cases, we also observed smaller tubules emerging from larger ones suggesting that the former were derived from the latter (Fig. 3H). Similar features and similar morphologies were also observed for amphiphysin, which is well known to induce membrane tubulation (Fig. 5, E–F).
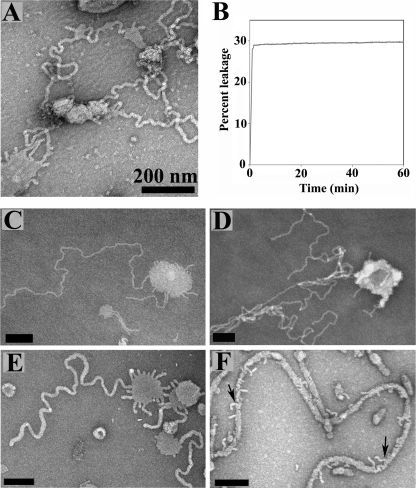
FIGURE 5.
Membrane tubulation of phospholipid vesicles in presence of apoA-1. A, negative stain EM image showing apoA-1 induced tubulation of POPG/POPC (1:4 molar ratio) vesicles at 1:100 P/L molar ratio.; B, leakage assay was performed with apoA-1 and POPG/POPC (1:4 molar ratio) vesicles at 1:200 P/L molar ratio. Large extruded 1 μm vesicles were used; C and D, POPG/POPC (1:4 molar ratio) vesicles at a α-synuclein to lipid (P/L) molar ratio of 1:10. E, N-BAR domain of amphiphysin with POPC/PE (porcine brain)/sphingomyelin/cholesterol (1:1:1:1.5 molar ratio) at a protein to lipid molar ratio of 1:300; F, N-BAR domain of amphiphysin with POPG/POPC (1:1 molar ratio) at a protein to lipid molar ratio of 1:40. Arrows show a smaller tubule coming out from a larger tubule. Large non-extruded vesicles were used for all EM studies. Scale bar is 200 nm.
The membrane remodeling data together with the circular dichroism suggest, but do not directly demonstrate, that α-synuclein is bound to tubules and small vesicular structures. To address this point, we gold-labeled α-synuclein. As shown in supplemental Fig. S5, the gold-labeled protein indeed localizes to tubules and small vesicular structures.
Vesicle Leakage during Membrane Remodeling
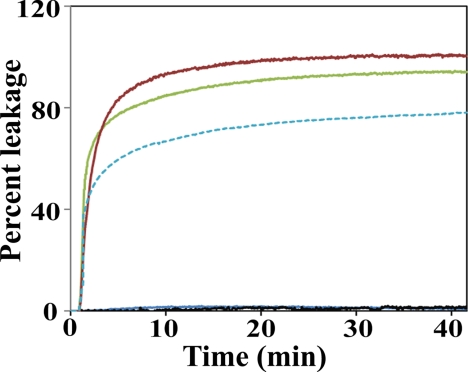
To test whether the rapid changes in shape and volume might be accompanied by disruption of membrane integrity, we performed membrane leakage assays for α-synuclein and β-synuclein. As a positive control we included amphiphysin. All proteins were assayed under conditions in which significant tubulation was observed by EM. POPG-containing vesicles were used for the synucleins. In the case of amphiphysin, similar results were obtained for POPG-containing as well POPG/POPE-containing vesicles. The data for the latter composition are shown since more homogenous tubulation was observed by EM. For all three proteins, membrane remodeling/tubulation was accompanied by rapid and pronounced vesicle leakage, demonstrating that tubulation is accompanied by significant disruptions of membrane integrity (Fig. 4). Although α-synuclein and β-synuclein were largely similar in this assay, the overall amount of leakage appeared to be slightly less in the case of β-synuclein (Fig. 4). These data again suggest that β-synuclein is a slightly less potent tubulator.
FIGURE 4.
Leakage of vesicles composed of negatively charged phospholipids in presence of α-synuclein and amphiphysin. Red, α-synuclein with POPG vesicles (1:20 P/L molar ratio); Broken blue, β-synuclein with POPG vesicles (1:20 P/L molar ratio); green, amphiphysin with POPG/POPE (2:1 molar ratio) vesicles (1:100 P/L molar ratio); black, control vesicles without α-synuclein or β-synuclein; blue, Control vesicles without amphiphysin N-BAR domain. Large extruded 1 μm vesicles were used.
Apolipoprotein A-I Tubulates Liposomes Containing Minor Proportions of Anionic Lipids
To test whether apolipoproteins can also promote tubulation, we incubated apoA-I with vesicles and monitored the resulting shapes by negative stain EM. Interestingly, apoA-I was able to tubulate POPG/POPC-containing (1:4 molar ratio) vesicles (Fig. 5A). The tubule diameters were on the order of ∼25 nm (Fig. 5A).
When compared with the tubules formed from α-synuclein under comparable conditions, the tubules formed by apoA-I showed significant similarities (Fig. 5, C and D), providing evidence that the membrane interactions of synucleins and apolipoproteins have some properties in common. Moreover, apoA-1 also induced membrane leakage under conditions where it shows tubulation (Fig. 5B). Thus, in all cases tested in this study, tubulation is accompanied by at least transient disruption of membrane.
Nevertheless there were also some differences with respect to the optimal lipid composition for membrane tubule formation. While α-synuclein generated tubules from POPG containing vesicles, we did not observe any apoA-1-mediated tubulation of such highly charged vesicles. In contrast, apoA-1 tubules appeared to be more stable under conditions of less negatively charged membranes. This trend mirrors the cellular localization of both proteins, considering that the extracellular apoA-1 is exposed to predominantly neutral lipids, whereas the cytosolic α-synuclein is predominantly exposed to negatively charged membranes (70).
DISCUSSION
Based upon their related 11 amino acid repeats, it has long been suspected that synucleins and apolipoproteins have similar lipid binding properties, but such similarities have proven difficult to identify. Here we find that α-synuclein, β-synuclein, and apoA-1 share the common ability to induce membrane curvature and cause tubulation or vesiculation of large vesicles of a large number of different lipid compositions. Considering that synucleins and apolipoproteins are known to interact with lipids and/or membranes in vivo, their ability to induce membrane curvature is likely to be of physiological relevance. For example, in the case of α-synuclein, functional roles have been proposed for a number of vesicle trafficking events (13, 14), including clathrin-mediated endocytosis (15). Our finding that α-synuclein can induce membrane shapes such as “nascent budding vesicles” (Fig. 3, F and G), suggests that this protein might be able to facilitate vesicle budding and induce membrane curvature in vivo. Although α-synuclein is capable of inducing membrane curvature in vesicles with physiologically relevant lipid composition (supplemental Fig. S4H), future work will be required to address these functional implications in vivo.
Vesicle leakage experiments for α-synuclein and amphiphysin, however, also show that induction of membrane curvature can be accompanied by a significant loss of membrane integrity. Thus, cellular mechanisms must be in place to prevent such potentially toxic membrane perturbations from occurring in vivo. Previous studies have hypothesized that membrane disruption of α-synuclein plays an important role in the pathology of PD (23–26). The present data suggest that uncontrolled induction of membrane curvature might be one of the mechanisms that cause membrane disruption in PD. Interestingly, tubule-like structures have been reported in the intracellular deposits of PD mouse models overexpressing α-synuclein disease mutant (19). In light of the present results, it is likely that these structures are caused by a direct membrane curvature effect generated by α-synuclein. Whereas future work will have to show whether α-synuclein aggregation promotes uncontrolled induction of membrane curvature, recent studies predict that the concerted action/aggregation of proteins strongly enhances induction of membrane curvature (71–73). Our present data are in good agreement with this notion. First, our optical imaging shows that membrane remodeling (blebbing) coincides with a local increase in α-synuclein concentration. Second, we find that the degree of curvature induction strongly depends upon the protein-to-lipid ratio (Fig. 3, B–D and supplemental Fig. S2). At low protein-to-lipid ratios, relatively wide tubules are observed and increasing amounts of protein lead to reduced tubule diameters, suggesting that curvature induction becomes stronger as more protein binds to the membranes (Figs. 3, B and C and 6A, form III and IV). Moreover, our data also show that smaller tubules can originate from larger ones in a stepwise manner (Fig. 3H), as we frequently observed smaller tubules emanating from larger ones. At the highest protein-to-lipid ratio used (1:10), even more highly curved and smaller rounded structures were formed (Figs. 3D and 6A, form V). Collectively, these data clearly show that increasing amounts of protein lead to a progressive increase in membrane curvature. Thus, limiting the local protein concentration may be an effective means for achieving a controlled and non-disruptive induction of membrane curvature.
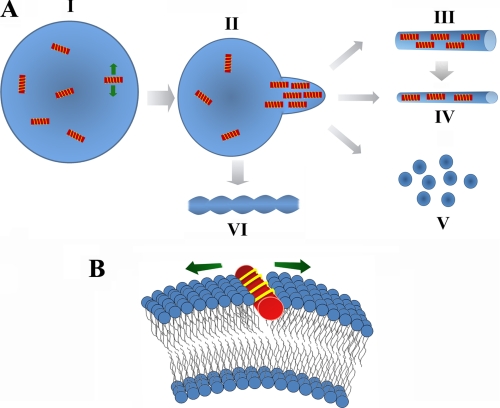
FIGURE 6.
Summary of α-synuclein-dependent membrane remodeling and curvature induction. A, α-synuclein molecules bind to a single vesicle (I). After attaining a critical concentration, the curvature strain causes initiation of a membrane tubule (II). The concentration and possibly orientation of the protein molecules bound on the membrane could determine the size (III or IV), and shape of tube (VI) with higher concentrations favoring more curved structures. The helices in II, III, and IV are schematically drawn parallel to the tubule axis in an orientation that would induce the maximal anisotropic curvature strain. However, slight deviation from this orientation cannot be excluded. Vesiculation or formation of smaller lipidic structures (V) could originate from smaller membrane tubes (IV) or directly from the large vesicles (II). B, insertion of the extended helical structure (red cylinder with yellow stripes) of α-synuclein on intact vesicles occurs at the phosphate level (31) and induces a highly anisotropic curvature strain. The green arrows indicate the direction of curvature strain.
A difference between amphiphysin and α-synuclein is that the latter does not possess a rigid scaffolding domain (BAR domain). Rather, curvature induction of α-synuclein is mediated by an α-helical structure that is induced upon membrane interaction. A number of different helical structures have been determined for α-synuclein; when stably bound to intact small unilamellar vesicles, α-synuclein takes up an extended helical structure (30–34). In contrast, when bound to non-bilayer or smaller micellar aggregates, α-synuclein forms structures containing two anti-parallel helices (31, 69, 74, 75). A common feature of all α-helical structures of α-synuclein is that all helices are amphipathic with their hydrophobic face inserting into the hydrophobic interior of the bilayer or micelle. In the case of vesicle-bound α-synuclein, the extended single-helical structure is located at the level of the phosphate (31). According to theoretical studies, this position results in a maximal wedge-like effect that pushes the headgroups apart (43, 76, 77) and, thereby, promotes membrane curvature (Fig. 6B). Moreover, in the known structures, α-synuclein forms relatively long helices (∼140 Å for the single helix structure and ∼50 Å and ∼70 Å in the two-helical structure). These helices should result in a highly anisotropic curvature strain, which might help to maintain tubule stability for days. However, future higher resolution structural analysis using cryo EM, site-directed spin labeling and other tools for investigating protein structure will be necessary to resolve the exact structural details of α-synuclein-dependent curvature induction. In fact, such studies may well reveal that the precise α-synuclein structures may vary for the different tubule types.
ApoA-1 exhibited a similar ability to tubulate as the synucleins with the slight distinction that it preferred less negatively charged lipid compositions. Future studies will have to show whether the ability of amphipathic segments to tubulate vesicles and progressively ratchet lipids may underlie its ability to generate matured HDL particles.
Supplementary Material
Acknowledgments
We thank Dr. Ulrich Baxa for helpful discussion and Dr. Tobias Ulmer, Dr. Jampani Nageswara Rao, and Dr. Jae-Eun Suk for providing β-synuclein expression vector (pET-41).
This work was supported, in whole or in part, by National Institutes of Health Grants GM063915 (to R. L.), R01 AG029246 (to J. C. V.), and the Intramural Research Program of NIAMS (to A. C. S.). This work was also supported by the Larry L. Hillblom foundation (to R. L.) and the Danish Research Councils, the University of Copenhagen and the Lundbeck foundation (to D. G. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PD
- Parkinson disease
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-RAC-(1-glycerol)]
- P/L
- protein/lipid ratio.
REFERENCES
- 1.Tofaris G. K., Spillantini M. G. (2007) Cell Mol. Life Sci. 64, 2194–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norris E. H., Giasson B. I., Lee V. M. (2004) Curr. Top Dev. Biol. 60, 17–54 [DOI] [PubMed] [Google Scholar]
- 3.Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. (1997) Nature 388, 839–840 [DOI] [PubMed] [Google Scholar]
- 4.Beyer K. (2007) Cell Biochem. Biophys 47, 285–299 [DOI] [PubMed] [Google Scholar]
- 5.Bisaglia M., Mammi S., Bubacco L. (2009) Faseb. J. 23, 329–340 [DOI] [PubMed] [Google Scholar]
- 6.Nemani V. M., Lu W., Berge V., Nakamura K., Onoa B., Lee M. K., Chaudhry F. A., Nicoll R. A., Edwards R. H. (2010) Neuron 65, 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abeliovich A., Schmitz Y., Fariñas I., Choi-Lundberg D., Ho W. H., Castillo P. E., Shinsky N., Verdugo J. M., Armanini M., Ryan A., Hynes M., Phillips H., Sulzer D., Rosenthal A. (2000) Neuron 25, 239–252 [DOI] [PubMed] [Google Scholar]
- 8.Cabin D. E., Shimazu K., Murphy D., Cole N. B., Gottschalk W., McIlwain K. L., Orrison B., Chen A., Ellis C. E., Paylor R., Lu B., Nussbaum R. L. (2002) J. Neurosci. 22, 8797–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy D. D., Rueter S. M., Trojanowski J. Q., Lee V. M. (2000) J. Neurosci. 20, 3214–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Withers G. S., George J. M., Banker G. A., Clayton D. F. (1997) Brain Res. Dev. Brain Res. 99, 87–94 [DOI] [PubMed] [Google Scholar]
- 11.Golovko M. Y., Barceló-Coblijn G., Castagnet P. I., Austin S., Combs C. K., Murphy E. J. (2009) Mol. Cell Biochem. 326, 55–66 [DOI] [PubMed] [Google Scholar]
- 12.Lee S. J., Jeon H., Kandror K. V. (2008) Acta Neurobiol. Exp. 68, 509–515 [DOI] [PubMed] [Google Scholar]
- 13.Outeiro T. F., Lindquist S. (2003) Science 302, 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willingham S., Outeiro T. F., DeVit M. J., Lindquist S. L., Muchowski P. J. (2003) Science 302, 1769–1772 [DOI] [PubMed] [Google Scholar]
- 15.Ben Gedalya T., Loeb V., Israeli E., Altschuler Y., Selkoe D. J., Sharon R. (2009) Traffic 10, 218–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosavi N., Lee H. J., Lee J. S., Patel S., Lee S. J. (2002) J. Biol. Chem. 277, 48984–48992 [DOI] [PubMed] [Google Scholar]
- 17.Fujita Y., Ohama E., Takatama M., Al-Sarraj S., Okamoto K. (2006) Acta. Neuropathol. 112, 261–265 [DOI] [PubMed] [Google Scholar]
- 18.Song D. D., Shults C. W., Sisk A., Rockenstein E., Masliah E. (2004) Exp. Neurol. 186, 158–172 [DOI] [PubMed] [Google Scholar]
- 19.Martin L. J., Pan Y., Price A. C., Sterling W., Copeland N. G., Jenkins N. A., Price D. L., Lee M. K. (2006) J. Neurosci. 26, 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meredith G. E., Totterdell S., Petroske E., Santa Cruz K., Callison R. C., Jr., Lau Y. S. (2002) Brain Res. 956, 156–165 [DOI] [PubMed] [Google Scholar]
- 21.Stichel C. C., Zhu X. R., Bader V., Linnartz B., Schmidt S., Lübbert H. (2007) Hum. Mol. Genet. 16, 2377–2393 [DOI] [PubMed] [Google Scholar]
- 22.Gai W. P., Yuan H. X., Li X. Q., Power J. T., Blumbergs P. C., Jensen P. H. (2000) Exp. Neurol. 166, 324–333 [DOI] [PubMed] [Google Scholar]
- 23.Lashuel H. A., Petre B. M., Wall J., Simon M., Nowak R. J., Walz T., Lansbury P. T., Jr. (2002) J. Mol. Biol. 322, 1089–1102 [DOI] [PubMed] [Google Scholar]
- 24.Lashuel H. A., Hartley D., Petre B. M., Walz T., Lansbury P. T., Jr. (2002) Nature 418, 291. [DOI] [PubMed] [Google Scholar]
- 25.Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 26.Sokolov Y., Kozak J. A., Kayed R., Chanturiya A., Glabe C., Hall J. E. (2006) J. Gen Physiol. 128, 637–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson W. S., Jonas A., Clayton D. F., George J. M. (1998) J. Biol. Chem. 273, 9443–9449 [DOI] [PubMed] [Google Scholar]
- 28.Nuscher B., Kamp F., Mehnert T., Odoy S., Haass C., Kahle P. J., Beyer K. (2004) J. Biol. Chem. 279, 21966–21975 [DOI] [PubMed] [Google Scholar]
- 29.Rhoades E., Ramlall T. F., Webb W. W., Eliezer D. (2006) Biophys. J. 90, 4692–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jao C. C., Der-Sarkissian A., Chen J., Langen R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8331–8336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jao C. C., Hegde B. G., Chen J., Haworth I. S., Langen R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19666–19671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgieva E. R., Ramlall T. F., Borbat P. P., Freed J. H., Eliezer D. (2008) J Am Chem Soc 130, 12856–12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trexler A. J., Rhoades E. (2009) Biochemistry 48, 2304–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreon A. C., Gambin Y., Lemke E. A., Deniz A. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 5645–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frost A., Unger V. M., De Camilli P. (2009) Cell 137, 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMahon H. T., Gallop J. L. (2005) Nature 438, 590–596 [DOI] [PubMed] [Google Scholar]
- 37.Drin G., Antonny B. (2009) FEBS Lett. 584, 1840–1847 [DOI] [PubMed] [Google Scholar]
- 38.Zimmerberg J., Kozlov M. M. (2006) Nat. Rev. Mol. Cell Biol. 7, 9–19 [DOI] [PubMed] [Google Scholar]
- 39.Gallop J. L., Jao C. C., Kent H. M., Butler P. J., Evans P. R., Langen R., McMahon H. T. (2006) EMBO J. 25, 2898–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda M., Takeda S., Sone M., Ohki T., Mori H., Kamioka Y., Mochizuki N. (2006) EMBO J. 25, 2889–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peter B. J., Kent H. M., Mills I. G., Vallis Y., Butler P. J., Evans P. R., McMahon H. T. (2004) Science 303, 495–499 [DOI] [PubMed] [Google Scholar]
- 42.Blood P. D., Voth G. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15068–15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campelo F., McMahon H. T., Kozlov M. M. (2008) Biophys. J. 95, 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farsad K., Ringstad N., Takei K., Floyd S. R., Rose K., De Camilli P. (2001) J. Cell Biol. 155, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jao C. C., Hegde B. G., Gallop J. L., Hegde P. B., McMahon H. T., Haworth I. S., Langen R. (2010) J. Biol. Chem. 285, 20164–20170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui H., Ayton G. S., Voth G. A. (2009) Biophys. J. 97, 2746–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bodner C. R., Dobson C. M., Bax A. (2009) J. Mol. Biol. 390, 775–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madine J., Hughes E., Doig A. J., Middleton D. A. (2008) Mol. Membr Biol. 25, 518–527 [DOI] [PubMed] [Google Scholar]
- 49.Necula M., Chirita C. N., Kuret J. (2003) J. Biol. Chem. 278, 46674–46680 [DOI] [PubMed] [Google Scholar]
- 50.Sparr E., Engel M. F., Sakharov D. V., Sprong M., Jacobs J., de Kruijff B., Höppener J. W., Killian J. A. (2004) FEBS Lett. 577, 117–120 [DOI] [PubMed] [Google Scholar]
- 51.Yip C. M., Darabie A. A., McLaurin J. (2002) J. Mol. Biol. 318, 97–107 [DOI] [PubMed] [Google Scholar]
- 52.Chirita C. N., Necula M., Kuret J. (2003) J. Biol. Chem. 278, 25644–25650 [DOI] [PubMed] [Google Scholar]
- 53.Davidson W. S., Thompson T. B. (2007) J. Biol. Chem. 282, 22249–22253 [DOI] [PubMed] [Google Scholar]
- 54.Luo C. C., Li W. H., Moore M. N., Chan L. (1986) J. Mol. Biol. 187, 325–340 [DOI] [PubMed] [Google Scholar]
- 55.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. (1992) J. Lipid Res. 33, 141–166 [PubMed] [Google Scholar]
- 56.Sung Y. H., Eliezer D. (2006) Protein Sci. 15, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uversky V. N., Li J., Souillac P., Millett I. S., Doniach S., Jakes R., Goedert M., Fink A. L. (2002) J. Biol. Chem. 277, 11970–11978 [DOI] [PubMed] [Google Scholar]
- 58.Brouillette C. G., Anantharamaiah G. M., Engler J. A., Borhani D. W. (2001) Biochim. Biophys. Acta 1531, 4–46 [DOI] [PubMed] [Google Scholar]
- 59.Silva R. A., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Der-Sarkissian A., Jao C. C., Chen J., Langen R. (2003) J. Biol. Chem. 278, 37530–37535 [DOI] [PubMed] [Google Scholar]
- 61.Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. (2007) J. Biol. Chem. 282, 9143–9149 [DOI] [PubMed] [Google Scholar]
- 62.Bhatia V. K., Madsen K. L., Bolinger P. Y., Kunding A., Hedegård P., Gether U., Stamou D. (2009) EMBO J. 28, 3303–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wimley W. C., Selsted M. E., White S. H. (1994) Protein Sci. 3, 1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vinson P. K., Talmon Y., Walter A. (1989) Biophys. J. 56, 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mishra V. K., Palgunachari M. N., Datta G., Phillips M. C., Lund-Katz S., Adeyeye S. O., Segrest J. P., Anantharamaiah G. M. (1998) Biochemistry 37, 10313–10324 [DOI] [PubMed] [Google Scholar]
- 66.Mishra V. K., Palgunachari M. N., Lund-Katz S., Phillips M. C., Segrest J. P., Anantharamaiah G. M. (1995) J. Biol. Chem. 270, 1602–1611 [DOI] [PubMed] [Google Scholar]
- 67.Hatzakis N. S., Bhatia V. K., Larsen J., Madsen K. L., Bolinger P. Y., Kunding A. H., Castillo J., Gether U., Hedegård P., Stamou D. (2009) Nat. Chem. Biol. 5, 835–841 [DOI] [PubMed] [Google Scholar]
- 68.Bendix P. M., Pedersen M. S., Stamou D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12341–12346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drescher M., van Rooijen B. D., Veldhuis G., Subramaniam V., Huber M. (2010) J. Am. Chem. Soc. 132, 4080–4082 [DOI] [PubMed] [Google Scholar]
- 70.Gennis R. B. (1989) Biomembranes: Molecular Structure and Function, Springer-Verlag, New York [Google Scholar]
- 71.Ayton G. S., Lyman E., Krishna V., Swenson R. D., Mim C., Unger V. M., Voth G. A. (2009) Biophys. J. 97, 1616–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ayton G. S., Blood P. D., Voth G. A. (2007) Biophys. J. 92, 3595–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ayton G. S., Voth G. A. (2009) Curr. Opin. Struct. Biol. 19, 138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ulmer T. S., Bax A., Cole N. B., Nussbaum R. L. (2005) J. Biol. Chem. 280, 9595–9603 [DOI] [PubMed] [Google Scholar]
- 75.Borbat P., Ramlall T. F., Freed J. H., Eliezer D. (2006) J. Am. Chem. Soc. 128, 10004–10005 [DOI] [PubMed] [Google Scholar]
- 76.Tytler E. M., Segrest J. P., Epand R. M., Nie S. Q., Epand R. F., Mishra V. K., Venkatachalapathi Y. V., Anantharamaiah G. M. (1993) J. Biol. Chem. 268, 22112–22118 [PubMed] [Google Scholar]
- 77.Segrest J. P. (1977) Chem. Phys. Lipids. 18, 7–22 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.