Abstract
Oral squamous cell carcinoma (OSCC) is a major health problem worldwide, and patients have a particularly poor 5-year survival rate. Thus, identification of the molecular targets in OSCC and subsequent innovative therapies are greatly needed. Prolonged exposure to alcohol, tobacco, and pathogenic agents are known risk factors and have suggested that chronic inflammation may represent a potential common denominator in the development of OSCC. Microarray analysis of gene expression in OSCC cell lines with high basal NF-κB activity and OSCC patient samples identified dysregulation of many genes involved in inflammation, wound healing, angiogenesis, and growth regulation. In particular IL-8, CCL5, STAT1, and VEGF gene expression was up-regulated in OSCC. Moreover, IL-8 protein levels were significantly higher in OSCC cell lines as compared with normal human oral keratinocytes. Targeting IL-8 expression by siRNA significantly reduced the survival of OSCC cells, indicating that it plays an important role in OSCC development and/or progression. Inhibiting the inflammatory pathway by aspirin and the proteasome/NF-κB pathway by bortezomib resulted in marked reduction in cell viability in OSCC lines. Taken together our studies indicate a strong link between inflammation and OSCC development and reveal IL-8 as a potential mediator. Treatment based on prevention of general inflammation and/or the NF-κB pathway shows promise in OSCCs.
Keywords: Gene Expression, Inflammation, Interleukin, Microarray, NF-κB, Anti-inflammatory, Biomarker, Oral Cancer, Meta-analysis
Introduction
Oral squamous cell carcinoma (OSCC)2 is the sixth most common cancer and an important public health concern worldwide (1, 2), with ∼405,000 new cases and 211,000 deaths reported annually (3). Patients diagnosed with oral cancer have a particularly low 5-year survival rate due to the compounding factors of late detection and lack of truly effective therapies according to the American Cancer Society and the online Facts page of The Oral Cancer Foundation. Therefore, development of early detection techniques and subsequent innovative therapies are greatly needed. Besides high mortality, OSCC is also often associated with eating difficulties, speech impairment, and general psychological distress (6, 7). Tobacco and alcohol consumption, betel quid chewing, and viral infections are some of the known risk factors for OSCC (8, 9). In addition, oral infections leading to periodontal diseases are also associated with OSCC (10, 11), About 20% of oral leukoplakia undergo malignant transformation and develop into OSCC (6).
Chronic inflammation is associated with the development of a variety of epithelial cancers such as colon and pancreatic cancers (12, 13), but whether it plays a significant role in development of oral cancers is unclear. It is believed that the chronic inflammatory environment causes genomic alterations that eventually lead to tumor development (8, 14). Essential components in this association are the cytokines produced by tumor cells themselves as well as by the innate immune cells activated during the inflammatory process (15, 16). In the present study we analyzed the roles of inflammatory genes in OSCC by gene expression screening in OSCC cell lines. In common with a variety of tumor types, these OSCC lines exhibited basally high activity of the NF-κB transcription factor, which is consistent with chronic activation of the inflammatory process. The data obtained by gene expression profiling were used to identify common molecular pathways affected in OSCC. Differentially regulated inflammatory genes, growth factors, and their receptors were subjected to further analysis to identify candidate genes that are potential biomarkers and molecular targets in OSCC. To define the clinical relevance of these candidate genes, we performed meta-analysis with a data base of a large number of human OSCC patient samples (www.ncbi.nlm.nih.gov). This analysis revealed common set of genes involved in inflammation, angiogenesis, and cell cycle. One important gene identified by this strategy was IL-8. High levels of IL-8 have been found in various human diseases and malignancies (17–21). IL-8 levels were found to be highly expressed in saliva of OSCC patients (22). Elevated levels of IL-8 correspond to an increased metastatic potential of melanoma (23), breast (24), renal (25), gastric (26), ovarian (27), pancreatic (28), and colorectal cancers (29). IL-8 overexpression is associated with disease progression of urogenital cancers, including transitional cell carcinoma of bladder (30) and prostate cancer (31, 32). IL-8 has been shown to play an important function in growth and tumor development under hypoxic conditions as well (28). These studies clearly signify a role of IL-8 in cancer development and its potential as a therapeutic target. Therefore, IL-8 expression was knocked-down using small-interfering RNA (siRNA) to examine the role of the IL-8 signaling pathway in OSCC. IL-8 knockdown significantly reduced the viability of OSCC cell lines. Furthermore, studies with an anti-inflammatory drug and an NF-κB inhibitor further indicated the importance of the inflammatory process in OSCC.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture
Human oral keratinocytes (HOK) were obtained from the ScienCell and grown in KBM-2 with supplements (Lonza, Walkersville, MD). Human oral squamous cell carcinoma cells SCC9, SCC15, and SCC25 (ATCC, Manassas, VA) were cultured in DMEM supplemented with 10% fetal calf serum, 100 μg/ml penicillin, and 100 μg/ml streptomycin. Cells were passaged twice a week.
NF-κB Activity Measurement
Nuclei isolated from HOK and OSCC cultures were extracted with buffer containing 20 mm HEPES (pH 7.9), 350 mm NaCl, 20% glycerol, 1% Nonidet P-40, 1 mm MgCl2, 0.5 mm EDTA, 0.1 mm EGTA, 0.5 mm DTT, 0.5 mm aprotinin, and 0.5 mm phenylmethylsulfonyl fluoride, and extracts were frozen and stored at −80 °C. For electrophoretic shift assay, the nuclear protein extracts (10–20 μg) were incubated with a 32P-labeled κB oligonucleotide probe (5′-TCA ACA GAG GGG ACT TTC CGA GAG GCC-3′) at 25 °C for 20 min and with anti-p50 or anti-p65 or a 50-fold excess of unlabeled oligonucleotide probe (cold) and separated on 4% non-denaturing polyacrylamide gels. NF-κB complexes were quantified by phosphorimaging.
RNA Isolation and cRNA Generation
Total RNA was isolated using TRIzol reagent (Invitrogen) and purified by the RNeasy MiniElute Cleanup kit (Qiagen, CA) and quantified on Agilent Bioanalyzer. High quality total RNA with a RIN (RNA integrity score) number of more than seven was used to generate cDNA and cRNA with the Illumina® TotalPrep™ RNA amplification kit (Ambion, Inc.). The procedure consisted of reverse transcription with an oligo(dT) primer bearing a T7 promoter using Array-Script™, a reverse transcriptase (RT) engineered to produce higher yields of first-strand cDNA than wild type enzymes. ArrayScript catalyzed the synthesis of full-length cDNA and ensured production of reproducible microarray samples. The cDNA then underwent second-strand synthesis and clean-up to become a template for in vitro transcription with T7 RNA polymerase and biotin UTP, which generates multiple copies of biotinylated cRNA. After purification, the purity and concentration of cRNA was checked using ND-1000 Spectrometer (NanoDrop). High quality cRNA was then used with the Illumina direct hybridization array kits.
Hybridization
0.75 μg of cRNA sample was hybridized on human HT-12 BeadChip for 16 h in a multiple step procedure according to the manufacturer's instructions. The chips were then washed, dried, and scanned on the Bead Array Reader (Illumina), and raw data were generated using GenomeStudio 3.4.0 (Illumina).
Microarray Data Analysis
Raw data were normalized with GenomeStudio (Illumina) using Quantile algorithm, and gene expression profiles were statistically compared using the “Differential Expression” feature of GenomeStudio with Mann-Whitney U test. Further analysis was performed using GeneSpringGX10.2 software (Agilent Technologies, Inc.). Statistical analysis on the data was performed by one-way analysis of variance and nonparametric t testing (Mann-Whitney rank test). Genes with a -fold change of ≥2.5 and ≤0.4 (p value ≤0.05) were considered significant. Further average linkage hierarchical clustering analysis was done using Euclidean distance. Differentially expressed genes were annotated using Gene Ontology Consortium. Molecular networks and pathways were generated using Pathways Analysis tool included in GeneSpringGX10.2 software. Further analysis was also done using EASE (The Expression Analysis Systematic Explorer) and DAVID (Data base for Annotation, Visualization, and Integrated Discovery) for proper functional annotations. Genes were clustered in EASE, and clusters were taken and analyzed in DAVID to determine -fold enrichment. Further gene networks and pathways were built based on KEGG and Biocarta and pathway analysis tool in GeneSpringGX10.2.
Comparison with Human OSCC Samples from Cancer Research Databases
The microarray data obtained from the OSCC cell lines was compared with the microarray data in the Gene Expression Omnibus on 16 human OSCC tumor samples from GEO-Data Set Record GDS1062 (www.ncbi.nlm.nih.gov). The cutoff rates were set at ≥2.5 and ≤0.4 (p value ≤0.05) for the genes to be called as significant, and the percentage of occurrence was calculated for every gene within 16 tumor samples as well as for the three OSCC cell lines. The most common differentially regulated genes were further analyzed with EASE and DAVID.
Quantitative Real Time PCR
Quantitative real time-PCR was performed on the iCyclerIQ detection system (Bio-Rad) using the iScript One-Step RT-PCR kit with SYBR Green (Bio-Rad) according to the manufacturer's instructions. The quantitative real time-PCR reaction parameters were as follows: cDNA synthesis at 50 °C for 20 min, iScript reverse transcriptase inactivation at 95 °C for 5 min, PCR cycling at 95 °C for 10 s, and 60 °C for 30 s for 40 cycles. The mRNA levels relative to β-actin and control were calculated using CT (cycle threshold) values. Primers used are listed in supplemental Table 1.
Knockdown of IL-8 with siRNA in OSCC Cells
The OSCC cells were seeded into 12-well plates at a density of 1 × 105 cells per well and allowed to attach overnight. SMARTpool siRNA (5 and 10 nm) and control non-targeting siRNA (5 nm) for IL-8 obtained from (Dharmacon, Lafayette, CO) were transfected into cells using 0.2 μl of Dharmafect transfection reagent (Dharmacon) according to the manufacturer's instructions. Methylthiazolyldiphenyltetrazolium bromide (MTT) assays (described below) were performed at 72 h post-transfection to determine cell viability and cell proliferation rate. IL-8 release in the cell culture media at 48 and 72 h post-transfection was detected by using IL-8 ELISA kit (sensitivity <25 pg/ml) according to the manufacturer's instructions (IL-8, Abcam).
Acetyl Salicyclic Acid and Velcade Treatment of OSCC Cells
Acetylsalicylic acid (Sigma) was added to tissue culture media at concentrations ranging from 1 to 20 mm dissolved in ethanol as vehicle (vehicle dilution of ethanol in DMEM/F-12 was 1:400 ratio (ethanol:DMEM/F-12) to solubilize acetylsalicylic acid). Velcade (Bortezomib® or PS-341, Millennium Pharmaceuticals, Cambridge, MA) was added at concentrations between 5 and 100 nm dissolved in DMEM/F-12 medium. After treating the cells with aspirin or bortezomib for 72 h, the cells were exposed to MTT solution (Sigma) for 3–5 h. Then cells were lysed in solvent (4 mm HCl, 0.1% Nonidet P-40 in isopropyl alcohol) for 30 min at room temperature with constant shaking, and color development was read at 595 nm with a Spectrophotometer (Ultrospec 2100 pro, GE Healthcare). The cell viability was calculated with the percentage of absorbance difference between treated and control samples.
RESULTS
Basal NF-κB Activation in OSCC Cell Lines
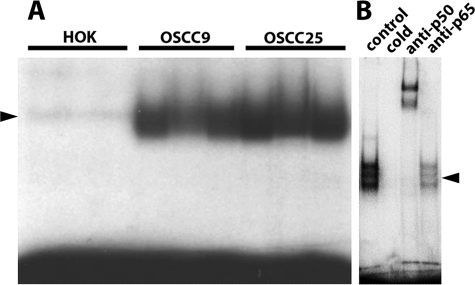
Nuclear extracts were prepared from three human oral squamous cell carcinoma cell lines (OSCC9, OSCC15, and OSCC25) and the normal HOK cell line and assayed for NF-κB activity by gel-shift assays using a consensus κB oligonucleotide probe. As shown in Fig. 1A, normal HOK showed little basal NF-κB activity, whereas high basal NF-κB activity was evident in the OSCC cell lines. High basal NF-κB activity has been observed in a variety of human cancer cell lines and tumor tissues and has been used to suggest that a particular cancer is associated with chronic inflammation. Super-shifted p50 and to a lesser extent p65 are present in the NF-κB complex (Fig. 1B). Excess cold NF-κB oligonucleotide probe competes out the formation of the NF-κB complex, demonstrating the specificity of NF-κB binding (Fig. 1B).
FIGURE 1.
Constitutive high NF-κB activity in OSCC cell lines. The gel shift was performed with nuclear extracts from both control and OSCC cells. The cells were incubated with 32P-labeled κB oligonucleotide probe. A, there is clear binding of the probe to the nuclear extracts from OSCC9 and OSCC25 cells but not in HOK cells. B, a gel-shift assay in OSCC25 is shown. p50, and to a lesser extent p65, are present in the NF-κB complex and can be super-shifted by antibodies specific to p50 and p65. The addition of excess cold NF-κB oligonucleotide probe competes out the formation of the NF-κB complex, demonstrating the specificity of NF-κB binding. Arrowheads mark the NF-κB complex.
Global Gene Expression Profiles in OSCC
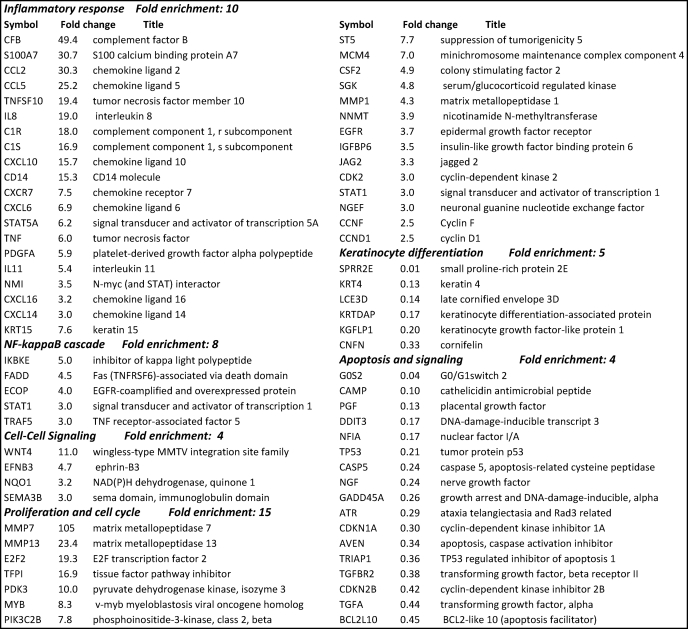
Total RNA was extracted from the OSCC and HOK cell lines, and cRNA probes were generated for hybridization to human-HT-12 BeadChips to investigate alterations in gene expression specific to OSCCs. Cluster analysis based on gene function showed up-regulation of genes in OSCC cell lines involved in inflammation (IL-8, CCL5, CXCL10, TLR6, and C3), cell proliferation (PDGF, CCND1, CDK6, CDK2, and CCNE2), and angiogenesis (VEGF, MMP7, MMP13, JAG2, and TNFSF12). In contrast, as shown in Table 1, genes down-regulated in OSCC were found to be involved in apoptosis regulation (BCL2L10, GADD45A, and CASP5), tumor inhibition (CDKN1A and TP53), and keratinization (KRT4, KGFLP1, LCE3D, etc.). The most highly up-regulated genes (>4-fold) encoded cytokines and growth factors and proteins involved in DNA replication and chromosome aggregation (Table 1), whereas down-regulated genes were involved in cell cycle checkpoints and keratinization (Table 1). These results suggest that inflammation, cell proliferation, and angiogenesis may play important roles in OSCC.
TABLE 1.
Partial list of genes significantly affected in OSCCs in comparison to human oral keratinocytes
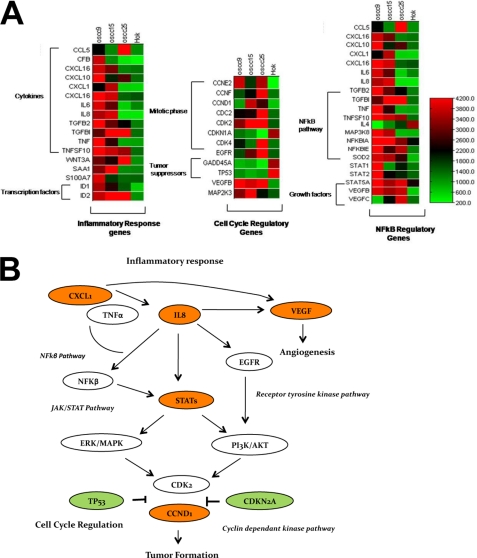
High expression of STAT1, TNFSF10, STAT5A, STAT3, and ID1 (Table 1 and Fig. 2A) might indicate their involvement in OSCC cell proliferation and tumor development. The data also suggest that NF-κB activation mediated by IL-8, TNF-α, and CCL5 under chronic inflammatory conditions might lead to OSCC development. Several genes in the NF-κB pathway were also up-regulated (NFKBIA, TRAF5, and FADD (FAS-associated death domain protein)), which might play a role in the high basal NF-κB activity in OSCC cell lines. Furthermore, signaling through the STAT pathway triggered by EGFR or VEGF might instigate proliferation and angiogenesis processes leading to cancer. Genes like WNT4, EFNB3, CFB, C1R, DEFB4, NMI, SAA1, SERPINA1, SERPINF2, APOL2, and TLR6 might indicate novel associations with inflammatory cancers.
FIGURE 2.
Global gene expression profiles in OSCC identify the dysregulation of important gene families. A, cluster analysis of genes in OSCC lines is shown. The heat maps show the differentially regulated genes and their clustering into functional groups such as cytokines, growth factors, cell cycle regulation, and transcription factors. Signal intensities for genes were averaged across all replicates of the three OSCC cell lines and HOK cells. A color scale in which red represents up-regulation and green represents down-regulation depicts the expression level of the genes. B, shown is an alteration in signal transduction pathways and gene networking in OSCC development. A schematic diagram illustrates signal transduction cascades after stimulation of IL-8 or inflammatory response genes. The IL-8 activation could promote activation of JAK/STAT pathway and the receptor tyrosine pathway, which in turn would regulate the cyclin-dependent kinase pathway. The genes in red represent oncogenes, whereas genes in green are tumor suppressors.
The alterations in gene expression suggest the involvement of IL-8, VEGF, EGFR, and STAT as important pathways in OSCCs. Other pathways such as androgen receptor and T-cell receptor seem to be correlated with tumor formation and growth in OSCC cells. These results suggest that signaling through IL-8 could trigger downstream targets such as STAT, NF-κB, and VEGF (Fig. 2B).
Comparison with Human OSCC Samples Dataset
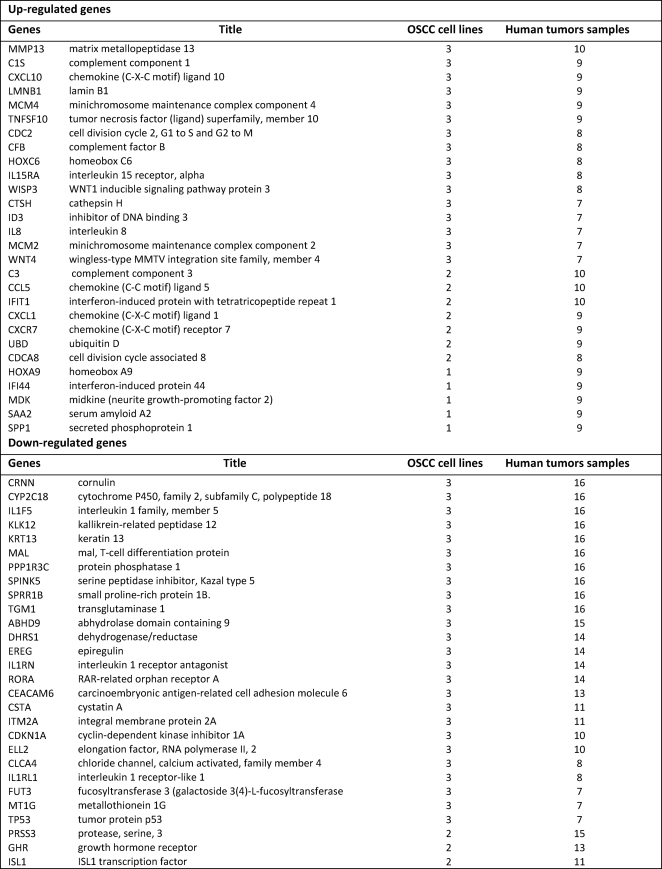
We next compared our findings on alterations in the gene expression patterns in the selected OSCC cell lines to the microarray data deposited in the human OSCC dataset from Gene Expression Omnibus (GEO). By calculating the prevalence of altered expression of these genes in 16 OSCC human patient samples and in the 3 OSCC cell lines, we found that there was an up-regulation of inflammatory genes such as IL-8, CCL5, TNFSF10, and CXCL10 as well as the VEGF growth factor in most of the OSCC patient samples (Table 2). MMP1, MMP13, STAT5A, C3, C1S, CFB, ID3, NMI, and WNT4 were highly up-regulated in the patient samples, indicating a novel association of these genes in the development and progression of OSCC (Table 2). Cell cycle regulators (CDKN1A), tumor suppressors (TP53), and keratins (KRT4, KRT13) were down-regulated in these patient samples, which was consistent with the data obtained with OSCC cell lines (Table 2). Thus, this analysis also showed that inflammatory genes are highly up-regulated in patient samples of OSCC and that the altered pattern of gene expression obtained with OSCC cell lines was similar to that obtained with patient samples.
TABLE 2.
List of genes significantly affected in OSCC cell lines (3) and human tumor samples (16) and their prevalence
Alterations in Gene Expression in OSCC Determined by Quantitative Real Time PCR
To further investigate and confirm the altered pattern of gene expression obtained by microarray analysis, we performed quantitative real time PCR on RNA obtained from the three OSCC cell lines as compared with HOK cells. We determined the expression levels relative to β-actin of a number of candidate genes in OSCC that were involved in inflammation (IL-8, IL-6, CCL5, and TNFSF10), growth factors (EGFR, VEGF), transcription factors (STAT1), and cell cycle regulators (CCND1, TP53, and CDKN2C). As shown in Fig. 3, EGFR, VEGF, STAT1, CCN1, TNSF10, IL-6, CCL5, and IL-8 were up-regulated in some of the OSCC cell lines, whereas TP53 and CDKN2C were down-regulated. Thus, the expression pattern for both up-regulated and down-regulated genes by quantitative real time PCR validated the microarray data.
FIGURE 3.
The mRNA expression levels of candidate genes in OSCC. Quantitative real time PCR was performed for several candidate genes in all the three OSCC cell lines to verify microarray expression data. The mRNA expression levels were compared with normal HOK cells and normalized to β-actin mRNA. The -fold changes represent the mean of triplicates and show validation of the microarray data.
Enhanced Expression of IL-8 in OSCC
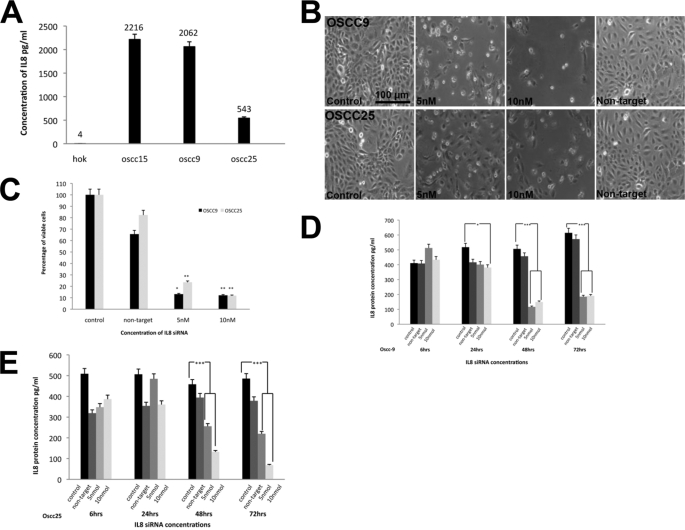
The gene expression profiles of human OSCC cell lines and tumor samples indicate that the IL-8 gene is significantly up-regulated. Therefore, we determined secreted IL-8 protein levels by ELISA in the tissue culture media of OSCC cell lines as compared with HOK cells. As shown in Fig. 4A, whereas IL-8 levels were barely detectable in the media from HOK cells (4pg/ml), IL-8 levels were significantly higher in all OSCC cells ranging from 540 to 2200 pg/ml.
FIGURE 4.
The role of IL-8 up-regulation in OSCC proliferation and viability. A, shown are IL-8 levels in OSCC cell lines. ELISA was used to estimate the levels of IL-8 in culture supernatants from the various cell lines. Normal HOK cells produced very low IL-8 amounts, whereas tumor cells produced significantly higher IL-8 levels ranging from 543–2216pg/ml of protein. B, shown are photomicrographs of representative OSCC cells transfected with IL-8 siRNA. The OSCC cells were seeded at 1 × 105 cells per tissue culture well and transfected with IL-8 siRNA (5 and 10 nm) or control non-target siRNA (5 nm) for 72 h. A significant change was observed in cell viability within 48 to 72 h of incubation. The concentration of transfection reagent used was 0.2 μm, and the data shown are representative of three independent experiments. C, shown is the viability of cells transfected with IL-8 siRNA. Cell viability was determined by the ability of the cells to metabolically reduce MTT to a formazan dye at 72 h after transfection with IL-8 siRNA. A decrease in cell viability after 48 h in transfected cells was noted at both 5 and 10 nm IL-8 siRNA. Significant change in cell viability in transfected cells was observed after 48–72 h at both siRNA concentrations. Statistical significance between control group and the siRNA-treated group was performed by one-way analysis of variance followed by Bonferroni's multiple comparison test (*, p < 0.05; **, p < 0.01). IL-8 levels in OSCC9 (D) and OSCC25 (E) cells transfected with IL-8 siRNA are shown. The levels of IL-8 secreted into the media from OSCC cell lines were estimated by ELISA at 6, 24, 48, and 72 h after transfection with siRNA (5 or 10 nm IL-8), and the non-target control pool was transfected with 5 nm scrambled siRNA. Significant changes in IL-8 levels were observed at 48–72 h after IL-8 knockdown. Statistical significance between control group and siRNA-treated cells was performed by one-way analysis of variance followed by Bonferroni's multiple comparison test (*, p < 0.05; **, p < 0.01).
Suppression of the IL-8 Signaling Pathway by IL-8 siRNA
Because IL-8 was markedly up-regulated at mRNA and protein levels, OSCC9 and OSCC25 cells were transfected with Smartpool siRNA directed against IL-8. A concentration of 5 and 10 nm siRNA against IL-8 significantly reduced IL-8 levels in the media by ∼85% at 48 h post-transfection as determined by ELISA (Fig. 4, D and E). Most interesting, IL-8 siRNA induced a significant decrease in the viability and proliferation of both OSCC lines (Fig. 4, B and C). Transfection reagent (0.25 μm) and non-target siRNA (5 nm) did not alter the cell growth or viability significantly. As shown in Fig. 4C, IL-8 siRNA knockdown drastically affected proliferation rate after 72 h. After 48 h, the cell viability in IL-8 knockdown experiments was reduced to 60% (Fig. 4, B and C) and at 72 h to about 20% as compared with the control knock-down experiments (Fig. 4C).
Effect of Anti-inflammatory Drugs on OSCC Cells
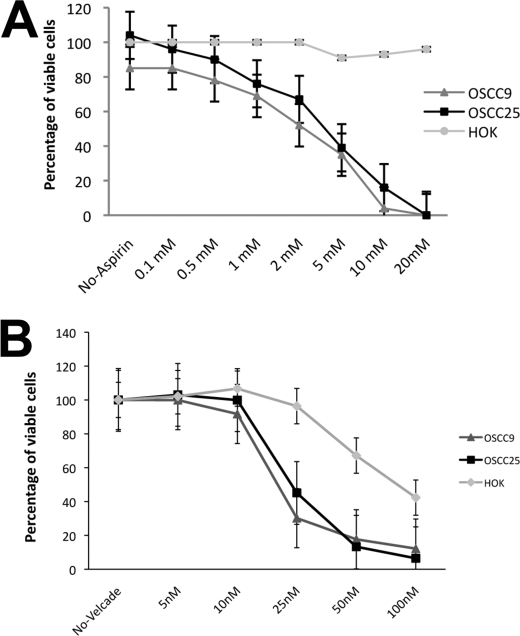
The inhibition of IL-8 signaling pathway dramatically reduced the proliferation and viability in OSCC cells. IL-8 is a major mediator of inflammatory response, and our gene expression profiling data indicated a strong association between inflammation and OSCCs. Therefore, we investigated the effect of anti-inflammatory drugs on OSCC cell proliferation. We assessed the effect of aspirin, which is an NSAID (non-steroidal anti-inflammatory drug) and Velcade, a proteasome/NF-κB inhibitor. Both aspirin and Velcade had a profound effect on OSCC cell proliferation/survival rate. Aspirin resulted in a dose-dependent decrease in the viability of OSCC9 and OSCC25 cells after 72 h of treatment (Fig. 5A) but did not show any significant effect of HOK cells. Velcade treatment of OSCC cells had no significant effect on cell viability at 10 nm but significantly decreased cell viability at 25 nm, and at 100 nm the number of viable OSCC cells was negligible (Fig. 5B). Thus, inhibition of inflammatory cascade by aspirin and Velcade dramatically reduced OSCC cell proliferation and viability. Aspirin had no effect on HOK cells, and Velcade treatment affected HOK cells only at the highest concentrations tested.
FIGURE 5.
Aspirin and Velcade reduce OSCC cell viability in a dose-dependent manner. OSCC cells were cultured in DMEM/F-12 supplemented with 10% FBS containing various doses of aspirin (A) or Velcade (B). The cells were plated at a density of 1 × 105 per well with drugs varying in concentrations from 0.1 to 20 mm for aspirin and 5 to 100 nm for Velcade. The cells showed significantly reduced viability at 2 mm aspirin and 25 nm Velcade. Cell viability was measured by MTT assay at 72 h and expressed relative to control cell viability. The control HOK cells did not show any effect with aspirin treatment. Velcade is a proteasome inhibitor and affects cell viability at high concentrations. The values represent the mean of multiple experiments.
DISCUSSION
Chronic inflammation leading to cancer is an example of dysregulation of an essential process becoming hazardous. Innate immunity, which is the first line of the host defense against a variety of insults, protects cells through the release of inflammatory mediators, such as cytokines, chemokines, matrix-remodeling proteases, and reactive oxygen species (33, 34). However, malfunctioning immune components could lead to chronic inflammation, generating a microenvironment that may initiate and promote carcinogenesis (15). The aim of the present study was to determine whether transcriptional alterations in oral cancers identifies a possible link between chronic inflammation and development of OSCC. Gene expression profiling was used to identify OSCC biomarkers that could be used as possible therapeutic targets or prognostic indicators.
We found that OSCC in common with a variety of forms of cancer was associated with high constitutive NF-κB activity, consistent with a chronic inflammatory state in the cancer cells. Differential gene regulation was observed in functional clusters such as inflammatory, wound healing, proliferation, angiogenesis, and apoptotic processes. Gene clustering indicated a strong link between inflammation, with an emphasis on wound healing processes and tumor development. The process of wound-healing involves a complex interplay of cells, mediators, growth factors, and cytokines (35) and is initiated by the recruitment of inflammatory cells to the site of infection and remodeling of collagen matrix (35). The highly over-expressed inflammatory genes in OSCCs were IL-8, CCL5, CXCL1, CXCL10, STAT5A, TLR6, and TNF-α. Wound healing-related genes were NMI, SAA4, SERPINF2, C3, APOL2, IRF7, TGFβ2, CD97, and CD14 indicating strong inflammatory response. Growth factors like VEGF and EGFR, which promoted cell proliferation and angiogenesis, were also over-expressed in OSCCs. Studies suggest that CXC chemokine family members such as IL-8 control the expression of growth factors in endothelial cells and trigger cell proliferation through NF-κB pathway (36). Also IL-8 signaling induces phosphorylation of VEGFR in endothelial cells (37), and IL-8 signaling can transactivate EGFR to further promote proliferation through MAPK signaling (27, 38). Thus, we propose that IL-8 might activate VEGF and possibly transactivated EGFR in some cases as VEGF is up-regulated in all OSCC cells where as EGFR is up-regulated in one cell line. Also in the present study we emphasize that the expression level of certain genes depends on the genotype of the individuals as well and CXC family members could be valuable targets for individualized medicine.
TGF-β family members, which are angiogenic factors and make the tumors more invasive (39, 40), were found to be over-expressed in OSCC samples. The cell cycle regulating genes CDK6, CDK2, and cyclin D1 were also highly elevated. Cyclin D1 accelerates the G1 phase of the cell cycle by binding to CDK4/CDK6 (41). Evidence indicated that Cyclin D1 expression associated well with clinical characteristics of tongue squamous cell carcinomas (41, 42). Thus, there is a potential association between OSCCs and inflammatory cytokines, wound healing genes, growth factors, and cell cycle genes. Interestingly, high expression of NMI, WNT4, WISP3, SAA41, OASL, IL11, TLR6, SERPINA1, and APOL2 might indicate a novel function, which could be explored in association with inflammatory neoplasms.
Genes in the WNT/β-catenin pathway have been associated with oral tissue development and disease (43, 44). Our analysis showed overexpression of WNT genes WNT4 and WISP (WNT1 signaling pathway inducible protein) along with some immune response genes such as C3 and CFB, suggesting a possible role of WNT signaling in OSCC. Growth factor signaling pathways such as insulin-like growth factor (IGF) might also be of importance in cancer development (45, 46). Our study also confirms that specific genes involved in IGF pathway such as IGF and IGFBP (IGF-binding protein) are highly up-regulated in OSCCs. The exact relevance of these pathways and their genes in inflammatory cancers will need to be explored in future studies.
Recent studies indicate a strong link between human papilloma virus (HPV) infection and oral cancers (47). Integrated HPV type 16 and loss of heterozygosity at 11q22 and 18q21 has been observed in oral carcinomas and derivative cell lines (48). Morphological and immunohistochemical evidence indicates the involvement of HPV in the dysplastic lesions of the uterine cervix (49). Evidence also suggests a clear association between HPV and oral cancers and also a subset of head and neck cancers (50–52). HPV-positive oral cancers have a better prognosis than HPV-negative oral cancers. HPV strains are responsible for almost 70% of cervical cancers and have been proposed to cause oral cancer (50). Interestingly, HPV-positive oral cancers may be considered a sexually transmitted disease, and thus, vaccination (Gardasil®) in sexually active young men and women has been implemented to not only prevent cervical cancer but also to reduce HPV-positive oral cancer by preventing chronic inflammation caused by HPV infection in the oral cavity. We examined HPV status in OSCC cell lines by performing a PCR screening for HPV16 and -18 sequences in OSCC genomic DNA (data not shown). The cells were negative for HPV16 and -18-specific amplicons, and it was brought to our attention that OSCC cell lines from ATCC are HPV negative.3 The HPV status in human tumor samples from GEO has not been documented but a comparative analysis with HPV-positive oral tumor profiles did not match the GEO set profiles, indicating that these patient samples used in our data analysis are HPV-negative.
Comparison of microarray data obtained from three OSCC cell lines and from 16 human patient samples identified gene dysregulation in functional categories as inflammation, wound healing, cell differentiation, cell proliferation, and DNA replication. Significant inflammatory genes such as IL-8, CXCL10, CCL5, TGFB2, TNFSF10, and growth factors like VEGF were highly up-regulated, suggesting that inflammatory genes, especially the CXC family members, might play a pivotal role in OSCC patients. The data further indicated that pro-inflammatory cytokines, especially IL-8, could activate VEGF and/or STAT signaling for proliferation and angiogenesis. Furthermore, IL-8 is an important modulator of inflammatory cancers, as tumor-derived IL-8 has a profound effect in the tumor microenvironment. Secreted IL-8 can enhance proliferation, angiogenesis, and survival rate of cancer cells. IL-8 signaling increases phosphorylation of the serine/threonine kinase and PKB/Akt (53) as well as the MAPK signaling cascade (54). Furthermore, IL-8 promotes transcriptional activity of STAT3 and β-catenin (53).
The DNA replication genes (MCM2, MCM4, and MCM5), complement pathway genes (C3, CIS, and CFB), matrix metalloproteinase (MMP1, MMP7), and other genes (WNT4, COL1A2, and SERPINA1) were also highly up-regulated in the OSCC patient samples, in agreement with the cell line data. Thus, taken together these results confirm not only active proliferation and angiogenesis but also involvement of other signaling pathways in inflammatory cancers in OSCC. Interestingly, most identified genes were common in both cell lines and patient samples, confirming the similarity of both in vitro and in vivo gene regulation patterns in OSCC development. The study also establishes the validity of the selected OSCC cell lines as models for exploring novel functional studies and association of genes and pathways involved in inflammatory cancers. The dysregulation of selected candidate genes (IL-8, IL-6, EGFR, VEGF, STAT1, CCND1, CCL5, TP53, etc.) by microarray analysis was validated by quantitative real time PCR for mRNA expression levels.
Pathway prediction analysis suggests that IL-8 could be an important signaling cascade deregulated in OSCC. Particularly the CXC family members IL-8, CXCL10, and CCL5 could promote tumor formation through activation of STAT and NF-κB signaling pathways (55, 56). Persistent NF-κB activation in cancer cells by the inflammatory microenvironment containing various cytokines, hypoxia, or genetic alterations is well established (57, 58). Thus, the present study strongly suggests that CXC family members, especially IL-8, activate NF-κB and STAT pathways. VEGF is highly up-regulated in the cell lines, suggesting a potential role for VEGF in inflammation leading to tumor development. There is ample evidence of cross-talk between growth factors such EGFR and STAT3 and NF-κB activation (59). Our results indicate a connection between IL-8 and STAT signaling and VEGF-dependent progression of tumors. IL-8 and IL-6 have been reported to be up-regulated in highly aggressive forms of cancer (5, 60, 61), and IL-8 is highly over-expressed in saliva of OSCC patients (4, 22). Our data clearly showed that IL-8 expression at both mRNA and protein levels in OSCC cells was significantly elevated compared with normal human oral keratinocytes. To further examine the biological activity of IL-8 in OSCC, we performed IL-8 gene knockdown by siRNA in OSCC cells. The IL-8 knockdown markedly inhibited OSCC cell proliferation and viability, indicating a potential role for IL-8 in OSCC development.
In addition, treatment of OSCC cell lines with the anti-inflammatory drug aspirin and the proteasomal inhibitor Velcade® (bortezomib), both of which inhibit NF-κB activity, provided important insights into potential novel therapies for OSCC. Relatively low concentrations of both aspirin and Velcade had significant effects on cell proliferation and viability of OSCCs. These assays also suggest another potential role of inflammation in OSCC development. Taken together, these findings establish chronic inflammation as a major cause of OSCC and IL-8 as an important regulator of inflammation-dependent oral cancers with potential in pharmacogenomics.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA133322 (NCI). This work was also supported by the College of Dentistry and Center for Integrative Cancer Research, University of Tennessee Health Science Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
This work is dedicated to the memory of Dr. Arun Bajpai, Dr. Nosrat′s first postdoctoral fellow at University of Tennessee Health Science Center who tragically passed away at the American Association for Dental Research meeting in 2008 in Dallas, TX, one day before he had the opportunity to present a poster on the initial OSCC microarray analysis data.
M. Gillison, personal communications.
- OSCC
- oral squamous cell carcinoma
- HOK
- human oral keratinocyte
- MTT
- methylthiazolyldiphenyltetrazolium bromide
- HPV
- human papilloma virus
- EGFR
- EGF receptor.
REFERENCES
- 1.Chen C., Méndez E., Houck J., Fan W., Lohavanichbutr P., Doody D., Yueh B., Futran N. D., Upton M., Farwell D. G., Schwartz S. M., Zhao L. P. (2008) Cancer Epidemiol. Biomarkers Prev. 17, 2152–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Swiahb J. N., Chen C. H., Chuang H. C., Fang F. M., Tasi H. T., Chien C. Y. (2010) Future Oncol. 6, 837–850 [DOI] [PubMed] [Google Scholar]
- 3.Parkin D. M., Bray F., Ferlay J., Pisani P. (2005) CA Cancer J. Clin. 55, 74–108 [DOI] [PubMed] [Google Scholar]
- 4.Li Y., St John M. A., Zhou X., Kim Y., Sinha U., Jordan R. C., Eisele D., Abemayor E., Elashoff D., Park N. H., Wong D. T. (2004) Clin. Cancer Res. 10, 8442–8450 [DOI] [PubMed] [Google Scholar]
- 5.Ondrey F. G., Dong G., Sunwoo J., Chen Z., Wolf J. S., Crowl-Bancroft C. V., Mukaida N., Van Waes C. (1999) Mol. Carcinog. 26, 119–129 [DOI] [PubMed] [Google Scholar]
- 6.Reibel J. (2003) Crit. Rev. Oral Biol. Med. 14, 47–62 [DOI] [PubMed] [Google Scholar]
- 7.Silverman S., Jr., Gorsky M., Lozada F. (1984) Cancer 53, 563–568 [DOI] [PubMed] [Google Scholar]
- 8.Pérez-Sayáns M., Somoza-Martín J. M., Barros-Angueira F., Reboiras-López M. D., Gándara Rey J. M., García-García A. (2009) Oncol. Rep. 22, 1277–1282 [DOI] [PubMed] [Google Scholar]
- 9.Lee C. H., Lee J. M., Wu D. C., Hsu H. K., Kao E. L., Huang H. L., Wang T. N., Huang M. C., Wu M. T. (2005) Int. J. Cancer 113, 475–482 [DOI] [PubMed] [Google Scholar]
- 10.Michaud D. S., Liu Y., Meyer M., Giovannucci E., Joshipura K. (2008) Lancet Oncol. 9, 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y. J., Viana A. C., Curtis K. M., Orrico S. R., Cirelli J. A., Mendes-Junior C. T., Scarel-Caminaga R. M. (2010) Clin. Chim. Acta 411, 1264–1268 [DOI] [PubMed] [Google Scholar]
- 12.Erreni M., Bianchi P., Laghi L., Mirolo M., Fabbri M., Locati M., Mantovani A., Allavena P. (2009) Methods Enzymol. 460, 105–121 [DOI] [PubMed] [Google Scholar]
- 13.Skipworth R. J., Moses A. G., Sangster K., Sturgeon C. M., Voss A. C., Fallon M. T., Anderson R. A., Ross J. A., Fearon K. C. (2010) Support Care Cancer, in press [DOI] [PubMed] [Google Scholar]
- 14.Mantovani A. (2010) Curr. Mol. Med. 10, 369–373 [DOI] [PubMed] [Google Scholar]
- 15.Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 16.Shinriki S., Jono H., Ota K., Ueda M., Kudo M., Ota T., Oike Y., Endo M., Ibusuki M., Hiraki A., Nakayama H., Yoshitake Y., Shinohara M., Ando Y. (2009) Clin. Cancer Res. 15, 5426–5434 [DOI] [PubMed] [Google Scholar]
- 17.Andia D. C., de Oliveira N. F., Casarin R. C., Casati M. Z., Line S. R., de Souza A. P. (2010) J. Periodontol., in press [DOI] [PubMed] [Google Scholar]
- 18.de la Iglesia N., Konopka G., Lim K. L., Nutt C. L., Bromberg J. F., Frank D. A., Mischel P. S., Louis D. N., Bonni A. (2008) J. Neurosci. 28, 5870–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerebours F., Vacher S., Andrieu C., Espie M., Marty M., Lidereau R., Bieche I. (2008) BMC Cancer 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunter M. J., Canzian F., Landi S., Chanock S. J., Sinha R., Rothman N. (2006) Cancer Epidemiol. Biomarkers Prev. 15, 1126–1131 [DOI] [PubMed] [Google Scholar]
- 21.Zabaleta J., Su L. J., Lin H. Y., Sierra R. A., Hall M. C., Sartor A. O., Clark P. E., Hu J. J., Ochoa A. C. (2009) Carcinogenesis 30, 1358–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagler R. M. (2009) Oral Oncol. 45, 1006–1010 [DOI] [PubMed] [Google Scholar]
- 23.Huang S., Mills L., Mian B., Tellez C., McCarty M., Yang X. D., Gudas J. M., Bar-Eli M. (2002) Am. J. Pathol. 161, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller L. J., Kurtzman S. H., Wang Y., Anderson K. H., Lindquist R. R., Kreutzer D. L. (1998) Anticancer Res. 18, 77–81 [PubMed] [Google Scholar]
- 25.Slaton J. W., Inoue K., Perrotte P., El-Naggar A. K., Swanson D. A., Fidler I. J., Dinney C. P. (2001) Am. J. Pathol. 158, 735–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X. Y., Chan W. Y., Whitney B. M., Fan D. M., Chow J. H., Liu Y., Ng E. K., Chung S. C. (2002) Diagn. Mol. Pathol. 11, 135–139 [DOI] [PubMed] [Google Scholar]
- 27.Venkatakrishnan G., Salgia R., Groopman J. E. (2000) J. Biol. Chem. 275, 6868–6875 [DOI] [PubMed] [Google Scholar]
- 28.Shi Q., Abbruzzese J. L., Huang S., Fidler I. J., Xiong Q., Xie K. (1999) Clin. Cancer Res. 5, 3711–3721 [PubMed] [Google Scholar]
- 29.Li A., Varney M. L., Singh R. K. (2001) Clin. Cancer Res. 7, 3298–3304 [PubMed] [Google Scholar]
- 30.Sheryka E., Wheeler M. A., Hausladen D. A., Weiss R. M. (2003) Urology 62, 162–166 [DOI] [PubMed] [Google Scholar]
- 31.Kim S. J., Uehara H., Karashima T., Mccarty M., Shih N., Fidler I. J. (2001) Neoplasia 3, 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veltri R. W., Miller M. C., Zhao G., Ng A., Marley G. M., Wright G. L., Jr., Vessella R. L., Ralph D. (1999) Urology 53, 139–147 [DOI] [PubMed] [Google Scholar]
- 33.Hussain S. P., Harris C. C. (2007) Int. J. Cancer 121, 2373–2380 [DOI] [PubMed] [Google Scholar]
- 34.Keibel A., Singh V., Sharma M. C. (2009) Curr. Pharm. Des. 15, 1949–1955 [DOI] [PubMed] [Google Scholar]
- 35.Clark R. A. (1985) J. Am. Acad. Dermatol. 13, 701–725 [DOI] [PubMed] [Google Scholar]
- 36.Martin D., Galisteo R., Gutkind J. S. (2009) J. Biol. Chem. 284, 6038–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petreaca M. L., Yao M., Liu Y., Defea K., Martins-Green M. (2007) Mol. Biol. Cell 18, 5014–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luppi F., Longo A. M., de Boer W. I., Rabe K. F., Hiemstra P. S. (2007) Lung Cancer 56, 25–33 [DOI] [PubMed] [Google Scholar]
- 39.Blobe G. C., Schiemann W. P., Lodish H. F. (2000) N. Engl. J. Med. 342, 1350–1358 [DOI] [PubMed] [Google Scholar]
- 40.Wahl S. M. (1992) J. Clin. Immunol. 12, 61–74 [DOI] [PubMed] [Google Scholar]
- 41.Mineta H., Miura K., Takebayashi S., Ueda Y., Misawa K., Harada H., Wennerberg J., Dictor M. (2000) Oral Oncol. 36, 194–198 [DOI] [PubMed] [Google Scholar]
- 42.Liu H. S., Lu H. H., Lui M. T., Yu E. H., Shen W., Chen Y. P., Chang K. W., Tu H. F. (2009) Oral Oncol. 45, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 43.Liu F., Millar S. E. (2010) J. Dent Res. 89, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Castro J., Gamallo C., Palacios J., Moreno-Bueno G., Rodríguez N., Feliu J., González-Barón M. (2000) Virchows Arch. 437, 599–604 [DOI] [PubMed] [Google Scholar]
- 45.Breuhahn K., Longerich T., Schirmacher P. (2006) Oncogene 25, 3787–3800 [DOI] [PubMed] [Google Scholar]
- 46.Declercq J., Van Dyck F., Van Damme B., Van de Ven W. J. (2008) Int. J. Oncol. 32, 1041–1047 [PubMed] [Google Scholar]
- 47.Jalouli J., Ibrahim S. O., Mehrotra R., Jalouli M. M., Sapkota D., Larsson P. A., Hirsch J. M. (2010) Acta Otolaryngol, in press [DOI] [PubMed] [Google Scholar]
- 48.Steenbergen R. D., Hermsen M. A., Walboomers J. M., Joenje H., Arwert F., Meijer C. J., Snijders P. J. (1995) Cancer Res. 55, 5465–5471 [PubMed] [Google Scholar]
- 49.Syrjänen K., Väyrynen M., Castrén O., Mäntyjärvi R., Pyrhönen S., Yliskoski M. (1983) Int. J. Gynaecol. Obstet. 21, 261–269 [DOI] [PubMed] [Google Scholar]
- 50.Gillison M. L., Koch W. M., Capone R. B., Spafford M., Westra W. H., Wu L., Zahurak M. L., Daniel R. W., Viglione M., Symer D. E., Shah K. V., Sidransky D. (2000) J. Natl. Cancer Inst. 92, 709–720 [DOI] [PubMed] [Google Scholar]
- 51.Psyrri A., Gouveris P., Vermorken J. B. (2009) Curr. Opin. Oncol. 21, 201–205 [DOI] [PubMed] [Google Scholar]
- 52.Chocolatewala N. M., Chaturvedi P. (2009) J. Cancer Res. Ther. 5, 71–77 [DOI] [PubMed] [Google Scholar]
- 53.Waugh D. J., Wilson C. (2008) Clin. Cancer Res. 14, 6735–6741 [DOI] [PubMed] [Google Scholar]
- 54.Knall C., Young S., Nick J. A., Buhl A. M., Worthen G. S., Johnson G. L. (1996) J. Biol. Chem. 271, 2832–2838 [DOI] [PubMed] [Google Scholar]
- 55.Pahl H. L. (1999) Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 56.Neiva K. G., Zhang Z., Miyazawa M., Warner K. A., Karl E., Nör J. E. (2009) Neoplasia 11, 583–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karin M. (2006) Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 58.Royds J. A., Dower S. K., Qwarnstrom E. E., Lewis C. E. (1998) Mol. Pathol. 51, 55–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Squarize C. H., Castilho R. M., Sriuranpong V., Pinto D. S., Jr., Gutkind J. S. (2006) Neoplasia 8, 733–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie Q., Thompson R., Hardy K., DeCamp L., Berghuis B., Sigler R., Knudsen B., Cottingham S., Zhao P., Dykema K., Cao B., Resau J., Hay R., Vande Woude G. F. (2008) J. Transl. Med. 6, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Efimova E. V., Liang H., Pitroda S. P., Labay E., Darga T. E., Levina V., Lokshin A., Roizman B., Weichselbaum R. R., Khodarev N. N. (2009) Int. J. Radiat. Biol. 85, 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.