Abstract
Schistosomiasis is the second most widespread human parasitic disease. It is principally treated with one drug, praziquantel, that is administered to 100 million people each year; less sensitive strains of schistosomes are emerging. One of the most appealing drug targets against schistosomiasis is thioredoxin glutathione reductase (TGR). This natural chimeric enzyme is a peculiar fusion of a glutaredoxin domain with a thioredoxin selenocysteine (U)-containing reductase domain. Selenocysteine is located on a flexible C-terminal arm that is usually disordered in the available structures of the protein and is essential for the full catalytic activity of TGR. In this study, we dissect the catalytic cycle of Schistosoma mansoni TGR by structural and functional analysis of the U597C mutant. The crystallographic data presented herein include the following: the oxidized form (at 1.9 Å resolution); the NADPH- and GSH-bound forms (2.3 and 1.9 Å, respectively); and a different crystal form of the (partially) reduced enzyme (3.1 Å), showing the physiological dimer and the entire C terminus of one subunit. Whenever possible, we determined the rate constants for the interconversion between the different oxidation states of TGR by kinetic methods. By combining the crystallographic analysis with computer modeling, we were able to throw further light on the mechanism of action of S. mansoni TGR. In particular, we hereby propose the putative functionally relevant conformational change of the C terminus after the transfer of reducing equivalents from NADPH to the redox sites of the enzyme.
Keywords: Electron Transfer, Enzyme Structure, Flavoproteins, Parasite, X-ray Crystallography, Schistosoma mansoni, Drug Target, Thioredoxin Glutathione Reductase
Introduction
Schistosomes are human platyhelminth parasites causing Schistosomiasis, a severe disease still classified among the major causes of mortality in tropical and subtropical countries, affecting more than 200 million people (1). The only drug employed to fight the disease is praziquantel, whose efficacy is restricted to the adult stages of the parasite and whose mechanism of action is still incompletely clarified (2, 3). Because this drug is administered to 100 million people every year, some less sensitive strains have already been isolated, and given the massive drug administration, resistance might become a serious problem (3).
Because of this, the search for a new drug against Schistosomiasis is a necessity and a priority according to the World Health Organization (4). The main evidence suggesting that targeting the thiol redox pathway of the parasite may represent a profitable starting point for rational drug design is as follows: (i) schistosomes, living in the human bloodstream, are subjected not only to endogenous reactive oxygen species but are also exposed to radicals produced by the host immune response; and (ii) the thiol redox pathway employed by the worms for the enzymatic reduction of the reactive oxygen species is different from its human counterpart (5, 6).
In mammals, two proteins belonging to the pyridine nucleotide disulfide oxidoreductase family, thioredoxin reductase (TR)3 and glutathione reductase (GR), are the head antioxidant enzymes shuttling electrons through the thioredoxin (Trx) and the GSH systems, respectively (5). Trx peroxidases in addition to Trxs are the other components of the Trx system; the GSH system involves GSH peroxidase (Gpx) and glutaredoxin (Grx), an oxidoreductase acting on S-glutathionylated substrates (7, 8). TRs and GRs are homodimeric NADPH-dependent flavoproteins that are structurally highly similar. The two proteins share the main fold and the architecture of the binding sites for NADPH and FAD. However, TRs differ from GRs because of the presence of a selenocysteine residue (Sec, U) located in a GCUG tetrapeptide at the C terminus of the protein. This additional C-terminal extension is required for the full catalytic activity of the enzyme (9–11). Thioredoxin glutathione reductase (TGR) is another selenoprotein belonging to the above-mentioned family of oxidoreductases. Characterization of mouse TGR demonstrated that the enzyme can reduce components of both mammalian Trx and GSH systems. In mammals, TGR exerts a wide range of catalytic activities though its selective overexpression in testis after puberty suggests its involvement in very specific functions related with sperm maturation (12).
A similar enzyme has been identified in Schistosoma mansoni (SmTGR) (6). The schistosomal antioxidant system lacks the typical mammalian TR and GR enzymes, which are both replaced by SmTGR; this peculiarity revealed that SmTGR may be a target of choice for the development of new antischistosomal drugs (5, 13). Moreover, TGR enzymes with similar roles have been isolated in other human parasites, such as Taenia (14) and Echinococcus (15). All these parasites can be killed, in vivo and ex vivo, by auranofin, a gold compound that specifically inhibits Sec- or Cys-dependent reductases (16).
Crystallographic studies on SmTGR have recently highlighted the structural basis for the multienzymatic function of this enzyme. SmTGR is a homodimeric flavoprotein, where each subunit results from the fusion of a typical TR domain with a glutaredoxin (Grx) domain, the latter being located at the N terminus of the enzyme. The two domains work either coupled or independently to catalyze the reduction of a large number of oxidized substrates. The C-terminal tail of each subunit, which is close to the FAD active site of the adjacent subunit, contains the Sec residue (17).
The catalytic mechanism of TGR has not been fully elucidated, although an hypothesis has been published for the mouse enzyme (18). Functional analysis performed on SmTGR and on different TGRs proved that the penultimate Sec is essential for the reduction of thioredoxin and the glutaredoxin domain (6, 12, 15). Reduction of glutathione under physiological conditions is probably dependent on the redox exchange with glutaredoxin and thus is also inhibited in Sec-lacking enzymes, even though we observed residual GR activity in a C-terminally truncated variant of SmTGR (17). This residual activity is only observed in the SmTGR variant that lacks the C-terminal arm and is not shared by the full-length U597C mutant (see supplemental material); thus, we believe that in the WT enzyme the GR activity is mostly accomplished by the Grx domain.
We present in this study a structural characterization of some key intermediates belonging to the catalytic cycle of SmTGR by means of x-ray crystallography and computer modeling, as described in Fig. 1. We provide evidence for the reactions in Fig. 1, showing from a structural point of view the steps that allow electron transfer from NADPH to S. mansoni thioredoxin (SmTrx) and to the Grx domain of SmTGR with the participation of the C terminus of the enzyme.
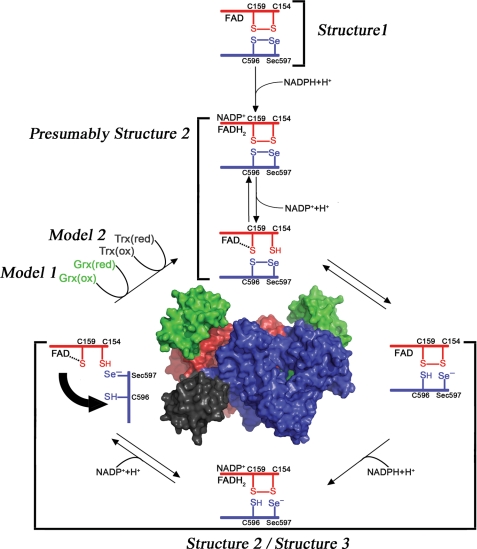
FIGURE 1.
Electron transfer in and out of the TR domain of SmTGR. The reaction scheme has been drawn following the experimental evidence reported in this work and elsewhere (18). When NADPH binds to the oxidized state of the enzyme (Structure 1), electrons are transferred from the nicotinamide ring to the FAD; reduced FADH2 donates electrons to the Cys154–Cys159 couple (in red) that is close to the isoalloxazine ring of the cofactor (as observed in Structure 2), and finally electrons reach the Cys596–Sec597 redox couple on the C-terminal segment of the other subunit (in blue, as observed in Structure 3). The C terminus acts as a flexible arm, which might donate electrons either internally to the Cys28–Cys31 redox active couple on the oxidized Grx domain (in green) or externally to various oxidized substrates such as SmTrx (in dark gray).
The evidence presented below includes the structural and functional characterization of the full-length U597C mutant of SmTGR (SmTGRfl, see Fig. 1). The enzyme was crystallized in the presence of the following: (i) CuSO4 to obtain a fully oxidized state (Structure 1 (1.9 Å)); (ii) NADPH (Structure 2 (2.3 Å)); (iii) reduced glutathione GSH (Structure 4 (1. 9 Å)). Moreover, we obtained the three-dimensional structure of SmTGRfl, in a different crystal form (Structure 3 (3.1 Å)), which contains one dimer, and not one protomer, in the asymmetric unit. The crystallographic analysis combined with the results obtained by computer modeling (Model 1 and Model 2) and by rapid kinetics experiments allows us to postulate and discuss the possible functionally relevant movements of the C terminus during the redox cycle of the enzyme.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of the SmTGR Mutant
The gene for full-length TGR, with the mutation U597C, was obtained starting from the gene of truncated SmTGR optimized for Escherichia coli expression (17), with appropriate DNA primers. The gene was cloned in a pGEX4T-1 commercial vector (GE Healthcare) and expressed in BL21(DE3) E. coli cells (Novagen). The best results, in terms of solubility and protein yield, were obtained by incubation of cells at 20 °C overnight with 20 μm FAD, without isopropyl 1-thio-β-d-galactopyranoside induction. Successful expression was confirmed by SDS-PAGE, highlighting a band at about 90 kDa. The protein was purified by standard affinity chromatography; the GST tag was cleaved by thrombin (Sigma) and removed according to the manufacturer's instructions. Protein purity was >98% as assessed by SDS-PAGE, and concentration was determined from the FAD absorption peak (ϵ463 = 11.3 mm−1 cm−1). SmTGRfl was dialyzed in 20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 100 μm FAD (to saturate the enzyme), concentrated to 10 mg/ml using an Amicon ultracentrifuge filter device (Millipore), and stored at −80 °C.
Protein Crystallization
Crystallization of Oxidized SmTGRfl
A crystal of the SmTGRfl, grown in 0.1 m Hepes, pH 7.4, 20% PEG 3350, 0.2 m KI, and 5 mm β-mercaptoethanol, was picked up and incubated in the same well solution with 2 μm CuSO4 and without β-mercaptoethanol. After 48 h, the crystal was frozen under liquid nitrogen, after adding 30% PEG 200 as cryoprotectant.
Crystallization of SmTGRfl-NADPH Complex
120 μm of purified SmTGRfl in 20 mm Tris, pH 7.4, 100 mm NaCl was initially incubated for 24 h with 300 μm plumbagin (Sigma), 400 μm NADPH, in anaerobiosis condition. The mixture was then concentrated to 10 mg/ml before crystallization trials were set up. The protein solution was mixed with an equal amount of the reservoir solution containing 0.1 m Hepes, pH 7.0, 20% PEG 3350, 0.2 m KSCN, and 5 mm GSH. Crystals grew for 1 month in the standard sitting drop method and were cryoprotected with the same reservoir solution adding 30% PEG 200.
Crystallization of the SmTGRfl-GSH Complex
Crystals were grown by sitting drop vapor diffusion. Crystals grew in conditions similar to those of oxidized SmTGRfl (see above). 5 mm GSH replaced β-mercaptoethanol. Crystals grew in 1 week and were cryoprotected with the same reservoir solution containing 30% PEG 200.
Crystallization of SmTGRfl with Reduced and Ordered C Terminus
Crystallization conditions were initially screened by the Phoenix nanoliter dispensing robot (ArtRobbins) and then manually refined. Crystallization was achieved at 293 K by the sitting drop vapor diffusion technique. 10 mg/ml protein samples were equilibrated in 1 mm NADPH in 20 mm Tris-HCl, 100 mm NaCl, pH 7.4. Drops of 2 μl were mixed with an equal amount of the reservoir solution containing 0.2 m magnesium formate, 20% PEG 6000, 0.1 m Hepes, pH 7.0–7.5, plus 5 mm β-mercaptoethanol. Crystals grew in 1–2 weeks and were cryoprotected with the same reservoir solution adding 30% PEG 200.
Data Collection, Processing, and Refinement
Oxidized Form, SmTGRfl-NADPH and SmTGRfl-GSH Complexes; Structures 1, 2, and 4
The best data sets for the oxidized and for the NADPH-bound form and for SmTGR(U597C) have been collected at BESSY (Berlin, German) at 100 K at 1.9 and 2.3 Å resolution, respectively; and for the GSH-bound form, the best data set was collected at ESRF at 1.9 Å resolution. Data analysis was performed with HKL2000 suite (19) indicating that all crystals belonged to the C2 space group, isomorphous to the SmTGR truncated form (17). Difference Fourier method was used to solve all structures, and refinements were performed using REFMAC5 (20). When present, NADPH and GSH were excluded from the first rounds of refinement and added only after convergence of R and Rfree factors. COOT (21) was used to build all the models of protein and ligands. The quality of the model was assessed with PROCHECK (22). Data collection and refinement statistics are summarized in Table 1. The structures were deposited at RCSB Protein Data Bank (PDB codes as follows: 2X8G for the oxidized form; 2X99 for the NADPH complex; and 2X8H for the GSH complex).
TABLE 1.
Crystallographic data collection and refinement statistics
| Structure 1 | Structure 2 | Structure 3 | Structure 4 | ||
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | C2 | C2 | P212121 | C2 | |
| Cell dimensions | |||||
| a, b, c | 141.6, 102.8, 59.2 Å | 141.6, 102.6, 60.0 Å | 84.5, 87.4, 185.5 Å | 141.6, 102.9, 59.1 Å | |
| α, β, γ | 112.4 | 112.8 | 112.6 | ||
| Resolution | 40.0/1.9 (2.0/1.9 Å)a | 40.0/2.3 Å (2.4/2.3 Å)a | 30.0/3.1 Å (3.2/3.1 Å)a | 40.0/1.9 Å (2.0/1.9 Å)a | |
| Rmerge | 0.07 (0.32)a | 0.1 (0.4)a | 0.1 (0.5)a | 0.1 (0.4)a | |
| I/σI | 12.2 (3.2)a | 18.4 (3.8)a | 11.8 (2.6)a | 9.6 (5.4)a | |
| Completeness | 90.1% (92.6%)a | 100% (99.9%)a | 99.3% (99.2%)a | 100% (99.7%)a | |
| Redundancy | 3.0 (2.8)a | 8.1 (7.8)a | 7.1 (7.0)a | 5.7 (5.7)a | |
| Refinement | |||||
| Resolution | 40.0/1.9 Å | 40.0/2.3 Å | 30.0/3.1 Å | 40.0/1.9 Å | |
| No. of reflections | 52,728 | 33,294 | 24,221 | 58,385 | |
| Rwork/Rfree | 0.19/0.21 | 0.19/0.23 | 0.22/0.29 | 0.18/0.19 | |
| No. of atoms | 4863 | 4936 | 9174 | 5008 | |
| Protein | 4502 | 4502 | 9031 | 4504 | |
| Ligand/ion | 106 | 249 | 143 | 139 | |
| Water | 255 | 185 | 0 | 365 | |
| B-Wilson | 19.0 | 35.1 | 63.4 | 14.6 | |
| B factors | |||||
| Overall | 18.6 Å2 | 22.5 Å2 | 43.5 Å2 | 14.6 Å2 | |
| Main chain | 17.6 Å2 | 20.1 Å2 | 43.3 Å2 | 13.0 Å2 | |
| Side chains | 19.6 Å2 | 24.7 Å2 | 43.7 Å2 | 16.1 Å2 | |
| Root mean square deviations | |||||
| Bond lengths | 0.008 Å | 0.011 Å | 0.009 Å | 0.007 Å | |
| Bond angles | 1.064° | 1.302° | 1.164° | 0.978° | |
| Model quality | |||||
| Ramachandran plot | |||||
| Most favored | 91.4% | 91.0% | 86.3% | 92.6% | |
| Additional allowed | 8.4% | 9.0% | 13.5% | 7.2% | |
| Generously allowed | 0.2% | 0% | 0.3% | 0.2% | |
a Values in parentheses refer to the highest resolution shell.
SmTGRfl with Reduced and Ordered C Terminus; Structure 3
Diffraction data were collected at 3.1 Å resolution at Bessy (Berlin, Germany) and analyzed using the HKL2000 suite (19). The three-dimensional structure was solved by molecular replacement (Phaser, 20) using as a search model one protomer of truncated SmTGR structure (PDB code 2v6o) (17). The structure was refined using REFMAC5 and fitted to generated electron density maps using Coot. Refinement was performed until a final R/Rfree of 22.0 and 29.3%, respectively, was reached. The final model showed one dimer in the asymmetric unit with a FAD molecule bound to each subunit. The quality of the model was assessed with PROCHECK (22). The statistics of crystallographic data collection and model refinement are shown in Table 1. The structure was deposited at RCSB protein data bank (PDB code 2X8C).
Modeling of the C Terminus of SmTGRfl onto the Grx Domain; Model 1
The C-terminal residues, Cys597 and Gly598, visible in Structure 3, were superimposed to the Cys and Gly residues of the GSH bound to the Grx domain in the GSH-bound form of the same SmTGRfl (Structure 4). The model was obtained mainly by rotation of the C-terminal arm at the level of Lys586 ψ angle. Small adjustments of the main chain were performed to obtain the best superposition of the last two residues of the protein with the analogue residues of the bound GSH, avoiding clashes between amino acid side chains. Idealization of the model (Refmac5, 20) was done at the end to ensure the absence of unfavorable contacts between atoms. The model, after optimization, was evaluated with PROCHECK (22).
Modeling of SmTGRfl-SmTrx Complex; Model 2
The structures of oxidized and reduced SmTrx (PDB codes 2XBI and 2XBQ)4 and Structure 3 of SmTGRfl (PDB code 2X8C) were docked using a rigid body method with shape complementarity to model a plausible complex. The structures were used as input for the SUMMA server (23). The server calculated the best complementary surface and was instructed to freely perform energy minimization only on SmTGRfl terminal arm (residues 509–598) and on the active site stretch of SmTrx (residues 33–38). Idealization of the model was automatically performed at the end to ensure the absence of unfavorable contacts between atoms. An S–S bond was built between SmTrx Cys34 and SmTGRfl Cys596 with rotamers compatible with the reduced SmTrx protein.
Reductive Half-reaction, Stopped-flow Experiments
Stopped-flow experiments were carried out using an Applied Photophysics (Leathered, UK) in aerobic conditions at 25 °C. Time-dependent spectra were followed using the diode array detector, and single wavelength kinetic traces were recorded with a photomultiplier. Solutions of SmTGRfl (2.5 and 7.5 μm) in 0.1 m NaCl, 0.1 m Tris, pH 7.4, and 1 mm EDTA were mixed with several NADPH solutions at concentrations ranging between 3.3 and 20 μm.
Oxidative Half-reaction, Spectrophotometric Experiments
SmTGRfl (3.0, 5, and 10 μm) were anaerobically reduced with 2 eq of NADPH to produce EH4, at 25 °C in 0.1 m NaCl, 0.1 m Tris, pH 7.4, 1 mm EDTA. The reduced enzyme samples were then mixed with 2 eq of SmTrx, and the reactions were followed until completion.
RESULTS
Structure 1, Oxidized SmTGRfl
The three-dimensional structure of SmTGRfl in the oxidized state was determined at 1.9 Å resolution. The statistics of the diffraction and refinement data are summarized in Table 1. The refined model consists of one protomer per asymmetric unit. The map gave the possibility to fit residues from 6 to 593. The overall architecture of Structure 1 is the same as that previously reported for the truncated form of the enzyme (PDB code 2v6o (17)). The catalytic sites, i.e. the Cys28–Cys31 couple of the Grx domain and the Cys154–Cys159 over the FAD of the TR domain, are both fully oxidized, with the distances between the two sulfurs being 2.0 Å in both cases. The position of Tyr296 is also consistent with this state, the aromatic ring being perpendicular with the isoalloxazine ring of FAD.
Structure 2, the SmTGRfl-NADPH Complex
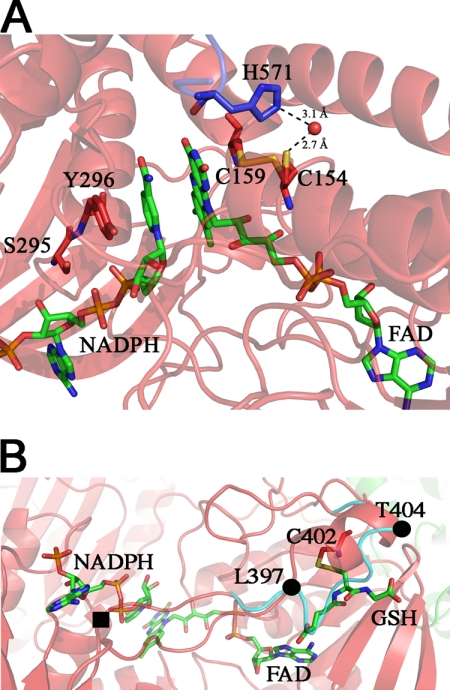
The structure of SmTGRfl in complex with NADPH and two molecules of GSH was determined at 2.3 Å resolution. The statistics of the diffraction and refinement data are summarized in Table 1. The refined model contains one protomer per asymmetric unit, one FAD, one NADPH, and two GSH molecules. The map gave the possibility to fit residues from 6 to 592. Overall, Structure 2 differs from Structure 1 mainly for slight side chain movements of some residues in the FAD and NADPH binding pockets. In particular, the two cysteine residues on the si-face of the isoalloxazine ring of FAD were found in both redox states due to a double conformation of Cys154 (each with 0.5 occupancy), although Cys159 maintains the same position. Thus, the sulfur-sulfur distance is 3.1 Å in the reduced state and 2.1 Å in the oxidized state. Moreover, Cys159 is at 3.2 Å from C4 and C10 of the isoalloxazine ring of FAD and might be implicated in a charge transfer complex, as already observed in human GR (PDB code 3djg) (10). The reduced Cys154 forms an H-bond with a water molecule, kept in place by His571, that may have a role in the stabilization of this conformation in the reduced state (Fig. 2, panel A) (24, 25).
FIGURE 2.
Structure 2, NADPH-binding site in SmTGRfl. Panel A, zoom in the FAD active site. The NADPH and FAD are shown as green sticks; Tyr296, Ser295, Cys154, and Cys159 are shown as red sticks, and His571 of the partner subunit is shown as blue sticks. Upon NADPH binding, the loop 295–297 changes conformation re-orienting the side chains of Ser295 and Tyr296 with respect to Structure 1 to make room for the reductant. The Cys159–Cys154 couple is partially reduced, and accordingly, Cys154 is found in a double conformation. In the reduced conformation (50% occupancy), the sulfur of Cys154 points toward the solvent and is in contact with a water molecule (2.7 Å, shown as red ball), which is kept in place by His571. Panel B, GSH-binding site on TR domain of Structure 2. The GSH (green sticks) is found in a position that in other TGR structures is usually occupied by the loop 397–404 (cyan ribbon). The ligand is found in a pocket formed by one α-helix (Leu397–Thr404), a β-strand (Lys227–Leu230), and the nucleotide moiety of the FAD. GSH makes a mixed disulfide with Cys402 and forces the loop 397–404 to adopt a α-helical secondary structure (red ribbon) by turning around two pivot residues Leu397 and Thr404 (shown as full black circles). One of the gates of the NADPH-binding site (full black square) is four residues upstream of Leu397.
The NADPH-binding site is mainly composed of three loops involving residues 292–297, 317–324, and 391–393. NADPH binds on the re-face of FAD and is stabilized by the following electrostatic interactions: a cation-π bond between Arg317 and the adenine; a salt bridge between Arg322 and the phosphate on ribose C2; and two H-bonds between Ser295-NADPH (AO2) and Ser318-NADPH (AOP2). The nicotinamide ring of NADPH is sandwiched between the isoalloxazine ring of FAD (at a distance of 3.8 Å) and the aromatic ring of Tyr296 (3.6 Å, Fig. 2A). Tyr296 swings upon NADPH binding and is stabilized in this new position by hydrophobic contacts between the phenol ring and the side chains of Pro324 and Val473 (as also reported for rat TR) (26). The occupancy of NADP+ is estimated at 80%, even though the B factor at the nicotinamide portion of the molecule is significantly higher than that of the protein.
GSH was found in two distinct sites of the structure. The first one lies in the Grx domain and will be described below in Structure 4. The second GSH is found in the TR domain, in a pocket formed by one α-helix (Leu397–Thr404), a β-strand (Lys227–Leu230), and the nucleotide moiety of the FAD. The Cys of GSH is engaged in a disulfide bond with Cys402 (2.1 Å). The rest of the molecule is stabilized by polar contacts with Lys227, Gly228, Arg229, and Leu230. Moreover the carboxylate group of the γ-glutaminyl end is at 2.9 Å from the FAD adenine amine group (Fig. 2, panel B).
The binding of one GSH molecule in the TR domain is associated with a clear change in the conformation of the region between residues 397 and 404, as compared with the unliganded structure (Fig. 2, panel B) (17). The presence of GSH forces the loop 397–404 to turn around two pivot residues (Leu397 and Thr404) and to acquire a 2-turn α-helix structure. As a result, the hydrophobic side chains of Val400 and Leu401 are exposed to the solvent, and GSH takes the place previously occupied by the loop (Fig. 2, panel B). The presence of NADPH/NADP+ in its binding site might be somehow correlated with the glutathionylation of Cys402, given the following: (i) attempts to co-crystallize the enzyme with NADPH in the absence of GSH were always unsuccessful; (ii) the change in conformation of the above mentioned loop is close to the gate of the NADPH-binding site (Fig. 2, panel B). At the moment, we are unable to propose a rationale to the concomitant presence of the two ligands, especially in view of the fact that Cys402 is not a member of a redox Cys couple.
Finally, in this structure, the Cys28–Cys31 couple on the Grx domain is reduced. This is probably due to the excess GSH present in the medium (5 mm).
Structure 3, SmTGRfl with Reduced and Ordered C Terminus
The structure of SmTGRfl in P212121 space group was solved at 3.1 Å resolution. The statistics of the diffraction and refinement data are summarized in Table 1. The main differences between this structure and the others reported so far (PDB codes 2v6o and 3h4k for Structures 1, 2, and 4) are due to the presence of the physiological dimer in the asymmetric unit and to the structuring of the reduced C-terminal arm, whose density is visible in one of the two subunits. The low resolution of Structure 3 does not permit an accurate estimate of the redox state of the two Cys couples at the FAD active site (Cys154–Cys159) and at the Grx redox site (Cys28–Cys31) because of the following. (i) The slight configurational change of Cys154 with respect to Cys159 upon reduction (∼1.0 Å, see Structure 2) would be hard to detect in view of the fact that the electron pair is shared between the FAD and the Cys154–Cys159 couple, leading to the latter being only partially reduced (as in Structure 2), consistent with the rapid kinetic experiments. (ii) Both the Grx domains are characterized by high mobility indicated by the high B factors (see below). Too weak electron density did not permit the assignment of the first five residues at the N terminus of both subunits, as well as of the last five residues at the C-terminal portion of subunit A. On the contrary, a stronger electron density allowed the building of the entire C-terminal segment of subunit B, implying a structural asymmetry in the organization of the two subunits. A second evidence of the same asymmetry comes from the different thermal factors of the two Grx domains (residues 7–104) as follows: the Grx portion of subunit A was rather less mobile (overall 〈B〉 = 57 Å2) than the Grx domain of subunit B (overall 〈B〉 = 88 Å2).
The terminal tetrapeptide GCCG, as it appears in the electron density map, is fully reduced, with the two cysteine residues (Cys596–Cys597) being in a trans position with respect to the main chain and pointing their sulfur atoms toward the solvent (Fig. 3, panel A). The positioning of the C terminus is permitted by a kink centered on Lys586. This conformation is stabilized by several H-bonds and salt bridges (listed in Table 2), mostly with residues of the B subunit but also with one residue of the A subunit. Hence the C terminus of the B subunit lies in proximity of the FAD-binding site of the A subunit, at 13 Å from the Cys154–Cys159 redox couple. This distance could be easily bridged by small rearrangements of the polypeptide chain to be made compatible with the electron transfer (Fig. 3, panel A).
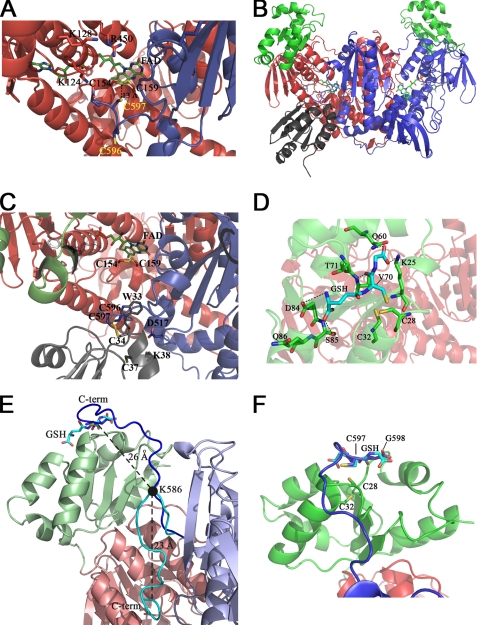
FIGURE 3.
Structure 3 and 4 and Models 1 and 2. The three-dimensional structure of the C-terminal region of SmTGRfl and the computed model of SmTGRfl-SmTrx complex are shown. The GSH-binding site of the Grx domain and a model of the C terminus pointing in its direction are shown. Panel A, Structure 3, ribbon representation of the C-terminal region of SmTGRfl. The two catalytic Cys belonging to the C-terminal arm (Cys596 and Cys597), shown as blue sticks, are both reduced; Cys597 is located about 13 Å from the Cys154–Cys159 couple (see text). The positively charged residues Lys124, Lys128, and Arg450 of one subunit involved in the stabilization of the C-terminal arm of the partner subunit are shown as red sticks. Panel B, Model 2, relative location of SmTrx and SmTGR in the modeled complex is shown. Components are as follows: SmTrx (gray); subunit A (red); subunit B of SmTGR (blue); Grx domains (green). Panel C, SmTrx-binding site on SmTGR in Model 2. The magnification allows us to visualize the relative topology of the redox sites (FAD, Cys154/Cys159, C terminus (Cys596/Cys597), and SmTrx (Cys34–Cys37)) involved in the electron transfer chain. The redox sites of both enzymes and the main side chains involved in the contact between SmTrx and SmTGR are shown as sticks. Panel D, GSH-binding site on the Grx domain. GSH (cyan sticks) is in a pocket above the redox site of the Grx domain (Cys28–Cys32, green sticks). Polar contacts are shown by dotted lines. The γ-glutamyl moiety of GSH interacts with Asp84, Ser85, and Gln86 (green sticks). The GSH sulfur points toward Cys28, and its position is stabilized by contacts with Thr70 and Val71. The carboxylate of the glutamic acid of GSH is H-bonded to Gln60 and Lys25. Panel E, superposition between Structure 3 and Model 1. The ribbon representation depicts the movement of the C-terminal arm. A rotation on the pivot residue Lys586 (visualized as a full black circle) brings the C terminus from the position found in Structure 3 (cyan) to the Grx domain (from Model 1, blue). The superposition between the last two residues of the C terminus (Cys597–Gly598, blue sticks) and the analogue residues of GSH (cyan sticks, as bound in Structure 4) is also shown. The distances between Cα of Lys586 and Cα of Cys597 and that between the former and the Cα of the GSH Cys are shown in the figure. Panel F, zoom of the superposition between Cys597–Gly598 (blue sticks) of the C-terminal arm and the GSH (cyan sticks) onto the Grx redox site (green sticks).
TABLE 2.
Polar contacts between the C-terminal arm and the TR domain of SmTGRfl, based on Structure 3
| C-terminal residue (subunit B) | Subunit A | Subunit B | Distance |
|---|---|---|---|
| Å | |||
| V583(O) | H502(N) | 3.0 | |
| T584(OG) | S587(OG) | 2.9 | |
| K585(N) | V500(O) | 2.9 | |
| K585(O) | G588(N) | 2.8 | |
| K585(NZ) | E327(OE1) | 2.8 | |
| K585(NZ) | Q327(OE1) | 2.8 | |
| P591(N) | N504(O) | 3.3 | |
| V593(N) | N518(OD1) | 2.7 | |
| G598(O) | K124(NZ) | 2.4 |
Structure 4, the SmTGRfl-GSH Complex
In the absence of NADPH, only one molecule of GSH per protomer was found located at the redox site on the Grx domain, as already quoted for Structure 2. The three-dimensional structure of SmTGRfl in complex with GSH was determined at 1.9 Å resolution. A summary of the statistics of the diffraction and refinement is presented in Table 1. The refined model consists of residues from 6 to 593 of one protomer per asymmetric unit.
The Cys154–Cys159 couple is partially reduced as it occurs in Structure 2, probably due to the presence of GSH in the crystallization conditions. We assume that the reversible electron transfer between FADH2 and Cys154–Cys159 occurs also in this structure, in the absence of NADP+. The distance between the sulfur atoms of Cys28 and Cys31 is 3.2 Å, suggesting that these Cys are also reduced. Electron density of the GSH molecule was detected in the proximity of Cys28 (Fig. 3, panel D). The average B factor of the GSH ligand is slightly higher (by ∼5 Å2) than that of surrounding residues. The GSH pocket is composed of three segments of the polypeptide chain, each capable of binding (noncovalently) one of the three residues of GSH. Amino acids from Asp84 to Glu86 contour the γ-glutamyl moiety, by clamping it with two H-bonds. The stretch 70–72 forms two H-bonds with GSH orienting the sulfur atom toward the redox active site of the Grx domain (3.4 Å distance from Cys28 sulfur). Finally, Gln60 and Lys25 bind the glycine moiety of the ligand (Fig. 3, panel D).
Model 1, C Terminus of SmTGRfl onto the Grx Domain
Using Lys586 as pivot residue and after superposition of Structures 3 and 4, we have modeled the possible movement of the C-terminal arm to internally shuttle electrons from FAD to the Grx domain. Without changing the conformation of the extended arm as refined in Structure 3, the last two residues of the C terminus Cys597–Gly598 were placed into the GSH-binding site of the Grx domain (Fig. 3, panels E and F). This was obtained by simple rotation on the ψ angle of Lys586 of subunit B, bringing the terminal arm onto the redox site of the Grx domain of subunit A (Fig. 3, panels E and F). The model clarifies the functional role of the TGR dimer. Indeed, the dimerization is made necessary by the fact that the Grx domain of each subunit is much closer to the C-terminal arm of the opposite subunit (∼20 Å) than to the C-terminal arm of the same subunit (∼65 Å; distances were measured between Cys28 of both subunits and Lys586 of subunit B as found in Structure 3).
Model 2, SmTGRfl-SmTrx Complex
Our interest was to find a docking surface for the external substrate SmTrx. Thus, we carried out a surface complementarity search between the two proteins, followed by automated cycles of restrained refinement to minimize the energy and regularize the geometry, avoiding possible clashes (see “Experimental Procedures”). SmTrx can be docked into SmTGRfl in a surface cleft at the bottom of the TR domain, opposite to the Grx domain (Fig. 3, panels B and C). This cleft, formed by residues belonging to the C terminus of subunit B and to the FAD-binding site of subunit A, has a complementary surface to the stretch from Trp33 to Lys38 of SmTrx, which contains the active site couple Cys34–Cys37. In particular, Trp33 of SmTrx fits into a hydrophobic pocket formed by residues of subunit A (Val155, Ile160, Leu164, and Leu208); Lys38 of SmTrx is engaged in a salt bridge with Asp517 of SmTGRfl subunit B; and Cys34 (the most external of the active site Cys couple) ends up at 2.0 Å distance from Cys596 of SmTGRfl subunit B (Fig. 3, panels B and C). The geometry of this mixed disulfide bond is superimposable to the mixed S–S bond found in the crystallographic complex of E. coli TR-Trx (PDB code 1F6M (27)), supporting the plausibility of this model. Another independent support to this model comes from the structures of Trx from other species, which show that the Trp adjacent to the catalytic site is engaged in hydrophobic contacts upon dimerization (28).
Functional Observations Relevant to the Structural Changes
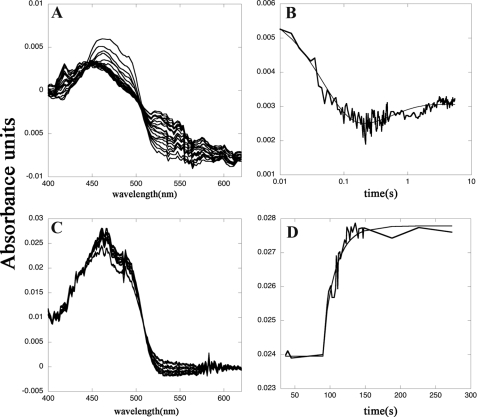
The catalytic cycle of SmTGR is complex, and the exploration of the individual reaction steps is not an easy task, except for the bimolecular reduction and oxidation reactions by substrates. The former reaction was studied by time-resolved spectroscopy, by systematically varying the concentrations of both the enzyme and NADPH. The reaction time courses recorded in this type of experiment are biphasic and span from the spectrum of the oxidized FAD to that of a partially reduced form. Indeed, despite the large difference in the reduction potentials of FAD and NADP+, we never succeeded in obtaining a complete reduction of the enzyme (Fig. 4, panel A). The time courses recorded at two significant wavelengths (460 and 560 nm) for five concentrations of NADPH and two concentrations of SmTGR were fitted to a two-step sequential mechanism using the integrated equation developed by Bateman (29). The wavelengths were selected as representative of the reduced FAD derivatives based on the better characterized homologues, GR and lipoamide dehydrogenase (30, 31). The two rate constants obtained were both second order, with values of 4 × 106 and 1 × 106 m−1 s−1 (Fig. 4, panel B). Absorbance changes at 680 nm, possibly indicative of the transient formation of a charge transfer complex between NADP+ and FADH2 (24), were looked for but not found; the time course at this wavelength could be interpreted in terms of the same two kinetic processes described above, indicating that the charge transfer complex does not accumulate in SmTGRfl.
FIGURE 4.
Reductive and oxidative half-reactions of SmTGRfl, spectrophotometric experiments. Panel A, spectra of 2.5 μm SmTGRfl reduced with 5 μm NADPH recorded in a stopped-flow apparatus. The decrease of the FAD peak at 460 nm and the increase of the absorbance around 550 nm due to the formation of the charge transfer complex are shown. Spectra were recorded every 5 ms from 5 to 500 ms and then every 50 ms up to 5500 ms. Panel B, time course recorded at 460 nm relative to the experiment reported in panel A fitted with a double exponential. The first process is assigned to the reduction of the FAD by NADPH (k = 4 × 106 m−1 s−1) to form the two-electron reduced species EH2; the second process is the perturbation of the cofactor spectrum due to the formation of EH4 (k = 1 × 106 m−1 s−1). These two processes are compatible with electrons transferred from NADPH to Structure 1 and from the reduced Cys154–Cys159 couple (as found in Structure 2) to the C-terminal redox center, allowing the formation of its reduced form as found in Structure 3. Panel C, spectra of reduced SmTGRfl (3.0 μm) oxidized with SmTrx (7 μm) were recorded in a spectrophotometer apparatus. The decrease of absorbance of the charge transfer complex around 550 nm and the increase of the FAD peak at 460 nm is shown. Panel D, time course at 460 nm relative to the experiment reported in panel C fitted to a single exponential. The second-order process is assigned to the oxidation of the FADH2 of SmTGRfl by SmTrx (k = 0.8 × 104 m−1 s−1). This step is compatible with the electron transfer from the reduced C terminus, as found in Structure 3, to SmTrx (see Fig. 1).
Our interpretation is the following. The first reaction step is the transfer of two electrons from NADPH to the primary redox center of the enzyme, constituted by FAD and the couple Cys154–Cys159, rate-limited by complex formation, and hence second order. Because the primary redox center of the enzyme has the ability to accept four electrons, rather than two, a charge transfer state is populated as the product, in which the electrons move back and forth between the FAD and the Cys couple, thus explaining the incomplete reduction of the former. This mixed valence state exchanges electrons with the C-terminal redox couple. The second reaction step again corresponds to the transfer of two electrons from a new molecule of NADPH to those enzyme molecules in which the primary redox center is fully oxidized because of the internal electron transfer, and it is thus second order. It entails little further reduction of the FAD, because it mainly occurs on those enzyme molecules in which the cofactor has been reoxidized by the C terminus. The observed spectroscopic changes (and the pertinent second-order rate constants) are thus assigned to the reaction steps leading from the oxidized enzyme (Structure 1) to the two-electron reduced intermediate (presumably in Structure 2), and from this to the four-electron reduced species (that may assume either Structure 2 or 3; see under “Discussion”). The time-resolved spectra and the overall behavior of the reaction are reminiscent of those recorded by Bauer et al. (32) under similar experimental conditions on TR from Drosophila melanogaster.
Re-oxidation of SmTGR (reduced by stoichiometric amounts of NADPH) by the physiological substrate SmTrx can also be followed by absorbance spectroscopy (Fig. 4, panel C). The U597C mutant used in this study yielded time courses well described by a second-order rate constant of 0.8 × 104 m−1 s−1 (Fig. 4, panel D). This value is approximately 2 orders of magnitude lower than that reported by Kuntz et al. (5) for the wild type enzyme, consistent with the important role played by Sec in the reduction of the physiological substrate. Both the oxidation and reduction half-cycles of SmTGR are rate-limited by second-order kinetic processes, and accumulation of a Michaelis type complex does not occur at the substrate concentrations used.
DISCUSSION
SmTGR is one of the most promising drug targets against Schistosomiasis (33, 34), the second most prevalent parasitic disease after malaria; thus the structural characterization of intermediates in the catalytic cycle is relevant. In this study, we solved several structures of this enzyme in complex with some of its substrates, and it would be highly desirable to assign these structures to the reaction intermediates. A partial description of the enzymatic cycle has been proposed for mouse TGR, mainly based on the better characterized cycle of TRs and on a three-dimensional model of mouse TGR (18, 35). Our structures fit the known or presumed catalytic intermediates of mouse TGR as follows; Structure 1 clearly corresponds to a unique species of SmTGR (the oxidized one). Structures 2 and 4 have been obtained in the presence of excess reductants, thus the species probably prevalent in the crystallization medium was the one called EH4 in TRs, with reducing equivalents stored on the FAD/Cys154–Cys159 site and the C terminus (in SmTGR two more electrons can be stored on the Grx domain, but we shall maintain the usual nicknames for clarity). Structures 2 and 4 are essentially identical to each other except for the presence or the absence of NADP+ and one GSH. Because there is no evidence that the overall structure of SmTGR is affected by the redox state of the disordered C-terminal residues, we believe that Structures 2 and 4 are also compatible with EH2, the two-electron reduced intermediate bearing the reducing equivalents on the FAD/Cys154–Cys159 site. Structure 3 is peculiar having one ordered C terminus per dimer, which is clearly reduced; thus this structure is compatible with EH4 and possibly also with the minor fraction of EH2 in which the electrons have been internally transferred from the FAD/Cys154–Cys159 to the C terminus (see Fig. 1).
Before discussing these assignments in greater detail, we should justify our decision to use a variant of the enzyme in which the C-terminal Sec597 has been replaced by cysteine (SmTGRfl) by the following: (i) the inherent difficulty to obtain suitable amounts of the highly purified Sec-containing wild type enzyme (35–37); (ii) the structure of a Sec-containing form of rat TR, the only reported so far (36), shows the stereochemistry of the entire C-terminal arm to be very similar to that found by us (see below); and (iii) the functional characterization of SmTGRfl, reported in the supplemental material, indicates that the enzyme has fully functional FAD and Grx redox sites and an active C-terminal arm, as demonstrated by the ability to reduce SmTrx. Hence, we believe that the catalytic mechanism we shall reconstruct is plausible and consistent with the Sec-containing wild type enzyme.
Electron Flow within the TR Domain, from NADPH to the C Terminus (Figs. 2 and 3)
The catalytic cycle starts with the transfer of two electrons from NADPH to fully oxidized SmTGR (Structure 1). The product of this reaction is usually called EH2. Kinetic experiments demonstrate that the reduction of FAD follows a bimolecular process with a rate constant k1 = 4 × 106 m−1 s−1 (Fig. 4, panels A and B). The reduction, as judged by the absorption spectra, is not complete because of the reversible internal electron transfer to Cys154–Cys159. The fact that less than 50% of the FAD is reduced agrees very well with the partial reduction of the Cys154–Cys159 couple observed in Structures 2–4.
Structure 2 clearly shows that binding of the reducing substrate to the enzyme occurs as described in previous studies on flavoenzymes from other organisms, where the nicotinamide and the isoalloxazine ring are oriented in a stacked position to favor electron transfer (Fig. 2, panel A) (26, 38). The recognized role of Tyr296 (39), acting as a “gate” for NADPH binding, is confirmed in our structure. In fact, the phenolic ring of Tyr296, which points toward the re-face of the isoalloxazine ring of FAD in a T-shape configuration (see Structure 1 and also Ref. 17 for the oxidized truncated form), swings out parallel to the FAD ring in the “open” conformation, allowing occupancy of the pocket by NADPH (Fig. 2, panel A).
The structure of EH2 is most likely similar or identical to our Structures 2 and 4 depending on whether the intermediate considered contains NADP+ or not. Reduction is associated with a shift of the Cys154 side chain, which moves from a position compatible with the formation of a disulfide bridge with Cys159 (2.0 Å in Structure 1) to a rotamer 3.1 Å distant from Cys159, compatible with the reduced state of the redox couple. The latter rotamer of Cys154 is also stabilized by a water molecule at 2.7 Å from the sulfur atom, in turn kept in place by His571 from the related symmetric subunit (Fig. 2, panel A); this residue is known to be relevant to the enzymatic activity of homologue enzymes (24, 25).
Once the Cys154–Cys159 couple has been reduced, electrons may be picked up by the oxidized C-terminal arm. We do not know whether this occurs before, after, or independently of the binding of the second molecule of NADPH. The reduced state of the C terminus may adopt an ordered conformation, at least in one protomer of the dimer, as it occurs in Structure 3 and in a structure of the Sec-containing TR1 from rat in its reduced state (36) (Fig. 3, panel A).
Even though the exact sequence of events is unknown, EH2 binds to a second molecule of NADPH and is converted to the four-electron reduced enzyme (EH4) in a bimolecular process with the apparent second-order rate constant k2 = 1 × 106 m−1 s−1 (Fig. 4, panel B). EH4, as populated at the end of the second reduction step, presents the reduced C terminus and may therefore adopt Structure 3. In agreement with Bauer et al. (32), Structure 3 is supposed to be ready to transfer reducing equivalents to its substrates, such as Trx and/or the Grx domain.
Electrons Exit from the TR Domain, from the C terminus to Either SmTrx or the Grx Domain (Fig. 3)
Based on Models 1 and 2 and on Structures 3 and 4, we describe in this section the major structural features involved in electron transfer sustaining the TR and Grx activities of the enzyme (see Fig. 1).
The TR activity of TGR follows the mechanism proposed for the mammalian enzymes, a key role being played by the protein C-terminal arm, which is postulated to shuttle electrons from Cys154–Cys159 to the incoming Trx (Fig. 1) (9, 36). Looking at Structure 3, this arm is solvent-exposed and ∼13 Å apart from the isoalloxazine ring, in a position suitable to supply reducing equivalents to SmTrx (Fig. 3, panel A). The C-terminal arm is probably quite mobile in solution, and one can assume that only a further slight movement is sufficient to shuttle electrons from Cys154–Cys159 to Trx.
Electrostatic interactions between the C-terminal arm of one subunit and a positive surface on the symmetric subunit (contributed by Lys124, Lys128, and Arg450, see Fig. 3, panel A) may suggest a mechanism whereby the motion of the former depends on its oxidation state. We presume that the C terminus is contacting the external surface of enzyme when reduced, although it slides back into the FAD-binding site upon oxidation (the net charge of the C terminus dropping from −2 to −1, Sec being completely ionized at physiological pH). Starting from the observation that the C-terminal is flexible, we propose a model (Model 2) for the complex between SmTGRfl and SmTrx (Fig. 3, panels B and C) whereby Trx is positioned in its putative binding site on SmTGR surface (as described previously by Cheng et al. (36) for human TR1). After energy minimization of the docked complex, Cys597 of SmTGRfl is 2.0 Å from Cys34 of SmTrx, thereby suggesting the formation of a mixed disulfide bond. The second-order rate constant for electron transfer from SmTGR to SmTrx is 0.8 × 104 m−1 s−1 (Fig. 4, panel D), compatible with the turnover of the U597C mutant but significantly lower than that estimated for the wild type enzyme (5).
The C-terminal arm of SmTGR is also involved in the reduction of the Grx domain and thus in the Grx activity of the enzyme as demonstrated by experiments on TGR from different organisms (6, 15, 35). In TGR, the Grx domain could be reduced either by GSH in vitro, as common to Grx proteins (8), or by the C terminus through the reducing equivalents supplied by FAD (see Fig. 1) (18). A computer model (35) predicted the Grx domain of TGR to be located in the same position as the Trx-binding site, which was not unreasonable because the Grx proteins belong to the Trx superfamily. However, our structural data strongly argue against this hypothesis. Until now, we crystallized three different variants of SmTGR in four different space groups (this paper and Refs. 13, 17).5 The data show different crystallographic contacts but always the same overall architecture, suggesting the structure in solution and in the crystal is the same. If the relative orientation of the Grx domain with respect to the TR domain is maintained, structural motions compatible with internal electron transfer are those bringing the mobile C terminus onto the Grx redox site. In fact, Structure 4 gives some hints on this movement. Here, GSH is bound to the Grx domain active site, with its –SH at 3.1 Å far from Cys28 of the protein (Fig. 3, panel D). We hypothesize that the C-terminal arm of SmTGR occupies the same site of GSH on the adjacent subunit (Fig. 3, panels E and F). To substantiate this hypothesis, we built a model (Model 1, Fig. 3, panels E and F) as follows: by rotating the C-terminal arm around the Cα–C bond of Lys586 (see “Experimental Procedures”), the Cys597–Gly598 pair of subunit B superimposes to GSH bound to the Grx domain of subunit A (Structure 4).
Consistent with this hypothesis, some structural features can be underlined as follows: (i) from Lys586 onward, the C terminus (as seen in Structure 3) is unstructured and anchored by only a few contacts with the rest of the protein (see “Results” and Table 2); (ii) the last two residues of SmTGR, i.e. Sec-Gly, resemble the Cys-Gly moiety of GSH (except for the selenium atom, Fig. 3, panels E and F); (iii) the distance between Lys586 Cα and Cys597 belonging to subunit B (∼23 Å) is comparable with that between Lys586 Cα and that of the GSH Cys bound onto the Grx domain of subunit A (∼26 Å, Fig. 3, panel E). Thus, a rigid body rotation on the pivot residue Lys586 is sufficient to bring the C terminus onto the GSH-binding site of the Grx belonging to the other domain (Fig. 3, panels E and F). In summary, we suggest that the reduced C-terminal arm is a GSH-like moiety pertaining to the enzyme and acting as a reducing agent, thereby mimicking free GSH.
Concluding Remarks
The data reported in this study describe the complete catalytic cycle of SmTGR, a complex enzyme, based on structural data and modeling. We describe the possible structure of the main intermediate species and assign at least two kinetic constants to the pertinent reduction steps. A crucial question is how the relevant oxidizing substrate, either the Grx domain or the external Trx, is selected; a detailed functional study of wild type SmTGR is required to settle this issue. This study defines some cornerstones on the following points: (i) the external Trx and the Grx domain do not compete for the same binding site; (ii) electrons are selectively delivered to their acceptor by the C-terminal arm shuttling between either of two alternative positions; (iii) it is possible, at least in theory, that TGR may transfer electrons from GSH to Trx (or vice versa) via the Grx domain and the C-terminal arm, thus bypassing NADPH.
The structural and evolutionary complexity of SmTGR suggests that the function of the enzyme is peculiar and that linking the two principal thiol-reducing pathways may offer selective advantages to the schistosome. TGR has been proposed to have evolved from the gene of TR1 (18, 40). Thus, we look at SmTGR as a modified TR, and we observe that most of the biochemical weaponry necessary for the complex and distinctive functions of TGR is already present in TRs, e.g. in both types of reductase the function of the C-terminal arm is that of reaching the redox site of Trx, but only in TGR does this appendage acquire the function of reducing the internal Grx domain.
A recent paper (41) reports the presence of TGR only in parasitic flatworms but not in the free-living species of the same phylum where separated GR and TR exist. A possible explanation for this finding is that the internal reduction of the Grx domain in TGR enzymes could be vital for parasites that spend all their live in the host organism, thus under constant oxidative attack. Under these conditions, sudden fluctuations of the GSH concentration may impair Grx activity and increase the percentage of protein glutathionylation, thus hindering some vital activities of the parasite (8, 42). The peculiar intramolecular electron pathway of TGR, from NADPH to the internal Grx domain through the C terminus, would ensure deglutathionylation of target proteins independently of GSH, hence helping the parasites to circumvent the host oxidative stress.
Assuming the above scenario to be reliable, it is natural to ask why the presence of TGR is associated with the absence of GR in parasitic flatworms? As SmTGR is not an efficient GR (5), one may wonder whether the schistosomes may derive part of their reduced GSH from the erythrocytes they ingest. It has been estimated that schistosome females consume about 330,000 erythrocytes per hour and males about 39,000 (43). Human erythrocytes contain about 3 mm reduced GSH, and thus a female schistosome could in principle obtain GSH from its diet at the remarkable rate of over 60 pmol/h (and should eliminate an equivalent amount of GSSG). It would be interesting to assess this hypothesis by direct measurement, as it may bear implications for therapy.
Supplementary Material
Acknowledgments
We are grateful to Prof. David L. Williams and Dr. Hsin-Hung Huang (Rush University, Chicago) for insightful suggestions and discussions. Use of the European Synchrotron Facilities of BESSYII (Berlin, Germany), ESRF (Grenoble, France), and Elettra (Trieste, Italy) are gratefully acknowledged.
This work was supported by Fondazione Roma (project title “Rational Approach to the Specific Inhibition of Plasmodium falciparum and Schistosoma mansoni”), Sapienza University of Rome Progetto Università 2007 and 2008 and Progetto Ateneo Federato 2007 and 2008, MIUR of Italy FIRB/Proteomica 2007-protRBRN07BMCT, and the European Community Seventh Framework Programme FP7/2007–2013, Agreement 226716.

The on-line version of this article (available at http://www.jbc.org) contains supplemental material and additional references.
The atomic coordinates and structure factors (codes 2X8G, 2X8H, 2X8C, 2X99, 2XBI, and 2XBQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
G. Boumis, F. Angelucci, A. Bellelli, M. Brunori, D. Dimastrogiovanni, and A. E. Miele, manuscript in preparation.
F. Angelucci, D. Dimastrogiovanni, G. Boumis, M. Brunori, A. E. Miele, F. Saccoccia, and A. Bellelli, unpublished results.
- TR
- thioredoxin reductase
- TGR
- thioredoxin glutathione reductase
- SmTGRfl
- S. mansoni TGR full length (U597C mutant)
- SmTrx
- S. mansoni thioredoxin
- GR
- glutathione reductase
- Grx
- glutaredoxin
- PDB
- Protein Data Bank
- Sec
- selenocysteine.
REFERENCES
- 1.Engels D., Chitsulo L., Montresor A., Savioli L. (2002) Acta Trop. 82, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angelucci F., Basso A., Bellelli A., Brunori M., Pica Mattoccia L., Valle C. (2007) Parasitology 134, 1215–1221 [DOI] [PubMed] [Google Scholar]
- 3.Doenhoff M. J., Cioli D., Utzinger J. (2008) Curr. Opin. Infect. Dis. 21, 659–667 [DOI] [PubMed] [Google Scholar]
- 4.Stothard J. R., Chitsulo L., Kristensen T. K., Utzinger J. (2009) Parasitology 136, 1665–1675 [DOI] [PubMed] [Google Scholar]
- 5.Kuntz A. N., Davioud-Charvet E., Sayed A. A., Califf L. L., Dessolin J., Arnér E. S., Williams D. L. (2007) PLoS Med. 4, e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alger H. M., Williams D. L. (2002) Mol. Biochem. Parasitol. 121, 129–139 [DOI] [PubMed] [Google Scholar]
- 7.Winyard P. G., Moody C. J., Jacob C. (2005) Trends Biochem. Sci. 30, 453–461 [DOI] [PubMed] [Google Scholar]
- 8.Johansson C., Lillig C. H., Holmgren A. (2004) J. Biol. Chem. 279, 7537–7543 [DOI] [PubMed] [Google Scholar]
- 9.Fritz-Wolf K., Urig S., Becker K. (2007) J. Mol. Biol. 370, 116–127 [DOI] [PubMed] [Google Scholar]
- 10.Berkholz D. S., Faber H. R., Savvides S. N., Karplus P. A. (2008) J. Mol. Biol. 382, 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnér E. S. (2009) Biochim. Biophys. Acta 1790, 495–526 [DOI] [PubMed] [Google Scholar]
- 12.Su D., Novoselov S. V., Sun Q. A., Moustafa M. E., Zhou Y., Oko R., Hatfield D. L., Gladyshev V. N. (2005) J. Biol. Chem. 280, 26491–26498 [DOI] [PubMed] [Google Scholar]
- 13.Angelucci F., Sayed A. A., Williams D. L., Boumis G., Brunori M., Dimastrogiovanni D., Miele A. E., Pauly F., Bellelli A. (2009) J. Biol. Chem. 284, 28977–28985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rendón J. L., del Arenal I. P., Guevara-Flores A., Uribe A., Plancarte A., Mendoza-Hernández G. (2004) Mol. Biochem. Parasitol. 133, 61–69 [DOI] [PubMed] [Google Scholar]
- 15.Bonilla M., Denicola A., Novoselov S. V., Turanov A. A., Protasio A., Izmendi D., Gladyshev V. N., Salinas G. (2008) J. Biol. Chem. 283, 17898–17907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cioli D., Valle C., Angelucci F., Miele A. E. (2008) Trends Parasitol. 24, 379–382 [DOI] [PubMed] [Google Scholar]
- 17.Angelucci F., Miele A. E., Boumis G., Dimastrogiovanni D., Brunori M., Bellelli A. (2008) Proteins 72, 936–945 [DOI] [PubMed] [Google Scholar]
- 18.Sun Q. A., Su D., Novoselov S. V., Carlson B. A., Hatfield D. L., Gladyshev V. N. (2005) Biochemistry 44, 14528–14537 [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 21.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 22.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 23.Summa C. M., Levitt M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3177–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H. H., Arscott L. D., Ballou D. P., Williams C. H., Jr. (2008) Biochemistry 47, 1721–1731 [DOI] [PubMed] [Google Scholar]
- 25.Huang H. H., Arscott L. D., Ballou D. P., Williams C. H. (2008) Biochemistry 47, 12769–12776 [DOI] [PubMed] [Google Scholar]
- 26.Sandalova T., Zhong L., Lindqvist Y., Holmgren A., Schneider G. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9533–9538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lennon B. W., Williams C. H., Jr., Ludwig M. L. (2000) Science 289, 1190–1194 [DOI] [PubMed] [Google Scholar]
- 28.Wahl M. C., Irmler A., Hecker B., Schirmer R. H., Becker K. (2005) J. Mol. Biol. 345, 1119–1130 [DOI] [PubMed] [Google Scholar]
- 29.Bateman H. (1910) Proc. Cambridge Phil. Soc. 15, 423–427 [Google Scholar]
- 30.Huber P. W., Brandt K. J. (1980) Biochemistry 19, 4568–4575 [DOI] [PubMed] [Google Scholar]
- 31.Argyrou A., Blanchard J. S., Palfey B. A. (2002) Biochemistry 41, 14580–14590 [DOI] [PubMed] [Google Scholar]
- 32.Bauer H., Massey V., Arscott L. D., Schirmer R. H., Ballou D. P., Williams C. H., Jr. (2003) J. Biol. Chem. 278, 33020–33028 [DOI] [PubMed] [Google Scholar]
- 33.Sayed A. A., Simeonov A., Thomas C. J., Inglese J., Austin C. P., Williams D. L. (2008) Nat. Med. 14, 407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rai G., Sayed A. A., Lea W. A., Luecke H. F., Chakrapani H., Prast-Nielsen S., Jadhav A., Leister W., Shen M., Inglese J., Austin C. P., Keefer L., Arnér E. S., Simeonov A., Maloney D. J., Williams D. L., Thomas C. J. (2009) J. Med. Chem. 52, 6474–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Q. A., Kirnarsky L., Sherman S., Gladyshev V. N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3673–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng Q., Sandalova T., Lindqvist Y., Arnér E. S. (2009) J. Biol. Chem. 284, 3998–4008 [DOI] [PubMed] [Google Scholar]
- 37.Hondal R. J. (2009) Biochim. Biophys. Acta 1790, 1501–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Argyrou A., Blanchard J. S. (2004) Prog. Nucleic Acids Res. Mol. Biol. 78, 89–142 [DOI] [PubMed] [Google Scholar]
- 39.Biterova E. I., Turanov A. A., Gladyshev V. N., Barycki J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novoselov S. V., Gladyshev V. N. (2003) Protein Sci. 12, 372–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otero L., Bonilla M., Protasio A. V., Fernandez C., Gladyshev V. N., Salinas G. (2010) BMC Genomics 11, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grant C. M. (2001) Mol. Microbiol. 39, 533–541 [DOI] [PubMed] [Google Scholar]
- 43.Basch P. F. (1984) Am. J. Trop. Med. Hyg. 33, 911–917 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.