Abstract
SUMO conjugation of cellular proteins is essential for proper progression of mitosis. PIASy, a SUMO E3 ligase, is required for mitotic SUMOylation of chromosomal proteins, yet the regulatory mechanism behind the PIASy-dependent SUMOylation during mitosis has not been determined. Using a series of truncated PIASy proteins, we have found that the N terminus of PIASy is not required for SUMO modification in vitro but is essential for mitotic SUMOylation in Xenopus egg extracts. We demonstrate that swapping the N terminus of PIASy protein with the corresponding region of other PIAS family members abolishes chromosomal binding and mitotic SUMOylation. We further show that the N-terminal domain of PIASy is sufficient for centromeric localization. We identified that the N-terminal domain of PIASy interacts with the Rod/Zw10 complex, and immunofluorescence further reveals that PIASy colocalizes with Rod/Zw10 in the centromeric region. We show that the Rod/Zw10 complex interacts with the first 47 residues of PIASy which were particularly important for mitotic SUMOylation. Finally, we show that depletion of Rod compromises the centromeric localization of PIASy and SUMO2/3 in mitosis. Together, we demonstrate a fundamental mechanism of PIASy to localize in the centromeric region of chromosome to execute centromeric SUMOylation during mitosis.
Keywords: Centromeres, Chromosomes, Mitosis, Sumoylation, Xenopus, Kinetochore, PIAS
Introduction
SUMOylation is a protein modification process conserved from yeast to vertebrates (1). The consequences of SUMO2 (small ubiquitin-like modifier) modification that have been elucidated over the past decade include modulation of gene transcription, DNA repair, protein translocation, protein/protein interaction, chromosomal organization, and sister chromatid segregation (2–4). Vertebrates have three SUMO isoforms and all three display roughly 50% identity with the single SUMO found in yeast (1). SUMO is conjugated to cellular substrates by an analogous pathway to that of ubiquitin. It has been reported that SUMOylation is mediated without E3 ligases in vitro (5, 6), but under physiological conditions, SUMO E3 ligases are essential to execute SUMOylation of cellular substrates (7–10). There are mainly two types of SUMO E3 ligases in vertebrates, RanBP2 (Nup358), and Siz/PIAS. RanBP2 has no homolog in yeast, and its ligase function is independent of either HECT or Ring finger-type ubiquitin E3 ligases (11). Siz/PIAS, on the other hand, initially identified in budding yeast, functions similar to Ring finger ubiquitin ligases (8). Vertebrates have four PIAS proteins (PIAS1, PIAS3, PIASx, and PIASy) that share important conserved functional domains (12). The SAP (scaffold attachment factor-A/B, acinus and PIAS) domain is positioned at the N terminus, and directly binds AT-rich regions of DNA (13–15). The SP-Ring domain is related to that of ubiquitin E3 ligase and is responsible for Ubc9 recruitment (16). The SUMO-interacting motif (SIM) is situated after the SP-Ring and redirects the Ubc9∼SUMO complex on substrate proteins, potentially contributing to SUMO paralogue specificity (17, 18). Domain analysis of Siz protein in vitro and in vivo suggests each domain contributes to SUMOylation with distinct functions. The N-terminal domain of Siz1 is involved in substrate recognition and the C-terminal domain in cell-cycle-dependent localization, which is critical for septin SUMOylation (19). Whether vertebrate PIAS proteins are organized in a similar manner is unknown.
Among PIAS family members, we have identified PIASy as crucial for SUMO2/3 modification of chromosome-associated proteins in mitosis (7). For example, DNA topoisomerase IIα (TopoIIα) and poly [ADP-ribose] polymerase I (PARP1) are each modified by SUMO2/3 in a PIASy-dependent manner during mitosis (9, 20). Immunodepletion of PIASy completely abolishes mitotic chromosomal SUMOylation in Xenopus egg extracts (XEE) and other PIAS family proteins fail to restore this defect, indicating a unique role of PIASy in mitotic chromosomal SUMOylation in XEE (7). Our initial domain analysis with mutated PIASy suggested that the N-terminal domain of PIASy is required for its association with mitotic chromosomes (7). The SUMO2/3 modification of chromosomal proteins is restricted not only to the early stages of mitosis but also to the centromeric regions, raising the question of how PIASy regulates mitotic SUMOylation in a temporal and regional manner (7, 9). Recent immunostaining has elucidated that PIASy is exclusively localized to centromeric regions and colocalizes with TopoIIα during mitosis, suggesting that the centromeric localization of PIASy is critical for the spatiotemporal regulation of mitotic SUMOylation.3 However, the molecular mechanism underlying this localization has remained unidentified.
Centromeres are specified regions of DNA where kinetochores are assembled to capture growing microtubules from spindle poles in mitosis (21). Kinetochores include multiple proteins whose functions are involved in mitotic checkpoints directly or indirectly, and each component is absolutely required for the accurate progression of the cell cycle including proper chromosome segregation (22). Rod (Rough deal) and Zw10 (Zeste white 10) are kinetochore proteins in higher eukaryotes (23). As a stable complex called RZZ (Rod/Zw10/Zwilch), Rod and Zw10 are localized to the kinetochore until anaphase commences (24). Rod and Zw10 are involved in mitotic checkpoint by recruiting Mad1/Mad2 and dynein/dynactin onto unattached kinetochores (23, 25, 26). Mutation of Rod, Zw10 or both Rod and Zw10 result in improper chromosome alignment and sister chromatid missegregation in Drosophila (27–29). Somatic mutations in Rod and Zw10 genes have been found in human colorectal cancers, implicating the complex in the progression of cancer (30).
To determine how PIASy distinctively executes mitotic SUMOylation in a spatiotemporal manner, we have identified the mechanism of PIASy recruitment onto mitotic chromosomes. Using a series of purified PIASy truncations, we found that the N-terminal region of PIASy is vital for chromosome localization and for consequent mitotic SUMOylation but is dispensable for the catalytic activity in vitro. Additionally, we demonstrated the ability of a PIASy N-terminal peptide to interact with chromosomes. Chimeric PIASy containing the N-terminal domain of other PIAS family proteins is defective in chromosome interaction. A PIASy N-terminal peptide fused with mCherry is localized to the centromeric region. Immunofluorescence microscopy combined with biochemical analysis and mass spectrometry revealed that Rod and Zw10 are unique binding proteins of PIASy N terminus among PIAS family proteins. Finally, we show that depletion of Rod proteins from XEE causes mis-localization of PIASy as well as loss of SUMO2/3 foci on the chromosomes. Taken together, these data reveal that PIASy is recruited onto the centromeric region of chromosomes through interaction of the PIASy N terminus with the Rod/Zw10 kinetochore complex, and this results in specific SUMOylation of the centromeric region.
EXPERIMENTAL PROCEDURES
Plasmids Construction
The C-terminal portion, encoding amino acids 970∼1060, of Rod DNA was cloned from Xenopus laevis tadpole cDNA (kindly provided by Drs. A. Arnaoutov and M. Dasso) using PCR amplification. For recombinant protein production, the C-terminal fragment of Rod cDNA was subcloned into pET28a and pGEX4T-1 with BamHI/XhoI restriction enzyme sites. PIASy truncations, shown in Figs. 1 and 5 were produced by PCR and subcloned into the pET28a vector using EcoRI/XhoI restriction sites. N-terminal fragments of PIAS family DNAs were amplified from original full-length cDNA of each PIAS gene (7) with additional EcoRI sites at the 5′-end and HinDIII sites at 3′-end, respectively, and were subcloned into the pET30a vector for recombinant protein production. For N-terminal-swapped chimeric constructs of PIASy, a DNA fragment of X. laevis PIASy without the N-terminal domain (amino acids 142∼501) was amplified by PCR creating HinDIII site at the 5′-end and XhoI site at 3′-end, respectively, and subcloned into pET28a. Subsequently, the N-terminal fragments of each PIAS equivalent of 1∼141 amino acids of PIASy was ligated to the PIASy C-terminal construct. In the case of the C-terminal PIAS-swapped chimeric constructs (y-1, y-3, y-x and y-yhs in Fig. 2), C-terminal fragment of PIAS DNAs equivalent to 410–501 amino acid of PIASy were amplified with additional BglII site at the 5′-end and XhoI sites at 3′-end, respectively. A DNA fragment of X. laevis PIASy lacking the C-terminal domain (1- 410 amino acids) was obtained by digesting full-length PIASy in pET28a with EcoRI and BamHI. The C-terminal fragments of PIAS proteins and the PIASy (1–410) fragment were ligated into pET28a EcoRI/XhoI sites. For mCherry fusion of N-terminal fragments of PIASx and PIASy, PCR-amplified mCherry cDNA (kindly provided by Dr. B. Oakley) with NotI and XhoI sites at 5′- and 3′-ends, respectively, was inserted into pET30a N-terminal PIAS constructs described above. cDNA of TopoIIα was subcloned into a pPIC 3.5K vector in which CBP-T7 Tag sequences were inserted. (The CBP and ZZ TAP tag plasmid was kindly provided from Dr. K. Gould.) All constructs were verified by DNA sequencing.
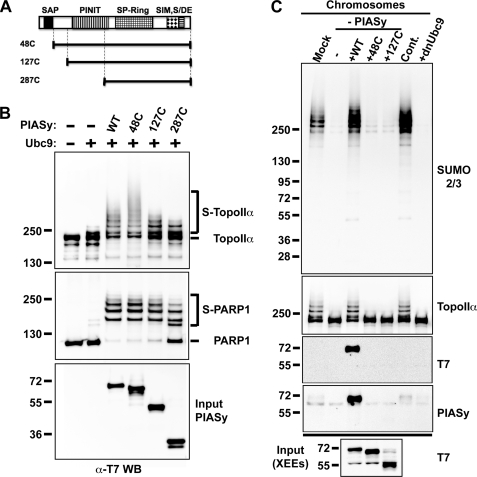
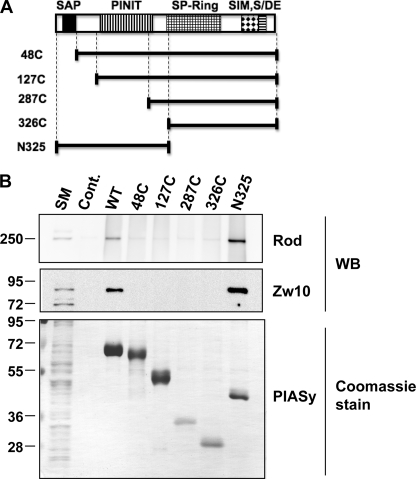
FIGURE 1.
N terminus of PIASy is dispensable in vitro but essential for mitotic SUMOylation in Xenopus egg extract assays. A, schematic diagram of PIASy truncations used in experiments in B and C. N-terminal deletions of PIASy are drawn to approximate scale and compared with full-length PIASy shown on the top. All truncation constructs and full-length PIASy contain a T7 tag at the N terminus. Conserved domains among PIAS family are indicated as follows: SAP in solid black, PINIT in vertical lines, SP-Ring in grid, SIM in diamond, and S/DE motif in horizontal lines. B, in vitro SUMOylation assay using N-terminal-truncated PIASy series. All constructs were expressed and purified as described under “Experimental Procedures.” Either TopoIIα or PARP1 was incubated with PIASy and Ubc9 as indicated in the presence of E1 and ATP for 1 h at 25 °C, and samples were analyzed by Western blotting with anti-T7 tag antibody, which detects recombinant TopoIIα and PARP1. SUMOylated and unmodified forms of proteins are specified by bracket and bar, respectively. The PIASy input is shown in the bottom panel. C, PIASy immunodepletion and addback experiments in Xenopus egg extracts (XEE). CSF-XEE was immunodepleted using IgG (Mock) or anti-PIASy antibody. Purified PIASy truncation proteins were added to the depleted XEE and 5,000 sperm chromatin/μl was incubated in the reactions at 25 °C for ∼1 h. The reactions without (Cont.) and with dnUbc9 were also prepared for positive and negative control of chromosomal SUMOylation. Isolated chromosome fractions were resolved on SDS-PAGE gel and analyzed by Western blot for indicated proteins. The input of PIASy proteins were analyzed in the bottom panel.
FIGURE 5.
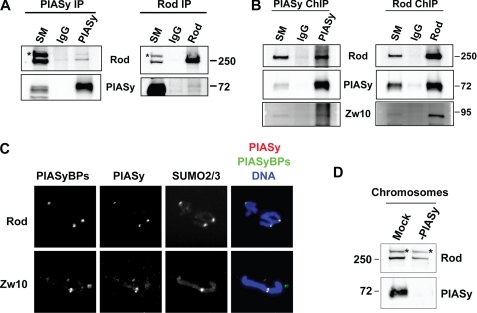
PIASy interacts with Rod/Zw10 complex on mitotic chromosomes. A, interaction of Rod and Zw10 proteins with endogenous PIASy in CSF-XEE. Left, IgG or anti-PIASy antibody cross-linked protein A beads were incubated with CSF-XEE. Immunoprecipitated proteins were resolved in SDS-PAGE and analyzed by Western blot for indicated proteins. Right, IgG or anti-Rod antibody cross-linked protein A beads were incubated in CSF-XEE in the same manner. The precipitants were resolved by SDS-PAGE followed by Western blot. SM stands for starting material. The band labeled with asterisk is a nonspecific protein cross-reacting with anti-Rod antibody in CSF-XEE. IP, immunoprecipitation. B, interaction of PIASy and Rod/Zw10 complex on chromosomes. Mitotic chromosomes prepared in CSF-XEE were subjected to chromatin immunoprecipitation as described under “Experimental Procedures,” and precipitated samples were analyzed as in A. C, localization of endogenous PIASy and binding proteins, Rod and Zw10. Replicated mitotic chromosome samples were prepared as in Fig. 3 followed by immunostaining for the indicated proteins. PIASyBPs, PIASy-binding proteins. D, depletion of PIASy does not eliminate the interaction of Rod/Zw10 complex to mitotic chromosomes. Mock-depleted by IgG (Mock) or PIASy-depleted CSF-XEE (-PIASy) were incubated in the presence of 5,000 sperm nuclei/μl for 1 h at 25 °C. Isolated chromosomes were resolved by SDS-PAGE and analyzed by Western blot. The antibody cross-reacting band is indicated with asterisk.
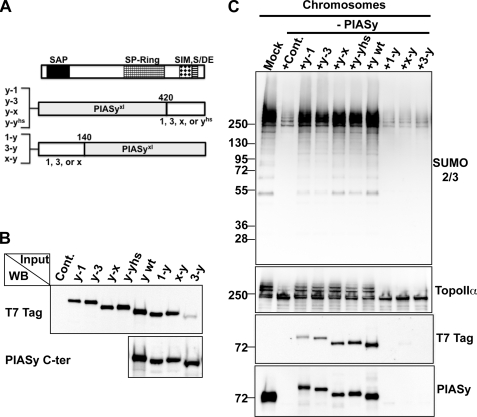
FIGURE 2.
Comparison of function of PIAS N-terminal domains in the XEE assay. A, schematic diagram of PIAS chimera proteins. The chimeric PIAS proteins with T7 tag at the N terminus were obtained by swapping conserved domains between PIAS family members as described under “Experimental Procedures.” The diagram indicates the positions of swapped domain between PIAS family proteins. 1, PIAS1; 3, PIAS3; x, PIASxα; yhs, human PIASy; and y, Xenopus PIASy. B, expression of the chimeric PIAS proteins in reticulocyte lysates. The chimera proteins constructed in A were expressed in rabbit reticulocyte lysates, and the expression level was analyzed by Western blot for T7 tag or anti-C terminus of PIASy antibody. The reticulocyte lysate containing the empty vector was prepared for the negative control (Cont.). C, immunodepletion and chimeric PIAS addback experiments in XEEs. CSF-XEE was immunodepleted by IgG (Mock) or anti-PIASy antibody. Reticulocyte lysates shown in B were added to the PIASy-depleted CSF extracts, and the extracts were incubated in the presence of 5,000 sperm chromatin/μl for ∼1 h at 25 °C. Isolated chromosomes were analyzed by Western blot for indicated proteins.
Recombinant Protein Expression and Purification and Preparation of Antibodies
For preparation of recombinant TopoIIα proteins, the plasmids were transformed into a GS115 strain of Pichia pastoris yeast and expressed according to the manufacturer's instructions (Invitrogen). Protein purification with CBP was performed according to TAP protocol (EMBL Heidelberg) with a slight modification of our needs. Briefly, frozen TopoIIα expressed in yeast cells were grinded with dried ice in a coffee mill and then mixed with lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm CaCl2, 1 mm MgCl2, 0.1% Triton X-100, 5% glycerol, 1mm DTT, and 10 mm PMSF). After centrifugation at 25,000 × g for 40 min, the supernatant was mixed with calmodulin-Sepharose resin (GE Healthcare) for 90 min at 4 °C to capture CBP-tagged TopoIIα. The resin was washed with lysis buffer, and TopoIIα was eluted with buffer containing 10 mm EGTA. The elution was further purified by Mono-Q anion-exchange chromatography (GE-Healthcare).
For preparation of the antigen for anti-Rod antibody, Both GST-fused and His6-tagged C-terminal fragment of Rod was expressed in BL21 (DE3) and His6-tagged C-terminal Rod polypeptide was purified from inclusion bodies under denaturing conditions (6 m urea) following the manufacturer's protocol (Clontech). GST fused C-terminal Rod polypeptide was solubilized by urea and directly conjugated to NHS-Sepharose beads. All N-terminal fragments of PIAS family and mCherry fusion proteins were expressed in BL21 (DE3) or Rossetta2 (DE3) (EMD Biosciences) and purified with Talone metal affinity resin (Clonthech) followed by ion-exchange chromatography. Preparation of E1 complex (Aos1/Uba2 heterodimer) (31), PIASy (7), Ubc9, and SUMO2 (32) were previously described.
Polyclonal antibodies against TopoIIα C terminus (1358–1579) were prepared in rabbits by injection with recombinant His-T7-fused fragments, and affinity-purified with antigens as described previously (20). Anti-PARP1, Anti-PIASy, and anti-SUMO2/3 antibodies used this study were reported previously (7, 9). Anti-Aurora B antibody was kindly provided by A. Arnaoutov and M. Dasso. Anti-Zw10 monoclonal antibody was purchased from Abcam. HRP-conjugated anti-T7 tag antibody and S protein were purchased from EMD Biosciences.
In Vitro SUMOylation Assay
The reactions were performed as previously described (9). In brief, the reaction contained 15 nm E1 Uba2/Aos1 heterodimer, 40 nm E2 Ubc9, 20 nm PIASy, 5 μm SUMO2-GG, 500 nm T7-tagged TopoIIα or PARP1, and 2.5 mm ATP. Reaction buffer consisted of 20 mm HEPES (pH7.8), 100 mm NaCl, 5 mm MgCl2, 0.05% Tween 20, 5% glycerol, 1 mm AEBSF, and 1 mm DTT. The reactions were incubated at 25 °C for 1 h and stopped by addition of one-half volume of 3× SDS-PAGE sample buffer. Samples were resolved on 8–16% Tris-HCl gradient gels (Invitrogen) by SDS-PAGE, and analyzed by Western blot with HRP-conjugated anti-T7 monoclonal antibody (EMD Biosciences) or other antibodies as indicated.
Xenopus Egg Extracts Assays, Pull-down, and Immunoprecipitation
Xenopus sperm chromatin and low speed cytostatic factor (CSF)-arrested XEE (CSF-XEE) or interphase XEE were prepared according to methods described by Kornbluth et al. (33) with slight modification (34). In order to obtain mitotic chromosomes for Western blot analysis, 5000 sperm chromatin/μl was incubated with freshly prepared CSF-XEE for 50 min at room temperature. Chromosomes were isolated as previously described (34).
For pull-down assays from XEE, S-protein-agarose beads (Novagen) or T7 antibody-conjugated-agarose beads (Novagen) were incubated with S-tagged PIAS N-terminal proteins or T7-tagged PIASy truncations, respectively, as specified in Figs. 4 and 6 overnight at 4 °C. Next day, protein-bound beads or non-protein-bound beads were blocked with 5% gelatin for 1 h. CSF extracts were diluted with two volumes of IP extract buffer (20 mm NaPi pH 7.8, 18 mm β-glycerol phosphate, pH 7.5, 5 mm MgCl2, 50 mm NaCl, 5% glycerol) followed by centrifugation at 25,000 × g for 20 min at 4 °C. Supernatants were mixed with an equal volume of ChIP buffer (20 mm NaPi, 18 mm β-glycerol phosphate, 50 mm NaCl, 5 mm MgCl2, 5% glycerol, 0.2% Tween 20, and 0.2% Triton X-100) and incubated with protein-bound or non-protein-bound beads for 2 h at 25 °C. Collected beads were washed thoroughly with ChIP buffer and PBS-T, and eluted in 1× SDS-PAGE buffer. Samples were, then, resolved in 8–16% gradient gels (Invitrogen) by SDS-PAGE, and analyzed by Western blot for the proteins indicated in Figs. 4–6.
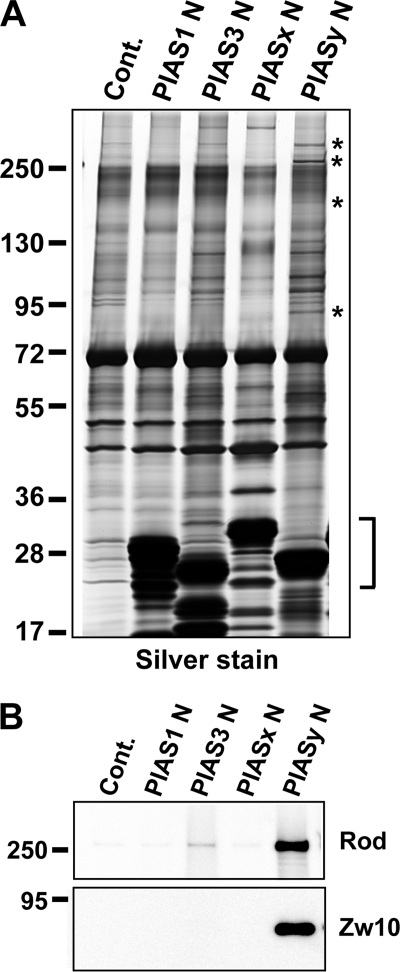
FIGURE 4.
Identification of N-terminal-binding proteins of PIASy. A, N terminus of PIASy has distinctive binding proteins. S-tagged N-terminal domains of PIAS family proteins were bound to S-agarose beads. Either S-agarose beads (Cont.) or PIAS N-terminal protein-bound bead preparations were incubated in CSF XEE. Precipitated proteins together with beads were resolved by SDS-PAGE followed by silver staining. Specific binding proteins for PIASy N-terminal domain are indicated with an asterisk. S-tagged PIAS N-terminal peptides used as bait are indicated with a bracket. B, confirmation of Rod and Zw10 binding to PIASy N terminus. Pull-down samples as in A were analyzed by Western blot for Rod and Zw10. The antibody against Rod, prepared as described under “Experimental Procedures,” or commercially available anti-Zw10 antibody (AbCam) were used to detect them in pull-down samples.
FIGURE 6.
Rod/Zw10 complex interacts with the first 47 residues of PIASy protein. A, schematic diagram of PIASy and its truncations used in B. PIASy and its truncations contain T7 tag at the N terminus. B, further identification of Rod/Zw10-interacting area on PIASy protein. Purified T7-tagged PIASy truncation proteins as well as full-length (WT) were bound to T7 antibody-conjugated agarose beads. Protein-bound beads or beads alone (Cont.) were incubated with CSF-XEE. The precipitated proteins together with the T7-antibody beads were resolved by SDS-PAGE and analyzed by Western blot for the indicated proteins or Coomassie Blue staining. SM, starting material.
Immunoprecipitation experiments were performed in a similar way to that described above. Briefly, protein A Dyna beads (Invitrogen) were incubated with either IgG (Cont.) or antibody against PIASy C terminus or Rod overnight at 4 °C according to the manufacturer's manual. Antibody-bound protein A beads were washed with PBS-T and blocked by 5% gelatin. CSF-XEE prepared as in the pull-down assay was incubated with the antibody-bound beads for 2 h at 25 °C. Collected beads were washed with ChIP buffer and PBS-T, and eluted in 1× SDS-PAGE sample buffer. Samples were resolved in 8–16% gradient gels (Invitrogen) by SDS-PAGE, and analyzed by Western blot for the proteins in Fig. 5A. For chromatin immunoprecipitation from mitotic chromosomes prepared in XEE in Fig. 5B, isolated mitotic chromosomes were subjected to sonication (30% Duty, 1.5 output) followed by micrococcal nuclease digestion with 1000 unit for 15–20 min at 37 °C following the manufacturer's protocol (Abcam). Digested chromosomal suspensions were subjected to immunoprecipitation with either anti-PIASy or anti-Rod antibodies immobilized on the protein A Dyna beads as above.
Immunodepletion was performed in a similar way to that previously described (7). Briefly, antibody-bound protein-A magnetic beads as above were blocked with CSF-XB buffer containing 5% BSA followed by mixing with freshly prepared CSF-XEE and depleted specified proteins described in the figures. For addback experiments, recombinant PIASy proteins or reticulocyte lysates expressing chimeric PIAS proteins were added to the PIASy-depleted CSF-XEE at a final concentration of 50 nm or 10% of the final reaction volume, respectively.
Immunofluorescence Analysis of Chromosomes
Immunofluorescence experiments were performed as previously described (7, 9). In brief, CSF-XEE was induced to undergo interphase by the addition of 0.6 mm CaCl2. 500 sperm/μl were incubated to replicate chromatins for ∼1 h at 25 °C. After checking the morphology of interphase nuclei, subsequently, mitosis was re-induced by the addition of one-half volume of fresh CSF-XEE. For Fig. 3, mCherry-fused PIAS N-terminal proteins were supplemented immediately before inducing mitosis at a final concentration of 50 nm. After additional incubation for 40 min at 25 °C, reactions were diluted three times with dilution buffer, and chromosomes were fixed by addition of an equal volume of 4% p-formaldehyde in dilution buffer. Samples were incubated for 5 min followed by spinning onto coverslips through a 35% glycerol cushion. After post-fixation with 1.6% paraformaldehyde, chromosome samples were subjected to immunostaining with indicated antibodies.
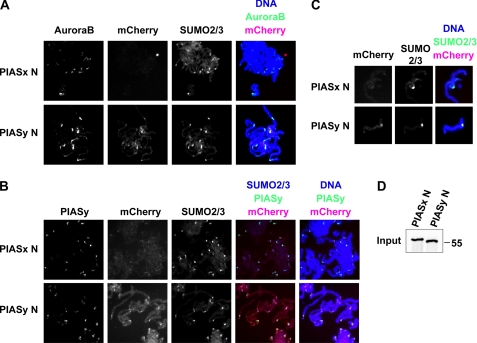
FIGURE 3.
N-terminal region of PIASy locates to centromere independent of its remaining residues. A and B, localization of mCherry-tagged N-terminal peptides of PIASx and PIASy. Replicated chromosomes were obtained by incubating 500 sperm nuclei/μl in interphase extract followed by re-entry into mitosis by CSF-XEE. N-terminal peptides of PIASx (PIASx N) or PIASy (PIASy N) that have mCherry fused at the C terminus were added before re-entry into mitosis. After re-induction into mitosis by fresh CSF-XEE, chromosomes were spun down on coverslips by centrifugation. Samples were immunostained for indicated proteins. C, centromeric localization of mCherry-fused N-terminal peptide of PIASy. Single sister chromatids are enlarged for the comparison of localization between PIASx N and PIASy N. D, input of PIASx N and PIASy N. The amount of PIASx N and PIASy N input was analyzed by Western blot with S-protein HRP (EMD Bioscience), which detects the S tag at the N terminus on both PIASx N and PIASy N peptides.
For immunofluorescence study with immunodepleted XEE (Fig. 7), anti-Rod antibody was cross-linked to protein A-Dyna beads with DMP (dimethyl pimelimidate·2HCl) according to company's instructions (Pierce), then CSF-XEE was depleted with the cross-linked anti-Rod antibody-bound bead as described above. Immunofluorescence experiments were performed as described above. Anti-rabbit Alexa 568, anti-guinea pig Alexa 684, anti-mouse Alexa 488, and anti-chicken Alexa 488 (Invitrogen) were used as secondary antibodies to visualize primary antibodies and DNA was visualized with Hoechst 33342 (1 μg/ml). Specimens were observed with Nikon TE2000-U microscope equipped with Plan Apo 100×/1.40 objective. Images were taken with a Retiga SRV CCD camera (Qimageing) operated by Volocity software (Improvision).
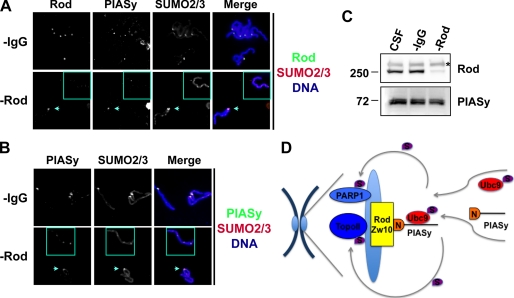
FIGURE 7.
Rod/Zw10 complex is required for localization of PIASy on the centromeric region as well as SUMO2/3 during mitosis. A and B, immunodepletion of Rod causes mislocalization of PIASy on mitotic chromosomes. A, CSF-XEE was immunodepleted for Rod and released into interphase by the addition of CaCl2. Sperm chromatin was incubated with the interphase extracts, and sister chromatids were obtained from the replicated chromatin by the addition of Rod-depleted CSF-XEE (−Rod). CSF-XEE was also mock-depleted using nonspecific IgG and processed as above (−IgG). Isolated chromosome samples were subjected to immunostaining for proteins as indicated. Affinity-purified anti-PIASy chicken IgY and anti-SUMO2/3 guinea pig IgG were used to visualize PIASy and SUMO2/3, respectively. B, samples in A were immunostained with affinity purified anti-PIASy antibody obtained from rabbit. The chromosome lacking Rod protein is shown in squares. The chromosome that still contains Rod in Rod-depleted samples is indicated with arrows. The centromere-specific localization of both PIASy and SUMO modification was compromised if Rod was completely eliminated. C, confirmation of Rod depletion. CSF-XEE before and after immunodepletion with anti-IgG or Rod antibody were analyzed for the indicated proteins. Asterisk shows background signals from antibody cross-reacting in CSF extracts. D, model for molecular mechanism of PIASy-dependent centromeric SUMOylation during mitosis. PIASy interacts with the Rod/Zw10 complex at the kinetochore through its N-terminal region (designated as N). Consequently, Ubc9-SUMO2/3 adduct is recruited onto centromeric regions via binding to PIASy and facilitates SUMO2/3 modification of substrates such as TopoIIα and PARP1 in a spatially regulated manner.
RESULTS
The N Terminus of PIASy Is Dispensable for in Vitro SUMOylation but Crucial for Mitotic SUMOylation in Xenopus Egg Extracts
We have previously shown that PIASy is required for mitotic SUMOylation (7). Because mitotic SUMOylation is not fully restored by other PIAS proteins in PIASy-depleted XEE, it is clear that PIASy has a distinct role as an E3 ligase during mitosis among PIAS family members (7). Domain analysis further suggested that this specificity is likely due to the unique ability of the N-terminal domain of PIASy to mediate mitotic chromosome binding (7). However, how the N-terminal domain facilitates the chromosome interaction of PIASy was not determined. One of our hypotheses was that PIASy might locate to chromosomes by the interaction of its N terminus with chromosomal substrates given that major substrates of PIASy, TopoIIα, and PARP1, are chromosome-resident proteins (9, 20). To address this question, we sought to examine whether N-terminal deletions of PIASy decrease SUMOylation activity in reconstituted SUMOylation assay in vitro. We constructed systematic truncations that lack a series of N-terminal region of PIASy (Fig. 1A). Purified truncated proteins were applied to in vitro reactions for SUMO2 modification of TopoIIα or PARP1. We observed that PIASy displayed robust activity to SUMOylate each substrate until deleted for amino acids 1–127 (127C) and the activity was reduced by further truncation (287C), indicating that the interaction between PIASy and each substrate was not dependent on the N-terminal 127 amino acids of PIASy but on the PINIT domain (Fig. 1B). This result also indicates that the N-terminal 127 amino acids, including the SAP domain, are dispensable for PIASy activity in vitro. Interestingly, PIASy that lacks the first 47 amino acids reproducibly exhibited stronger SUMOylation activity than full-length PIASy on TopoIIα but not on PARP1 (Fig. 1B). Further investigation is required for a clear explanation of this phenomenon.
The function of the N-terminal domain of PIASy under physiological conditions was examined in a XEE cell free assay system. Cytostatic factor-arrested XEE (CSF-XEE) was immunodepleted for PIASy and recombinant truncated PIASy proteins were used to supplement the extract in the presence of sperm chromatin. Elimination of over 95% of PIASy resulted in near abolition of mitotic SUMOylation, and the addition of recombinant PIASy wild type fully restored the SUMOylation (Fig. 1C, lanes 1, 2, 3, and 6). Consistent with our previous studies using reticulocyte lysate (7), truncated PIASy that lacks either 47 (48C) or 127 (127C) amino acids from the N terminus completely failed to re-establish SUMOylation of mitotic chromosomal proteins (Fig. 1C, lanes 4 and 5). Only full-length PIASy but not PIASy with N-terminal truncations was able to interact with chromosomes (Fig. 1C, T7 and PIASy Western blot), verifying that the N terminus is involved in the recruitment of PIASy onto mitotic chromosomes. Together, these data indicate that N-terminal domain of PIASy is not engaged in the interaction with its substrates and not important for SUMOylation in vitro, but facilitates chromosomal interaction and is essential for mitotic SUMOylation.
Functional Comparison of PIAS N-terminal Domain in XEE Assay
PIAS family proteins are categorized by several conserved domains: a SAP domain resides at the N terminus, a SP-Ring (Siz/PIAS-Ring) domain locates in the middle, followed by a SIM (SUMO-interacting motif) (12). The SAP domains of mammalian PIAS family members (PIAS1, -3, -xα/β, and -y) are ∼60% identical and ∼90% similar in amino acid sequence. The SAP domain has been determined to bind scaffold or matrix attachment DNA (13, 15) and, in fact, all PIAS proteins have been shown to bind interphase chromatin (7). Yet, only PIASy and a minor fraction of PIASx are recruited to chromosomes during mitosis in XEE assays, suggesting there are unique features of the PIASy N terminus that direct mitotic chromosome binding. To test this hypothesis, we constructed chimeras of PIAS family proteins. The first group of constructs was composed of SAP and SP-Ring domains from X. laevis PIASy and the C-terminal SIM, S/DE domains from other PIAS proteins (y-1, y-3, y-x, and y-yhs). The second group of constructs contained the N-terminal ∼140 amino acids of either PIAS1, -3, or -x protein linked with the remaining domains of X. laevis PIASy (1-y, 3-y and x-y) (Fig. 2A). The chimera constructs were transcribed and translated in rabbit recticulocyte lysates (Promega), and their expression levels were verified by Western blot with anti-T7 tag and anti-PIASy antibodies. The PIAS3-PIASy chimeric protein (3-y) was barely detectable with anti-T7 tag antibody, probably due to the degradation of the T7 tag, but Western blot with anti-PIASy (C terminus) antibody indicated a similar level of expression of all proteins (Fig. 2B, lower panel). These reticulocyte lysates were supplemented into the PIASy-immunodepleted CSF-XEE and incubated with sperm chromatin to assemble mitotic chromosomes. Western blot analysis of isolated chromosomes indicated that each chimeric protein containing the N terminus of PIASy was recruited to mitotic chromosomes (Fig. 2C, lower two panels), suggesting N terminus of PIASy can uniquely facilitate chromosome binding. In contrast, constructs containing the N terminus of either PIAS1, -3, or -x failed to bind to chromosomes and execute TopoIIα SUMOylation, confirming that the N terminus of other PIAS family members cannot mediate chromosome interaction. Taken together, these results indicate that the N terminus of PIASy is a key element to achieve the unique role of PIASy for mitotic chromosomal SUMOylation by allowing interaction of PIASy to mitotic chromosomes.
The N Terminus of PIASy Is Sufficient for Localization to the Centromeric Region
Because removal of the N terminus PIASy or replacement with the N terminus of other PIAS members resulted in failure to bind chromosomes, we sought to identify whether the N-terminal domain of PIASy simply is sufficient for chromosomal localization in XEE assay. To test this, fluorescent protein, mCherry, was fused to the N-terminal peptide of PIASx (PIASx N) or PIASy (PIASy N) and expressed in bacteria. PIASx was previously shown to partially restore mitotic SUMOylation in the absence of PIASy but not to the same extent as PIASy (7). Purified proteins were added when DNA replication of sperm chromatin was completed in interphase XEE, then mitosis was induced by addition of CSF-XEE to obtain chromosomes. The amounts of PIASx N and PIASy N added to the extracts were shown to be equivalent by Western blot analysis (Fig. 3D). In Fig. 3A, immunostaining indicated that PIASy N not only binds chromosomes but also accumulates at centromeric regions, resulting in colocalization with both SUMO2/3 and Aurora B, markers of inner-centromere, meanwhile PIASx N was sparsely distributed and barely detectable. In particular, PIASy N resembles the behavior of full-length PIASy on chromosomes localizing at the centromeric region during mitosis (Figs. 3B and 5C),3 indicating that the N-terminal ∼140 amino acids of PIASy is sufficient to direct the localization of PIASy to the centromeric region (Fig. 3, B and C).
Rod and Zw10 Bind to PIASy N Terminus
It has been shown that N-terminal domains of PIAS proteins are involved in protein interactions for multiple purposes (35, 36). Therefore, we speculated that the N-terminal region of PIASy might interact with specific protein(s) for centromeric localization. In order to identify protein(s) that specifically interact with the N terminus of PIASy but not with other PIAS family members, we performed pull-down assay with N-terminal fragments of PIAS family proteins. The 1–139 amino acid fragment of PIASy and the equivalent residues from other PIAS proteins were fused to an S tag at the N terminus and expressed in Escherichia coli. Purified proteins were captured on S tag affinity beads and the beads were mixed with XEE to isolate binding proteins. Associated proteins were separated by SDS-PAGE and analyzed by silver staining. As shown in Fig. 4A, several bands (indicated with asterisks) were reproducibly observed to associate with the PIASy N-terminal peptide but not with the N termini of any other PIAS protein. Bands of ∼250 and 90 kDa were further identified as Rod and Zw10, respectively, by tandem mass spectrometry. To confirm the interaction of PIASy with Rod and Zw10, we prepared a polyclonal antibody against Rod and obtained commercially available anti-Zw10 antibody. Western blot analysis of samples from pull-down assays as shown in Fig. 4A revealed that Rod and Zw10 specifically interact with the PIASy N-terminal region and not with other PIAS proteins (Fig. 4B). We next investigated whether the interaction of PIASy and the identified proteins takes place under physiological conditions. To test this, endogenous PIASy proteins were immunoprecipitated from CSF-XEE with antibody against C terminus of PIASy, and co-precipitants were analyzed with Western blot. We observed that PIASy co-precipitated with Rod, confirming that they interact in XEE bona fide. Yet, only a minor fraction of Rod interacts with PIASy, indicating that limited portion of PIASy interact to Rod or their interaction is highly transient in XEE (Fig. 5A, left). In a reciprocal immunoprecipitation performed with an antibody against Rod, Rod was able to precipitate a small but detectable amount of PIASy reproducibly (Fig. 5A, right).
Rod and Zw10 are components of a stable complex called RZZ (Rod/Zw10/Zwilch) (26, 37). The RZZ complex is localized at kinetochores from pro/metaphase to metaphase in somatic cells (24). PIASy was also observed near centromeres in XEE assays (Fig. 3B).3 Thus, we sought to determine whether Rod and Zw10 interact with PIASy on mitotic chromosomes using chromatin immunoprecipitation and immunofluorescence approaches. Chromatin immunoprecipitation of mitotic chromosomes prepared from XEE showed that PIASy co-precipitates Rod and Zw10 (Fig. 5B, left). Reciprocal analysis with an anti-Rod antibody also showed co-precipitation of both PIASy and Zw10 (Fig. 5B, right). Moreover, immunofluorescence data indicated that Rod and Zw10 colocalize with PIASy at centromeric regions together with SUMO2/3 as expected (Fig. 5C), supporting the idea that PIASy binds to the Rod/Zw10 complex for the execution of mitotic SUMOylation. It is also possible that PIASy directs the Rod/Zw10 complex to chromosomes. Immunodepletion of PIASy in CSF-XEE, however, did not eliminate chromosomal association of Rod/Zw10 (Fig. 5D), arguing against PIASy-dependent chromosomal localization of the RZZ complex. Taken together, we conclude that Rod and Zw10 bind to the N terminus of PIASy at the kinetochore, and their colocalization with PIASy and immunodepletion analysis suggest that Rod/Zw10 might direct the centromeric localization of PIASy.
Depletion of Rod Causes Mislocalization of PIASy during Mitosis
Deletion of amino acids 1–47 of PIASy (48C) eliminates its chromosome binding (Fig. 1C), suggesting that a protein interacting with the first 47 residues is critical for chromosome localization. Thus, we sought to further characterize where Rod and Zw10 interact within the N-terminal 140 amino acid region of PIASy, which we used as bait in pull-down assay (Fig. 4). To this end, serial truncation constructs of PIASy that contain T7 tag at the N terminus were prepared (Fig. 6A). Purified truncation proteins were bound to anti-T7 tag antibody-conjugated beads followed by incubation in CSF-XEE. Western blot analysis of pull-downed proteins by the beads indicated that Rod and Zw10 interact with full length of PIASy similar to data shown in Fig. 4. Strikingly, those interactions were no longer observed with PIASy that 1–47 amino acids were deleted (48C) as well as with further truncations (Fig. 6B). Consistent with pull-down result in Fig. 4, PIASy (N325) with a C-terminal deletion retained the interaction with Rod and Zw10, confirming that the Rod/Zw10 complex binds to the N terminus, but not the C terminus of PIASy. This result reveals that the interaction of the Rod/Zw10 complex with PIASy is restricted to 1–47 amino acids of the N terminus, the region critical for chromosome localization of PIASy (Fig. 1C).
It has been shown that PIASy-dependent SUMOylation is confined to the centromeric region of chromosomes in mitosis (7, 9). Given that the Rod/Zw10 complex is localized at the kinetochore, we hypothesized that the Rod/Zw10 complex is required for PIASy to localize to the centromeric region. To address this question, we immunodepleted endogenous Rod and examined the localization of PIASy and of SUMOylation. The efficiency of Rod depletion was ∼80% (Fig. 7C), and a Rod signal was detected by immunofluorescence on some chromosomes prepared from Rod-depleted XEE (-Rod), albeit its signal was much weaker than the one in Mock-depleted samples (-IgG). Nonetheless, it was evident that the centromeric localization of Rod was completely linked to that of PIASy: chromosomes containing Rod showed the centromeric localization of PIASy without exception while ones without Rod had mislocalized PIASy on the chromosomes (Fig. 7A, compare chromosomes indicated with arrows and squares). Rod-deficient chromosomes never showed clear PIASy foci, rather they showed dispersed and weak signals of PIASy. Chromosome samples were also stained with another anti-PIASy antibody for further confirmation. We consistently observed mislocalized PIASy on the Rod-depleted chromosomes, but rarely on Mock-depleted chromosomes (Fig. 7B). It was notable that Rod-deficient chromosomes showed loss of intensive SUMO2/3 foci presumed to locate near centromeres, following mislocalization of PIASy (Fig. 7, A and B and supplemental Fig. S1). All together, our data indicate that the Rod/Zw10 complex is required for the centromeric localization of PIASy and spatial regulation of SUMO2/3 modification near the centromere.
DISCUSSION
Biological Significance of the PIASy N-terminal Domain
We have thoroughly characterized PIASy, an essential SUMO E3 ligase of mitotic substrates, using both systematic in vitro domain analysis and Xenopus egg extract cell-free assays. It was previously demonstrated that the PIASy N terminus, mostly composed of a SAP domain, plays a key role in chromosome interaction (7). Recent studies have shown that Siz1, a Siz/PIAS SUMO E3 ligase in budding yeast, interacts with substrates via its N-terminal domain (16, 19), suggesting that the N-terminal domain of Siz/PIAS is responsible for substrate binding. Given that major substrates of PIASy are chromosome-resident proteins in mitosis, we hypothesized that the N terminus of PIASy achieves chromosome binding through the interaction with its substrates. In vitro analysis using purified N-terminal PIASy truncations revealed that the first 127 amino acids of PIASy are dispensable for SUMOylation in vitro, with further deletion (including PINIT domain (16, 36)) starting to affect SUMOylation (Fig. 1B). This finding suggests that N terminus anterior to the PINIT domain is not involved in substrate binding. Conversely, XEE assays have shown that the first 47 amino acids of PIASy, including the SAP domain, is absolutely required for mitotic SUMOylation (Fig. 1C), strongly suggesting that the PIASy N terminus binds specific molecule(s) to mediate chromosomal localization, and that the interaction between PIASy, and these partners is critical for mitotic SUMOylation.
A number of studies have described a discrepancy between in vivo and in vitro substrate specificity of Siz/PIAS proteins of SUMO E3 ligases (19, 38, 39). It has been suggested that subcellular localization of SUMO E3 ligases is crucial for substrate specificity in vivo (19, 38, 39). Our XEE assays have shown that chimera PIASy proteins containing the N terminus of PIAS1, -3, or -x fail to interact with chromosomes and fail to facilitate mitotic SUMOylation (Fig. 2). We have also shown that the N terminus of PIASy is sufficient to direct localization to the centromeric region (Fig. 3), arguing that PIAS proteins display substrate specificity by means of subcellular localization. The SAP domain was characterized as a putative A/T-rich DNA binding domain, yet proteins such as p53, Msx1, and Oct-4 interact with specific PIAS proteins through their SAP domain (15, 40, 41). We have identified that the N terminus of PIASy containing the SAP domain interacts with kinetochore-associated proteins, Rod and Zw10 (Figs. 4 and 6), raising the possibility that the SAP domain contributes to protein interactions for regulating subcellular localization of PIAS proteins. It will be of interest to investigate whether the SAP domain-mediated localization is a common mechanism to regulate the localization of other PIAS family proteins in cells. This information will provide insight into the in vivo substrate specificity of mammalian PIAS family proteins.
Molecular Mechanism of PIASy Recruitment to the Kinetochore
PIASy-dependent SUMOylation becomes detectable in prometaphase, peaks in metaphase, and disappears with the onset of anaphase (20). This mitotic phase-specific SUMOylation is further restricted to the inner centromere (7). Here we show that PIASy binds to kinetochore proteins, Rod and Zw10; components of the RZZ complex, through its N terminus (Figs. 4–6) and that elimination of Rod compromises PIASy localization to kinetochores (Fig. 7). These results suggest that the Rod/Zw10 complex has a critical role in the centromeric recruitment of PIASy and, thus, in the SUMOylation of chromosomal substrates near the centromere. Previous studies in Drosophila embryos revealed that GFP-Rod enters the nucleus by the time that nuclear envelope breaks down, accumulates on kinetochores, and disperses as sister chromatids are separated (24). This aspect of Rod and Zw10 distribution during mitosis highly resembles PIASy distribution, leading to an explanation for how PIASy executes centromeric SUMOylation during mitosis. We propose that PIASy accumulates on kinetochores by docking with Rod/Zw10 complex that is concentrated on kinetochores when the nuclear envelope disassemble and then PIASy facilitates SUMO2/3 modification of its substrates (TopoIIα and PARP1) near the centromere (Fig. 7D). As Rod and Zw10 are shed from kinetochores, PIASy falls off and SUMO modification decreases. One puzzling observation is that PIASy is mislocalized but does not fail to interact with chromosomes deficient in Rod/Zw10 complex. This does not fit with the observation that PIASy lacking first 47 residues failed to interact with chromosomes (Fig. 1B), implying that there might be another protein(s) delivering PIASy close to chromosomes. Further analysis of PIASy movement dynamics in mitosis may elucidate this question.
The RZZ complex delivers dynein/dynactin onto unattached kinetochores (42). The complex also functions in activating spindle assembly checkpoint signaling by bringing two checkpoint components, Mad1 and Mad2 (25, 43). In Drosophila and C. elegans, the phenotypes of mutation in Rod, Zw10, or both cause as premature chromosome segregation with anaphase bridges (28, 37). Components of the SUMO pathway are clearly connected to mitosis. Mutations in SUMO (smt3, pmt3), Aos1, and Ubc9 show defects in mitotic progression in budding and fission yeast (44–47). Defects in SUMO modification also show chromosomal missegregation in XEE (7, 20). Given that the Rod/Zw10 complex is required for proper centromeric localization of PIASy and SUMO modification (Fig. 7 and supplemental Fig. S1), it is reasonable to consider that the RZZ complex could regulate two completely different pathways: metaphase checkpoint and centromeric SUMOylation. If so, the abnormal chromosome segregation shown in Rod/Zw10 mutants might be the dual outcome from defects in both the mitotic checkpoint and the SUMO pathway.
In summary, we have demonstrated the molecular mechanism of PIASy-dependent SUMOylation in mitosis by showing that PIASy interacts with the Rod/Zw10 complex, a kinetochore component, through its SAP domain in the N terminus. Our results suggest that the SAP domain of PIAS family proteins could show binding specificity that plays a critical role in determination of substrate specificity in vivo. Together, our studies reveal the likely molecular mechanism of PIASy-dependent spatiotemporal regulation of SUMO modification and a novel role of the RZZ kinetochore complex on chromosomal SUMOylation.
Supplementary Material
Acknowledgments
We thank Dr. Kathleen Gould for the gift of the plasmid for CBP tag and Dr. Frauke Melchior for the plasmids for E1 expression. We thank Drs. Alexei Arnaoutov and Mary Dasso for the gift of X. laevis cDNA and anti-Aurora B antibody and for discussion of our results. We are grateful to Dr. Kristi Neufeld and Dr. Berl Oakley for critical reading and for providing editorial assistance of this manuscript. Protein identification was done at the Harvard Microchemistry and Proteomics Analysis Facility.
This work was supported, in whole or in part, by National Institutes of Health/NIGMS Grant GM80278 and a start-up grant from the Dept. of Molecular Biosciences at the University of Kansas.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
H. Ryu, M. Furuta, D. Kirkpatrick, S. P. Gygi, and Y. Azuma, submitted manuscript.
- SUMO
- small ubiquitin-like modifier
- SIM
- SUMO-interacting motif
- SAP
- scaffold attachment factor-A/B, acinus, and PIAS domain
- SP-Ring
- Siz/PIAS-Ring
- CSF-XEE
- cytostatic factor-arrested Xenopus egg extracts.
REFERENCES
- 1.Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 2.Bossis G., Melchior F. (2006) Cell Div. 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dasso M. (2008) Cell Div. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hay R. T. (2005) Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 5.Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 6.Yunus A. A., Lima C. D. (2006) Nat. Struct. Mol. Biol. 13, 491–499 [DOI] [PubMed] [Google Scholar]
- 7.Azuma Y., Arnaoutov A., Anan T., Dasso M. (2005) EMBO J. 24, 2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson E. S., Gupta A. A. (2001) Cell 106, 735–744 [DOI] [PubMed] [Google Scholar]
- 9.Ryu H., Al-Ani G., Deckert K., Kirkpatrick D., Gygi S. P., Dasso M., Azuma Y. (2010) J. Biol. Chem. 285, 14415–14423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J. M. (2008) Cell 133, 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pichler A., Knipscheer P., Saitoh H., Sixma T. K., Melchior F. (2004) Nat. Struct. Mol. Biol. 11, 984–991 [DOI] [PubMed] [Google Scholar]
- 12.Palvimo J. J. (2007) Biochem. Soc. Trans. 35, 1405–1408 [DOI] [PubMed] [Google Scholar]
- 13.Aravind L., Koonin E. V. (2000) Trends Biochem. Sci. 25, 112–114 [DOI] [PubMed] [Google Scholar]
- 14.Suzuki R., Shindo H., Tase A., Kikuchi Y., Shimizu M., Yamazaki T. (2009) Proteins 75, 336–347 [DOI] [PubMed] [Google Scholar]
- 15.Okubo S., Hara F., Tsuchida Y., Shimotakahara S., Suzuki S., Hatanaka H., Yokoyama S., Tanaka H., Yasuda H., Shindo H. (2004) J. Biol. Chem. 279, 31455–31461 [DOI] [PubMed] [Google Scholar]
- 16.Yunus A. A., Lima C. D. (2009) Mol. Cell 35, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekiyama N., Ikegami T., Yamane T., Ikeguchi M., Uchimura Y., Baba D., Ariyoshi M., Tochio H., Saitoh H., Shirakawa M. (2008) J. Biol. Chem. 283, 35966–35975 [DOI] [PubMed] [Google Scholar]
- 18.Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I. (2006) J. Biol. Chem. 281, 16117–16127 [DOI] [PubMed] [Google Scholar]
- 19.Reindle A., Belichenko I., Bylebyl G. R., Chen X. L., Gandhi N., Johnson E. S. (2006) J. Cell Sci. 119, 4749–4757 [DOI] [PubMed] [Google Scholar]
- 20.Azuma Y., Arnaoutov A., Dasso M. (2003) J. Cell Biol. 163, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torras-Llort M., Moreno-Moreno O., Azorín F. (2009) EMBO J. 28, 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santaguida S., Musacchio A. (2009) EMBO J. 28, 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karess R. (2005) Trends Cell Biol. 15, 386–392 [DOI] [PubMed] [Google Scholar]
- 24.Basto R., Scaerou F., Mische S., Wojcik E., Lefebvre C., Gomes R., Hays T., Karess R. (2004) Curr. Biol. 14, 56–61 [DOI] [PubMed] [Google Scholar]
- 25.Buffin E., Lefebvre C., Huang J., Gagou M. E., Karess R. E. (2005) Curr. Biol. 15, 856–861 [DOI] [PubMed] [Google Scholar]
- 26.Williams B. C., Li Z., Liu S., Williams E. V., Leung G., Yen T. J., Goldberg M. L. (2003) Mol. Biol. Cell 14, 1379–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y., Wang Z., Ge L., Chen N., Liu H. (2009) Cell Struct. Funct. 34, 31–45 [DOI] [PubMed] [Google Scholar]
- 28.Scaërou F., Aguilera I., Saunders R., Kane N., Blottière L., Karess R. (1999) J. Cell Sci. 112, 3757–3768 [DOI] [PubMed] [Google Scholar]
- 29.Basto R., Gomes R., Karess R. E. (2000) Nat. Cell Biol. 2, 939–943 [DOI] [PubMed] [Google Scholar]
- 30.Wang Z., Cummins J. M., Shen D., Cahill D. P., Jallepalli P. V., Wang T. L., Parsons D. W., Traverso G., Awad M., Silliman N., Ptak J., Szabo S., Willson J. K., Markowitz S. D., Goldberg M. L., Karess R., Kinzler K. W., Vogelstein B., Velculescu V. E., Lengauer C. (2004) Cancer Res. 64, 2998–3001 [DOI] [PubMed] [Google Scholar]
- 31.Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 32.Azuma Y., Tan S. H., Cavenagh M. M., Ainsztein A. M., Saitoh H., Dasso M. (2001) Faseb J. 15, 1825–1827 [DOI] [PubMed] [Google Scholar]
- 33.Kornbluth S., Yang J., Powers M. (2001) in Current Protocols in Cell Biology (Yamada K. ed), pp. 11.11.11–11.11.13, John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 34.Azuma Y. (2009) Methods Mol. Biol. 582, 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rytinki M. M., Kaikkonen S., Pehkonen P., Jääskeläinen T., Palvimo J. J. (2009) Cell Mol. Life Sci. 66, 3029–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duval D., Duval G., Kedinger C., Poch O., Boeuf H. (2003) FEBS Lett. 554, 111–118 [DOI] [PubMed] [Google Scholar]
- 37.Scaërou F., Starr D. A., Piano F., Papoulas O., Karess R. E., Goldberg M. L. (2001) J. Cell Sci. 114, 3103–3114 [DOI] [PubMed] [Google Scholar]
- 38.Takahashi Y., Toh-E. A., Kikuchi Y. (2003) J. Biochem. 133, 415–422 [DOI] [PubMed] [Google Scholar]
- 39.Kotaja N., Karvonen U., Jänne O. A., Palvimo J. J. (2002) Mol. Cell Biol. 22, 5222–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolkunova E., Malashicheva A., Parfenov V. N., Sustmann C., Grosschedl R., Tomilin A. (2007) J. Mol. Biol. 374, 1200–1212 [DOI] [PubMed] [Google Scholar]
- 41.Lee H., Quinn J. C., Prasanth K. V., Swiss V. A., Economides K. D., Camacho M. M., Spector D. L., Abate-Shen C. (2006) Genes Dev. 20, 784–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starr D. A., Williams B. C., Hays T. S., Goldberg M. L. (1998) J. Cell Biol. 142, 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kops G. J., Kim Y., Weaver B. A., Mao Y., McLeod I., Yates J. R., 3rd, Tagaya M., Cleveland D. W. (2005) J. Cell Biol. 169, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biggins S., Bhalla N., Chang A., Smith D. L., Murray A. W. (2001) Genetics 159, 453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka K., Nishide J., Okazaki K., Kato H., Niwa O., Nakagawa T., Matsuda H., Kawamukai M., Murakami Y. (1999) Mol. Cell Biol. 19, 8660–8672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.al-Khodairy F., Enoch T., Hagan I. M., Carr A. M. (1995) J. Cell Sci. 108, 475–486 [DOI] [PubMed] [Google Scholar]
- 47.Shayeghi M., Doe C. L., Tavassoli M., Watts F. Z. (1997) Nucleic Acids Res. 25, 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.