Abstract
Protein translocation across the endoplasmic reticulum membrane occurs at the Sec61 translocon. This has two essential subunits, the channel-forming multispanning membrane protein Sec61p/Sec61α and the tail-anchored Sss1p/Sec61γ, which has been proposed to “clamp” the channel. We have analyzed the function of Sss1p using a series of domain mutants and found that both the cytosolic and transmembrane clamp domains of Sss1p are essential for protein translocation. Our data reveal that the cytosolic domain is required for Sec61p interaction but that the transmembrane clamp domain is required to complete activation of the translocon after precursor targeting to Sec61p.
Keywords: Membrane Biogenesis, Protein Secretion, Protein Targeting, Protein Translocation, Trafficking
Introduction
The translocon is a highly conserved protein complex that forms a channel for the translocation of proteins across the endoplasmic reticulum (ER)2 membrane. It is a heterotrimer consisting of Sec61p/Sec61α, Sss1p/Sec61γ, plus the nonessential Sbh1p/Sec61β subunit and is both structurally and functionally homologous to the plasma membrane SecYEG and SecYEβ complexes found in eubacteria and archaea, respectively (1). Analysis of SecYEβ and SecYEG complexes at high resolution has identified a number of key structural features and led to a general model for translocon function (2, 3). In this model, the SecY/Sec61p/Sec61α subunit (α-subunit) forms a transmembrane protein-conductive channel with a “plug” domain located on the luminal side of a central narrow constriction in the closed state, which upon channel activation is predicted to move toward the periplasm/ER lumen. A lateral gate formed by TM domains 2b and 7 of the α-subunit is proposed to interact with signal sequences to allow their partition into the membrane bilayer. The SecE/Sss1p/Sec61γ subunit (γ-subunit) is located at the channel periphery and has a long curved TM domain that traverses the membrane diagonally. It contacts both the N (TM1–5) and C (TM6–10) halves of the α-subunit and has been proposed to act as a “clamp” to hold both halves of the SecY/Sec61 together, thus regulating lateral access to the translocation channel (2). This elegant model predicts an important role for Sss1p in regulating the yeast ER translocon, and we have sought to test this both in vivo and in vitro. We have constructed a series of novel mutations in Sss1p and used these to examine the roles of both the cytosolic and TM clamp domains in translocon function. The phenotypes of these mutants show that both the cytosolic domain and the TM domain are essential for protein translocation. In addition we show that the TM domain is required for Sss1p membrane association but that the cytosolic domain is essential for interaction with Sec61p and specifically with Sec61p TM9. Substitution of the TM clamp domain with the TM region of another ER tail-anchored protein resulted in a Sss1p mutant that could associate efficiently with Sec61p but was unable to support translocation in vivo. This translocation defect was reconstituted in vitro where we used site-specific cross-linking to reveal that precursor protein was efficiently targeted to Sec61p in this context but was unable to progress beyond this step. Our results demonstrate that Sss1p is required after signal sequence interaction with the translocon to complete channel activation and that the clamp domain of Sss1p is a key regulator of translocon function.
EXPERIMENTAL PROCEDURES
Growth of Yeast Cells
Yeast strains (Table 1) were grown at 30 °C in YP medium (2% peptone, 1% yeast extract) containing 2% glucose and 0.02% adenine (YPAD) or in minimal medium (0.67% yeast nitrogen base) with 2% glucose or galactose with appropriate supplements for selective growth. For glucose repression, cells were grown in minimal galactose medium to an A600 nm 0.2, harvested, and resuspended in YPAD at the same density. For methionine repression, cells were similarly transferred from methionine-free to medium containing 2 mm methionine. Solid media were supplemented with 2% agar. Yeast transformation and 5-fluoroorotic acid (5-FOA) counterselection of URA3 plasmids have been previously described (7). All growth medium was purchased from Difco Laboratories. Geneticin and 5-FOA were from Melford Laboratories.
TABLE 1.
Yeast strains used in this article
| Strain | Genotype | Reference |
|---|---|---|
| FKY198 | MATα, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, sss1::URA3, pep4::LEU2, p[GAL10::SSS1, ADE2] | 4 |
| W303-2n | MATa/MATα, ade2-1/ade2-1, ura3-1/ura3-1, his3-11,-15/his3-11,-15, leu2-3,-112/leu2-3,-112, trp1-1/trp1-1, CAN1-100/CAN1-100 | 5 |
| BWY12 | MATα, Δsec61::HIS3, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, [pBW7] | 6 |
| BWY529 | MATa/MATα, SSS1/Δsss1::KanMX4, ade2-1/ade2-1, ura3-1/ura3-1, his3-11,-15/his3-11,-15, leu2-3,-112/leu2-3, trp1-1/trp1-1, CAN1-100/CAN1-100 | This study |
| BWY530 | MATα, Δsss1::KanMX4, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, [FKp53] | This study |
| BWY531 | MATa, Δsss1::KanMX4, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, [FKp53] | This study |
| BWY875 | MATa, Δsss1::KanMX4, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, [pCM214] | This study |
| BWY886 | MATa, sss1Δ12::KanMX6, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, [pCM214] | This study |
| CMY5 | MATα, Δsss1::KanMX4, Δsec61::HIS3, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, [pCM203] | This study |
| CMY28 | MATα, Δsss1::KanMX4, Δsec61::HIS3, ade2-1, ura3-1, his3-11,-15, leu2-3,-112, trp1-1, CAN1-100, [pCM214/pCM108] | This study |
Construction of SSS1 Mutations
SSS1 was cloned on a 1136-bp SalI/SspI fragment from FKp52 (4) into SalI/EcoRV-digested pRS313 (8). The resulting plasmid (pJKB2) was used for site-directed mutagenesis (QuikChange, Stratagene) to create the G57L mutation (primers SSS1-G1, G1a: see supplementary Table S1 for primer sequences). The G62L mutation (primers SSS1-G2, G2a) was introduced into the G57L plasmid, and this G57L/G62L double mutation was used in turn to add the G65L mutation (primers SSS1-G3, G3a) and hence generate the G57L/G62L/G65L triple mutation. A stop codon was inserted after the SSS1 Lys52 codon (primers SSS1-ΔC, ΔCa) in pJKB2 to produce a construct expressing the Sss1p TM deletion (Sss1ΔCp). A product including the UBC6 promoter and the cytosolic domain sequences (residues Met1–Ser232) followed by a BamHI site was generated by PCR (primers UBC6–1, -3) from Saccharomyces cerevisiae genomic DNA. A fragment encoding the C-terminal region of SSS1 (residues 53–80) and transcription termination sequences preceded by a BamHI site was produced by PCR (primers; SSS1–1, 4) from FKp52. The two PCR products were digested with BamHI, ligated, end-filled, and subsequently ligated into SmaI-digested pRS313 generating a plasmid encoding the USCp fusion. A fragment encoding the TM of Ubc6p (residues Met233–Met249) preceded by a BamHI site and followed by SSS1 sequences starting from the Lys69 codon was generated by PCR using FKp52 (primers USSS1C, Ca). The product was digested with BamHI and Ssp1, ligated into BamHI/Ssp1-digested pJKB2 derivative with a BamHI site inserted immediately after the K52 codon to generate the NSUp fusion plasmid pBW236. The sequences of all mutations and constructs generated by PCR were verified by DNA sequencing. A 774-bp SalI/SmaI fragment from pBW236 encoding the NSUp fusion was cloned into pRS316 (8) to provide low copy expression on a URA3 vector (pBW252). Similarly, a 1140-bp SalI/EcoRI SSS1 encoding a fragment from pJKB2 was cloned into pRS316 to generate pBW249. These pRS316 based plasmids were used for CMY28 (Table 1) transformation.
Other SSS1 Plasmids
A plasmid expressing SEC61 and SSS1 (pCM203; CEN, URA3) was generated by ligation of a 7.7-kb Sma1/XhoI fragment of pBW7 (6) with a 1.8-kb SalI/SmaI SSS1-encoding fragment of FKp52 (4). The MET3-SSS1 plasmid (pCM214; CEN, TRPI) was constructed as follows. A 1.8-kb SalI/EcoRI fragment encoding SSS1 was isolated from FKp52 and ligated into SalI/EcoRI-digested pRS314 (8). A unique NdeI site was then inserted immediately upstream of the SSS1 ATG start codon (primers SSS1-NdeIF, NdeIR) by site-directed mutagenesis. The resulting plasmid was digested with NdeI and end-filled by Klenow treatment. This product was SalI-digested to excise the SSS1 promoter, which was replaced with a MET3 promoter generated by BamHI digestion, end-filling with Klenow and then SalI digestion of pJT31 (9).
Construction of the Sec61-tmd9CCC Plasmid
The three endogenous cysteine codons of SEC61 (Cys121, Cys150, and Cys373) were changed to alanine codons by site-directed mutagenesis (primers SEC61-121-SEC61-373a) of a pBW11 derivative (CEN, SEC61, LEU2) plasmid containing a BamHI site at nucleotides 532–537 of the SEC61 ORF (10). This was used as a template to convert the three consecutive codons Ala423, Phe424, and Gly425 in TM9 (primers SEC61tm9C, Ca) to cysteine codons to generate the functional sec61-tmd9CCC allele expressed on plasmid pCM108.
Yeast Strain Construction
The Δsss1::KanMX4 null mutation was made by PCR amplification (primers-Δsss1-1, 1a) of the pFA6a-kanMX4 module (11), and transformation of strain W303-2n (Table 1) to Geneticin resistance. Correct integration of Δsss1::kanMX4 was confirmed by PCR analysis of genomic DNA. The SSS1/Δsss1 strain, BWY529, was transformed with the SSS1, URA3 plasmid FKp53 (4), sporulated, and tetrad-dissected on YPAD medium. Geneticin-resistant Ura+ colonies consistent with rescue of the lethal Δsss1 phenotype by the plasmid were obtained. One of these, BWY530, was used for the shuffle of sss1 mutant alleles on 5-FOA medium. A MATa equivalent, BWY531, was crossed with BWY12, and the resulting diploid was transformed with pCM203, sporulated, and tetrad-dissected on YPAD medium to yield the Δsss1/Δsec61 strain CMY5. CMY28 was derived by transformation of CMY5 jointly with pCM108 and pCM214, followed by passage on 5-FOA medium. The sss1Δ12::kanMX6 allele was made by PCR amplification of a pFA6a-GFP(S65T)-kanMX6 module (11) designed to truncate Sss1p after residue Ile68. It was used to transform W303-2n (Table 1) to Geneticin resistance. The resulting SSS1/sss1Δ12::kanMX6 transformant was confirmed by PCR analysis of genomic DNA. It was transformed with FKp53 (4), sporulated, and tetrad-dissected on YPAD medium. Geneticin-resistant Ura+ colonies were unable to grow on 5-FOA medium. One of these colonies and BWY530 were transformed with pCM214, followed by growth on 5-FOA medium lacking methionine to generate strains BWY875 (Δsss1) and BWY886 (sss1Δ12) rescued by the MET3-SSS1 allele.
Cell Extract Preparation and Immunoblotting
Yeast whole cell extracts were prepared from 5.0 A600 nm units of exponentially growing cells by suspension in sample buffer with 0.5-mm glass beads and disruption at 6.5 m/s for 30s (Hybaid Ribolyser). The samples were incubated for 5 min at 65 °C, then 5 min at 95 °C. After SDS-PAGE, proteins were electroblotted to PVDF membranes (Millipore), and antibody incubations were performed as previously described (7). Antisera recognizing the C terminus of Sec61p (12), Sbh1p (13), DPAP B (7), Kar2p (9), and Sec62p, Sec63p, and ppCPY (14) have been described. Antisera against Sss1p and Ubc6p were raised in sheep by immunization using antigens consisting of residues 1–52 and 1–228, respectively, which had been purified by His tag affinity purification.
Cellular Fractionation and Carbonate Extraction of Membranes
For cellular fractionation, 50 A600 nm units of exponentially growing cells were harvested, washed in distilled water, and resuspended in 0.5 ml of ice-cold lysis buffer (0.2 m sorbitol, 20 mm HEPES, pH 7.4, 50 mm KOAc, 5 mm EDTA, 1 mm DTT, 10 μg/ml each leupeptin/aprotinin, 5 μg/ml each chymostatin/pepstatin). The cells were broken by glass bead lysis. The lysate was removed from the beads and subjected to centrifugation at 100,000 × g for 30 min at 4 °C with the supernatant providing the cytosolic fraction and the pellet providing the membrane fraction. For carbonate extraction, membrane preparations (10) were mixed with 4 volumes of 125 mm Na2CO3, pH 11.5, and left on ice for 30 min. The samples were then subjected to centrifugation at 100,000 × g for 30 min at 4 °C. The pellet that was solubilized in SDS-PAGE sample buffer provided the carbonate-resistant fraction, whereas the soluble fraction that was subjected to precipitation with 10% trichloroacetic acid providing the carbonate-soluble fraction.
Phenanthroline Oxidation
Phenanthroline was activated by mixing 1 volume of 240 mm CuSO4 with 2 volumes of 360 mm 1,10-phenanthroline (prepared in 50% ethanol) followed by 12-fold dilution in aqueous solution. Membranes were treated with 2 mm Cu2+(phe)3 at 30 °C for 20 min, with quenching by the addition of neocuproine to 25 mm for 5 min. Membranes were prepared as previously described (7).
Radiolabeling and Immunoprecipitation
[35S]Methionine/cysteine (PerkinElmer Life Sciences) labeling of yeast cells was performed as described previously (7). Briefly, 5.0 A600 nm equivalents of cleared cell extract were added to saturating amounts of antiserum followed by rotation for 2 h at room temperature. Immune complexes were bound to protein A-Sepharose beads (Sigma) at room temperature for a further 2 h. Beads were washed three times with immunoprecipitation buffer, and antigens were disassociated in SDS-PAGE sample buffer. 14C-Amino acid (PerkinElmer Life Sciences) labeling of yeast cells was similarly performed (14). After SDS-PAGE, dried gels were analyzed with a Fuji BAS3000 PhosphorImager.
In Vitro Translation, Membrane Translocation, and Photocross-linking
A lysine codon was substituted at position 14 of a K5 ppαF construct (15). The resulting K5/K14-ppαF species was transcribed (Promega T7 express) and then translated in reticulocyte lysate (Promega) for 1 h at 30 °C in the presence of [35S]methionine and 4-(3-trifluoromethyldiazarino) benzoic acid (TDBA)-lysyl-tRNA (15) in the dark. After the addition of cycloheximide to 1 mm, ribosomes were removed by centrifugation at 400,000 × g for 20 min before use. For translocation reactions, 10 μl of a K5/K14-ppαF translation were mixed with 0.2 A280 nm units of yeast membranes in a final volume of 20 μl with a 20-min incubation at 20 °C. For the targeting assay, 100 μl of K5/K14-ppαF translation and 2.0 A280 nm units of membranes were ATP-depleted (15), mixed with a 20-min incubation at 20 °C. The membranes were pelleted at 20,000 × g for 10 min followed by resuspension in 100 μl of membrane buffer. Two-thirds were UV-irradiated (UVP spot cure emitting 254–365nm, 800 mW/cm2) on ice for 2 s, the remaining third kept on ice in the dark.
RESULTS
Cytosolic and Transmembrane Domains Are Essential for Sss1p Function
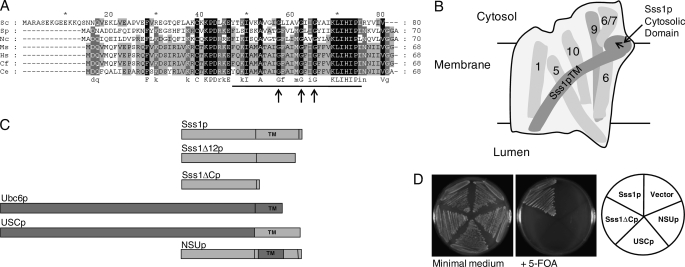
Sss1p is a small C-terminal anchor protein and in the simplest analysis consists of an N-terminal cytosolic domain (residues Met1–Asp46) and a C-terminal TM domain of 28 residues (Tyr47–ILe75, Fig. 1A). Structural analysis has shown that the cytosolic domain is in close proximity to the C-terminal half of Sec61p and has a specific contact with the cytosolic region of Sec61p between TM6 and TM7, whereas the Sss1p TM contacts TM 1, 5, 6, and 10 of Sec61p (Fig. 1B). The role of the TM was initially addressed by mutation of conserved TM residues identified by inspection of Sss1p/Sec61γ protein sequences from various species (Fig. 1A). Three highly conserved glycine residues equating to glycine residues 57, 62, and 65 of Sss1p were changed to leucine to assess their role in Sss1p function. However, no growth or ER translocation defects were evident in cells bearing these mutations (see supplemental Fig. S1).
FIGURE 1.
Sequence alignment of Sss1p/Sec61γ homologs and the structure and function of Sss1p domain mutants. A, S. cerevisiae (Sc) Sss1p aligned with its homologs from Schizosaccharomyces pombe (Sp), Neurospora crassa (Ns), Mus musculus (Mm), Homo sapiens (Hs), Canis familaris (Cf), and Caenorhabditis elegans (Ce). Alignments were performed using CLUSTAL_X (16), with black, dark gray, and light gray shading representing 100%, 80%, and 60% amino acid conservation. The TM domain (2) is underlined, and the highly conserved glycine residues are indicated by arrows. B, diagram of the Sec61-translocon in the ER membrane as viewed from the back of the structure where the Sss1p TM domain is situated (see Ref. 1), illustrating the key features of the Sec61p-Sss1p interface. The long curved TM domain of Sss1p has contacts with Sec61p TM domains 1, 5, 6, and 10 (numbered, behind the Sss1p TM in this view). The cytosolic domain is orientated at ∼90° to its TM domain (direction of the arrow), having contact with the cytosolic loop between Sec61p TM 6 and 7 (6/7), and further toward the N terminus is in close proximity to Sec61p TM9 (9, behind the 6/7 loop in this view). C, structure of Sss1p domain mutants. Sss1p (8.9 kDa) and Ubc6p (28.3 kDa) are ER tail-anchored membrane proteins with TM sequences as indicated. The Sss1ΔCp (5.9 kDa) is truncated after residue Lys52 of Sss1p, and the Sss1Δ12p (7.5 kDa) is truncated after residue Ile68. USCp (29.6 kDa) consists of the cytosolic domain of Ubc6p (Met1–Ser232) fused to the C-terminal region of Sss1p (Ala53–Val80). NSUp (9.5 kDa) consists of the cytosolic domain of Sss1p (Met1–Lys52) fused to the TM of Ubc6p (Met233–Met249) with Sss1p residues Lys69–Val80 at the C terminus. D, functional analysis of Sss1p domain mutants. Vector and plasmids encoding SSS1, the Sss1ΔC, USC, and NSU mutants were transformed into the SSS1 plasmid shuffle strain BWY530. All transformant strains grew normally after 3 days on minimal medium with selection for the SSS1, URA3 plasmid (FKp53), and the HIS3 (pRS313)-transformed plasmids. After 3 days on 5-FOA containing minimal medium, the vector-only strain was unable to grow due to the lethality of the Δsss1 mutation upon FKp53 counterselection, whereas the wild-type plasmid (SSS1) transformant grew by providing Sss1p function. However, the Sss1ΔCp-, NSUp-, and USCp-encoding plasmids were unable to support growth, indicating the lethality of deletion and partial substitution of the Sss1p TM domain and the substitution of the Sss1p cytosolic domain, respectively.
The lack of phenotypes upon mutation of residues in the TM raised the possibility that this domain was dispensable for Sss1p function. This was addressed by generating a deletion mutation expressing the N-terminal 52 residues of Sss1p (Sss1ΔCp) thus lacking the TM (Fig. 1C). However, this TM deletion mutation was unable to complement the lethal Δsss1 mutation (Fig. 1D), indicating that the cytosolic domain alone was nonfunctional either due to a defect in ER membrane localization or because the TM domain has a function in addition to that of a membrane anchor. This was tested by replacing 16 residues (Lys53–Ile68) of the Sss1p TM, including the conserved glycine residues and also encompassing the region interacting with Sec61p TM5 and TM10 (Fig. 1B) with the TM domain of the ER C-terminal anchor protein, Ubc6p. The resulting fusion protein “NSUp,” containing the N terminus of Sss1p and the Ubc6p TM (Fig. 1C), was also unable to complement the null mutation (Fig. 1D), suggesting a critical role for the Sss1p TM domain. Perhaps not surprisingly, when the function of the Sss1p cytsosolic domain was tested by analyzing a fusion of the cytosolic domain of Ubc6p to the Sss1p C terminus, “USCp” (Fig. 1C), it was also unable to complement (Fig. 1D), showing that this domain is also essential for Sss1p function.
Expression of Sss1p Mutant Proteins
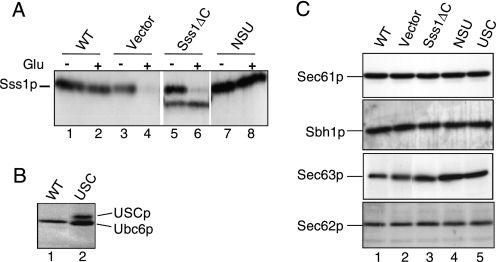
We next tested the expression and membrane association of the Sss1p domain mutant proteins in the yeast strain FKY198 which enables the analysis of nonfunctional sss1 mutations. This strain, which is grown in galactose medium but is inviable upon glucose repression of the GAL10-SSS1 allele (4), was transformed with control and mutant plasmids. Immunoblot analysis of total cell extracts from transformants grown in galactose and glucose medium was performed (Fig. 2A). The expression of Sss1p was largely abolished after 6 h in glucose medium in the vector-transformed control cells (Fig. 2A, lanes 3 and 4), compared with the continued expression of Sss1p in cells transformed with the wild-type SSS1 plasmid (Fig. 2A, lane 2). Cells carrying the Sss1ΔCp plasmid expressed a novel faster migrating species corresponding to the truncated derivative of Sss1p (Fig. 2A, lanes 5 and 6). The NSU protein was expressed as a species migrating very similar in size to Sss1p (Fig. 2A, lanes 2 and 8), whereas the USC protein was detected as a novel species migrating slower than Ubc6p (Fig. 2B, lane 2).
FIGURE 2.
Expression of mutant Sss1p derivatives and SEC translocon subunits. A, expression of Sss1p mutants. Whole cell extracts prepared from FKY198 cells transformed with vector, wild-type SSS1 (WT), Sss1ΔC, and NSU plasmids grown in galactose (−Glu) and after transfer to glucose medium for 6 h (+Glu) were subjected to 12.5% SDS-PAGE and immunoblot analysis with Sss1p antiserum. The panels were edited from the same immunoblot. B, whole cell extracts prepared from FKY198 cells after transfer to glucose medium for 6 h bearing wild-type and USC plasmids as indicated were analyzed by 10% SDS-PAGE and immunoblotting with Ubc6p antiserum. C, glucose-repressed extracts from above (A) were analyzed by 12.5% SDS-PAGE and immunoblotting with antiserum against Sec61p, Sec62p, Sec63p, and Sbh1p as indicated. These data are representative of two independent experiments.
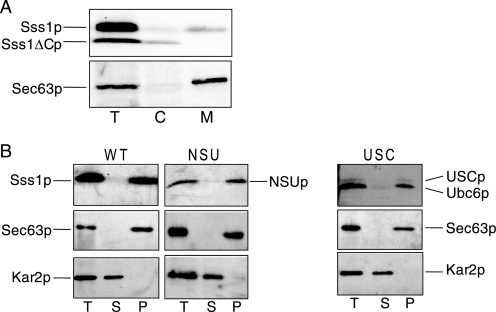
The membrane association of Sss1ΔCp was examined by fractionation of extracts prepared from Sss1ΔCp-expressing cells. Not all of the Sss1p and Sss1ΔCp could be accounted for, but the presence of some Sss1ΔC protein in a cytosolic fraction (Fig. 3A) suggests that the cytosolic domain of Sss1p may not be stably associated with the ER membrane in the absence of the TM domain. The NSU and USC proteins were found in membrane fractions, and both were found to be resistant to carbonate extraction, demonstrating that they are stably membrane-integrated (Fig. 3B).
FIGURE 3.
Membrane association of Sss1p domain mutants. A, FKY198 cells bearing the Sss1ΔCp plasmid were grown exponentially in galactose-containing medium allowing expression of both Sss1p and Sss1ΔCp. These cells were fractionated into crude cytosolic (C) and membrane (M) fractions. 2.0 A600 nm cell equivalents of each fraction and a total sample (T) were analyzed by 8–16% SDS-PAGE and immunoblotting with Sss1p (upper panel) and Sec63p (lower panel) antisera. The Sss1 protein and the ER integral membrane protein Sec63p (17) were found largely in the membrane fraction (third lane) as expected. However, some Sss1ΔCp was found in the cytosolic fraction (second lane). B, membranes prepared from FKY198 cells transformed with Sss1p (WT) NSUp- and USCp-expressing plasmids grown for 6 h in glucose medium were subjected to carbonate extraction. Total (T), carbonate-soluble (S), and carbonate-resistant pellet (P) samples were analyzed by 12.5% SDS-PAGE and immunoblotted with antisera against Kar2p, Sec63p, and Sss1p and Ubc6p as appropriate. Kar2p and Sec63p provide carbonate-extractable and -resistant ER protein species, respectively (17). These data are representative of two independent experiments.
Translocon Subunit Stability Is Unaffected by Sss1p Mutant Protein Expression
The nonfunctionality of the Sss1p mutant proteins could be explained if their expression resulted in the destabilization of translocon components. Although overexpression of Sss1p can stabilize mutant Sec61p species (18), depletion of Sss1p might conversely destabilize Sec61p or components of the larger heptameric SEC complex (19). This was addressed by immunoblotting after depletion of Sss1p in vector-transformed FKY198 cells indicating no obvious reduction in the expression levels of Sec61p, Sec62p, Sec63p, or Sbh1p subunits (Fig. 2C, lanes 1 and 2). No reduction was observed either in cells expressing the various Sss1p mutant proteins (Fig. 2C, lanes 3–5). Therefore, Sss1p is not required to maintain the stability of the translocon subunits tested, and furthermore Sss1p mutant expression does not cause their instability.
NSUp Is Assembled into Sec61 Complexes
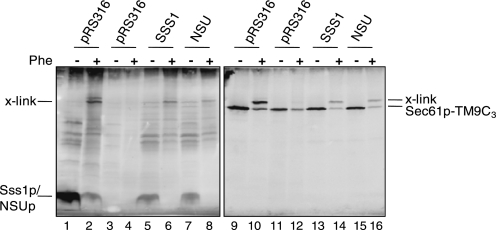
Although NSUp is expressed at the same level as Sss1p (Fig. 2A) and does not perturb translocon subunit stability (Fig. 2C), its phenotypes could be explained by a failure to interact with Sec61p. This was analyzed by utilizing the finding that the sole cysteine residue of Sss1p which is located in the cytosolic domain (residue Cys39) can form a disulfide bridge with a Sec61p species containing three cysteine residues in TM9 (Sec61p-TM9C3). Yeast strain CMY28, which expresses Sec61p-TM9C3 as the only source of Sec61p and also expresses Sss1p under the control of a MET3 promoter, was transformed with control and NSUp-expressing plasmids. Phenanthroline treatment of membranes from the vector-transformed strain before Sss1p depletion or the SSS1 plasmid transformed strain after Sss1p depletion showed a cross-link detected with both Sec61p and Sss1p antisera (Fig. 4, lanes 2, 6, 10, and 14). This was absent upon Sss1p depletion in the vector strain (Fig. 4, lanes 4 and 12). This Sec61p-Sss1p cross-link is consistent with the close juxtaposition of the Sss1p cytosolic domain and Sec61p TM9 predicted by structural analysis (2, 3). A cross-link was also evident between NSUp and Sec61p-TM9C3 (Fig. 4, lanes 8 and 16), demonstrating that NSUp assembles into a complex with Sec61p.
FIGURE 4.
NSUp interaction with Sec61p. Membranes prepared from CMY28 cells transformed with the pRS316-based SSS1/NSU plasmids or vector before (lanes 1, 2, 9, and 10) and after 10-h growth in 2 mm methionine medium (lanes 3–8, 11–16) were mock-treated or treated with 2 mm Cu2+(phe)3 (Phe). After 13% nonreducing SDS-PAGE, immunoblotting with Sss1p (left panel) and reprobing of the same blot with Sec61p (right panel) antiserum was performed. The adduct formed between Sec61p-TM9C3 and Sss1p/NSUp is marked as x-link. These data are representative of two independent experiments.
Cytosolic and Transmembrane Domains of Sss1p Are Essential for Protein Translocation
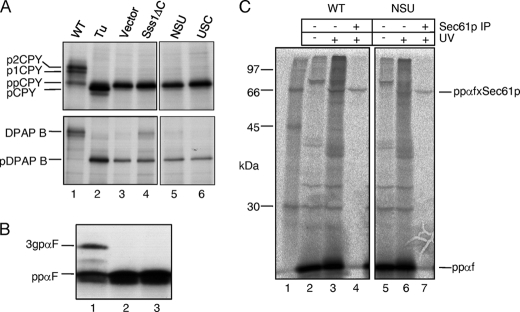
The impact of deleting or substituting Sss1p domains on protein translocation was analyzed after Sss1p depletion in cells expressing the various mutations. Radiolabeled cell extracts of FKY198 transformants were immunoprecipitated with antisera directed against ppCPY and DPAP B, which are translocated by the post- and co-translational pathways, respectively (20). The accumulation of both ppCPY and pDPAP B precursors upon Sss1p depletion confirmed the requirement for Sss1p in both translocation pathways (Fig. 5A, lane 3). Cells expressing Sss1ΔCp, NSUp, or USCp were also found to accumulate both ppCPY and pDPAP B precursors following Sss1p depletion (Fig. 5A, lanes 3–5), thus showing that the cytosolic and TM domains of Sss1p are essential for both the post- and co-translational modes of protein translocation.
FIGURE 5.
Protein translocation phenotypes of Sss1p domain mutants. A, FKY198 cells containing control or Sss1p mutant plasmids were grown in glucose medium for 6 h prior to 35S labeling for 5 min. Whole cell extracts were immunoprecipitated with ppCPY- or DPAP B-specific antiserum, and the precipitates were resolved by 10 and 7.5% SDS-PAGE, respectively (panels edited from the same gels). Upon ER translocation, ppCPY is signal-cleaved and modified by N-linked glycan addition in the ER (p1CPY) and the Golgi (p2CPY). The type II membrane protein, DPAP B, acquires N-linked glycans upon correct ER membrane integration. Tunicamycin (Tu) treatment yielded signal-cleaved, but unglycosylated pro-CPY (lane 2) and the unglycosylated pre-form of DPAP B (pDPAP B). B, post-translational translocation in vitro. Membranes from FKY198 cells containing Sss1p (lane 1), vector (lane 2), or the NSUp-expressing plasmids (lane 3) after 6 h in glucose were incubated with a 35S/TDBA-labeled K5/K14-ppαf substrate, with visualization after 12.5% SDS-PAGE. The 3gpαf translocated and glycosylated form is indicated. C, analysis of precursor targeting to Sec61p. Membranes prepared from FKY198 cells containing Sss1p (WT) or the NSUp-expressing plasmids after 6 h of growth in glucose medium were incubated with the 35S/TDBA-labeled K5/K14-ppαf after ATP-depletion. Two-thirds of each reaction was UV-irradiated, and half of this was precipitated with Sec61p-specific antiserum. The reaction products were visualized after 12.5% SDS-PAGE, and cross-links between ppαf and Sec61p are indicated (ppαfxSec61p). Lane 1 contains radiolabeled protein standards. The data in A and B are representative of two independent experiments, and the data in C are representative of three independent experiments.
Sss1p Is Required after Polypeptide Interaction with the Translocon
The role played by Sss1p was further characterized by examining the targeting of secretory polypeptides to the translocon. Initially, translocation competence in vitro was tested using a ppαf substrate containing photoreactive lysine residues at positions 5 and 14 of the signal sequence (K5/K14-ppαf). Incubation of this substrate with wild-type membranes generated the larger N-glycosylated 3gpαf species, indicating successful translocation (Fig. 5B, lane 1), whereas membranes from NSUp-expressing cells were completely defective in translocation (Fig. 5B, lane 2). These results demonstrate that the Sss1p TM domain is also essential for protein translocation in vitro. The same precursor was then used to assay translocon targeting, by monitoring cross-linking to Sec61p under conditions of ATP depletion, which prevents translocation to the ER lumen after translocon targeting. In wild-type membranes, a UV-dependent cross-linked adduct could be immunoprecipitated with Sec61p antiserum (Fig. 5C, lane 4), consistent with published data (15, 21). In the case of NSU mutant membranes, significant cross-linking of the ppαf signal sequence to Sec61p was also detected (Fig. 5B, lane 7) from which we conclude that the Sss1p TM domain is not essential for polypeptide targeting to Sec61p.
Extreme C Terminus Is Essential for Sss1p Function
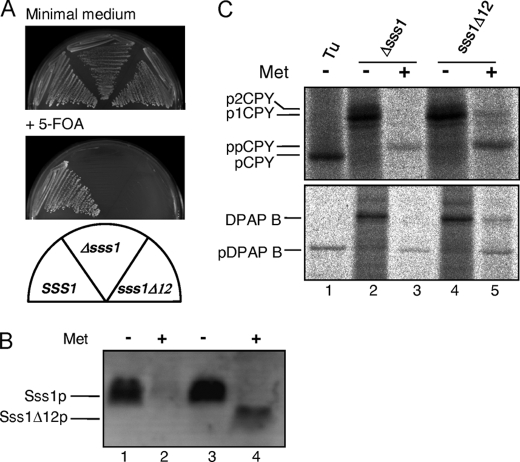
The NSUp fusion contains 12 amino acid residues of the extreme C terminus of Sss1p. The importance of this well conserved region of Sss1p (Fig. 1A, residues Lys69–Val80) retained in the NSUp fusion was addressed by analyzing a truncated allele lacking these residues (sss1Δ12). This allele was constructed by integration of a kanMX6 termination cassette immediately after the Ile68 codon of SSS1 in diploid strain W303-2n (see “Experimental Procedures”). Haploid sss1Δ12 spores were only obtained upon tetrad dissection in the presence of the SSS1, URA3 plasmid FKp53. The viability of this mutation was then checked by testing growth upon 5-FOA plasmid counterselection. It was found that the sss1Δ12 strain was unable to grow upon 5-FOA counterselection of the wild-type SSS1 plasmid, demonstrating that it is unable to provide Sss1p function (Fig. 6A). Expression of the truncated protein was examined in a strain expressing SSS1 under the control of a MET3 promoter both before and after methionine repression. The Sss1 protein was found to be largely depleted by methionine repression (Fig. 6B, lanes 2 and 4), but a faster migrating species corresponding to the truncated Sss1Δ12p was specifically detected in the sss1Δ12 strain only after Sss1p depletion (Fig. 6B, lane 4). This indicates that Sss1Δ12p is only stable in the absence of Sss1p, suggesting that the extreme C-terminal region may be required for efficient interaction with Sec61p. Defects in the translocation of DPAP B and ppCPY were also detected in the SssΔ12p-expressing strain after Sss1p depletion (Fig. 6, lane 5), indicating the requirement for the extreme C terminus of Sss1p in ER translocation.
FIGURE 6.
Expression and phenotypes of the sss1Δ12 mutation. A, haploid yeast strains bearing genomic wild-type SSS1, Δsss1, and sss1Δ12 alleles also containing a SSS1, URA3 plasmid (FKp53) were grown on minimal medium lacking uracil for plasmid selection and on 5-FOA medium for plasmid counterselection for 3 days. All strains grew normally on minimal medium with plasmid selection. After 3 days on 5-FOA medium, the wild-type strain grew as expected, and the Δsss1 strain was lethal upon plasmid counterselection as expected. The sss1Δ12 strain was also unable to grow on this medium, indicating the lethality of removing the C-terminal 12 residues of Sss1p. B, whole cell extracts prepared from the MET3-SSS1 strains BWY875 (Δsss1) and BWY886 (sss1Δ12) grown in minimal medium without methionine or in the presence of 2 mm methionine for 8 h (to deplete Sss1p) were subjected to 16% SDS-PAGE and immunoblot analysis with Sss1p antiserum. C, strains BWY875 and BWY886 grown as above were labeled with 14C-amino acids for 5 min. The sample in lane 1 was provided by BWY875 cells grown in the absence of methionine and treated with tunicamycin (Tu). Whole cell extracts were then immunoprecipitated with ppCPY- or DPAP B-specific antiserum, and the precipitates were resolved by 10 and 7.5% SDS-PAGE, respectively. For a description of ppCPY and DPAP B ER processing, see Fig. 5 legend. The data in B and C are representative of two independent experiments.
DISCUSSION
The Sss1 protein is an essential subunit of the ER protein translocon, but its function has been poorly understood. In this study we have analyzed the function of Sss1p by constructing a series of mutant derivatives based on sequence conservation and structural data. Particular attention has been paid to the role of the Sss1p TM domain, which has been shown to span the membrane diagonally to clamp the pseudo-symmetrical halves of Sec61p (2). Initially and somewhat surprisingly, mutation of three highly conserved glycine residues in the Sss1p TM domain did not result in growth or protein translocation phenotypes. However, the analysis of yeast cells expressing Sss1ΔCp demonstrated that deletion of the Sss1p TM resulted in cell inviability and protein translocation defects. The Sss1ΔC protein did not co-fractionate with membranes, indicating the requirement of the TM for membrane localization of Sss1p and thus explaining the phenotypes associated with its deletion. The possibility that the Sss1p TM domain acts as a clamp on the Sec61 translocon and thus suggesting a function in addition to that of a membrane anchor was analyzed by construction of the NSU TM transplacement allele. The NSU protein was stably membrane-integrated and maintained Sec61p interaction, but could not suppress the lethality of the Δsss1 mutation, and cells expressing the protein were accordingly unable to translocate secretory proteins across the ER membrane.
The Sss1p cytosolic domain was similarly analyzed by replacement with the topologically equivalent domain of Ubc6p to yield the USC protein. As with NSUp, USCp was unable to suppress the Δsss1 mutation, and cells expressing the protein were deficient in ER protein translocation. This demonstrated that the Sss1p cytosolic domain like the Sss1p TM domain is also essential for Sss1p function, cell viability, and protein translocation. The ability of NSUp to interact with Sec61p also strongly indicates a key role for the Sss1p cytosolic domain in the assembly of Sss1p into the translocon. Analysis of the Sss1Δ12p truncation showed that the extreme C terminus of Sss1p retained in NSUp is also essential for Sss1p function. This region of Sss1p is proposed to interact with the luminal end of Sec61p TM1, and thus this interaction may play a key role in the assembly of the functional translocon. The oxidative cross-linking observed between the Sss1p cytosolic domain and the cytosolic end of Sec61p TM9 is in agreement with structural data (2, 3). The depletion of Sss1p or the expression of domain mutations did not result in the instability of other translocon subunits. It is therefore concluded that the observed lethality and translocation defects are a direct reflection of the role played by Sss1p in ER protein translocation.
This study shows that the cytosolic and TM clamp domains of Sss1p are required for both the post- and co-translational modes of translocation in vivo. This strongly suggests that both domains are required at a crucial step during protein translocation irrespective of the mode of precursor targeting. This rules out roles in Signal Recognition Particle receptor or ribosome binding, and indeed, the structures of Sec61 complexes bound to nontranslating and translating ribosomes have not suggested any specific contacts with the γ-subunit of the translocon (22, 23). Sss1p-depleted membranes were found to be completely defective in the translocation of ppαf in vitro, consistent with the severe translocation phenotypes evident upon Sss1p depletion in vivo found in this study and in a previous report (4). NSUp-containing membranes were also unable to translocate ppαf in vitro. However, intriguingly, it was found that ppαf polypeptides were targeted efficiently to Sec61p in NSUp-containing membranes as detected by photocross-linking analysis. These findings indicate that the key role of Sss1p occurs after signal sequence targeting to the translocon.
Evidence has emerged that signal sequence insertion into the translocon channel leads to displacement of the plug domain and partial opening of the lateral gate (23–25). Our data demonstrate that signal sequence targeting to Sec61p can be completed in NSUp membranes from which we can conclude that the clamp domain is not essential for this initial targeting step. However, translocation cannot progress beyond this stage, and so the clamp domain is required to activate the precursor-translocon complex to initiate polypeptide translocation. The simplest model that would account for our results would involve a rearrangement of the Sss1p transmembrane clamp domain relative to the hinge domain within Sec61p that would promote lateral opening of the channel to allow the translocation substrate to fully access the translocon channel. Such a model would enable translocation to proceed and would give Sss1p an important role in restricting channel opening to prevent lipid flooding and perhaps to restrict small molecule leakage. Interestingly, structural models predict that as the lateral gate opens, the α-subunit TM 7, 8, and 9 domains would move outward, with TM9 in close proximity to the γ-subunit cytosolic domain (24). Our cross-linking data support this model because we see very efficient cross-linking between specific cysteine residues in TM9 of Sec61p with the cytosolic domain of Sss1p. Thus, then the two domains of Sss1p would act co-ordinately to complete channel opening and translocon activation.
Supplementary Material
Acknowledgments
We thank Adabella van der Zand for producing the Ubc6p antiserum. We also thank Stephen High, Tom Rapoport, and Sirrka Keränen for the TBDA-lysyl-tRNA, the K5ppαf plasmid, and the Sbh1p antiserum, respectively.
This work was supported by the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
- ER
- endoplasmic reticulum
- DPAP B
- dipeptidylaminopeptidase B
- 5-FOA
- 5-fluoroorotic acid
- ppαf
- pre-pro-α-factor
- ppCPY
- pre-pro-carboxypeptidase Y
- TBDA
- 4-(3-trifluorodiazirino) benzoic acid
- TM
- transmembrane.
REFERENCES
- 1.Osborne A. R., Rapoport T. A., van den Berg B. (2005) Annu. Rev. Cell Dev. Biol. 21, 529–550 [DOI] [PubMed] [Google Scholar]
- 2.van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 3.Mitra K., Schaffitzel C., Shaikh T., Tama F., Jenni S., Brooks C. L., 3rd, Ban N., Frank J. (2005) Nature 438, 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esnault Y., Blondel M. O., Deshaies R. J., Scheckman R., Képès F. (1993) EMBO J. 12, 4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas B. J., Rothstein R. (1989) Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson B. M., Esnault Y., Craven R. A., Skiba F., Fieschi J., Képès F., Stirling C. J. (1997) EMBO J. 16, 4549–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson B. M., Tyson J. R., Reid P. J., Stirling C. J. (2000) J. Biol. Chem. 275, 521–529 [DOI] [PubMed] [Google Scholar]
- 8.Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tyson J. R., Stirling C. J. (2000) EMBO J. 19, 6440–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson B. M., Critchley A. J., Stirling C. J. (1996) J. Biol. Chem. 271, 25590–25597 [DOI] [PubMed] [Google Scholar]
- 11.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 12.Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. (1992) Mol. Biol. Cell 3, 129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toikkanen J., Gatti E., Takei K., Saloheimo M., Olkkonen V. M., Söderlund H., DeCamilli P., Keränen S. (1996) Yeast 12, 425–438 [DOI] [PubMed] [Google Scholar]
- 14.Young B. P., Craven R. A., Reid P. J., Willer M., Stirling C. J. (2001) EMBO J. 20, 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plath K., Mothes W., Wilkinson B. M., Stirling C. J., Rapoport T. A. (1998) Cell 94, 795–807 [DOI] [PubMed] [Google Scholar]
- 16.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 17.Feldheim D., Yoshimura K., Admon A., Schekman R. (1993) Mol. Biol. Cell 4, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esnault Y., Feldheim D., Blondel M. O., Schekman R., Képès F. (1994) J. Biol. Chem. 269, 27478–27485 [PubMed] [Google Scholar]
- 19.Panzner S., Dreier L., Hartmann E., Kostka S., Rapoport T. A. (1995) Cell 81, 561–570 [DOI] [PubMed] [Google Scholar]
- 20.Ng D. T., Brown J. D., Walter P. (1996) J. Cell Biol. 134, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plath K., Wilkinson B. M., Stirling C. J., Rapoport T. A. (2004) Mol. Biol. Cell 15, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ménétret J. F., Hegde R. S., Aguiar M., Gygi S. P., Park E., Rapoport T. A., Akey C. W. (2008) Structure 16, 1126–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker T., Bhushan S., Jarasch A., Armache J. P., Funes S., Jossinet F., Gumbart J., Mielke T., Berninghausen O., Schulten K., Westhof E., Gilmore R., Mandon E. C., Beckmann R. (2009) Science 326, 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmer J., Nam Y., Rapoport T. A. (2008) Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.du Plessis D. J., Berrelkamp G., Nouwen N., Driessen A. (2009) J. Biol. Chem. 284, 15805–15814 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.