Summary
Non-small cell lung cancer (NSCLC) is still the leading cause of cancer-related deaths. The effect of the PI3K/PTEN/AKT/mTOR signaling pathway on cancer treatment, including NSCLC, has been well documented. In this study, we analyzed associations between genetic variations within this pathway and clinical outcomes following platinum-based chemotherapy in 168 patients with stage IIIB (wet) or stage IV NSCLC. Sixteen tagging SNPs in five core genes (PIK3CA, PTEN, AKT1, AKT2, and FRAP1) of this pathway and identified SNPs associated with development of toxicity and disease progression. We observed significantly increased toxicity for patients with PIK3CA:rs2699887 (OR: 3.86, 95% CI: 1.08 – 13.82). In contrast, a SNP in PTEN was associated with significantly reduced risk for chemotherapeutic toxicity (OR: 0.44, 95% CI: 0.20 - 0.95). We identified three SNPs in AKT1 resulting in significantly decreased risks of distant progression in patients carrying at least one variant allele with HRs of 0.66 (95% CI: 0.45 - 0.97), 0.52 (95% CI: 0.35 - 0.77), and 0.62 (95% CI: 0.42 - 0.91) for rs3803304, rs2498804, and rs1130214, respectively. Furthermore, these same variants conferred nearly two-fold increased progression-free survival times. The current study provides evidence that genetic variations within the PI3K/PTEN/AKT/mTOR signaling pathway are associated with variation in clinical outcomes of NSCLC patients. With further validation, our findings may provide additional biomarkers for customized treatment of platinum-based chemotherapy for NSCLC.
Keywords: lung cancer, chemotherapy, platinum-agents, AKT, clinical outcomes
Introduction
Lung cancer is the leading cause of cancer mortality in the United States with over 160,000 deaths estimated in 2009 [1]. Approximately 80% of lung cancer cases are non-small cell lung cancer (NSCLC) and of these, a majority present with advanced stage [2]. The prognosis for these patients is poor with few options for treatment – including chemotherapy and radiation [3]. Because of this, there is a need for better, and more individualized treatment options for advanced NSCLC.
Platinum-based chemotherapeutic agents, such as cisplatin and carboplatin, are used to treat various types of cancers including lung cancer. Unfortunately, although platinum-based combination chemotherapy has enhanced overall survival and quality of life for lung cancer patients, the 1-year survival rate is still only 29% [4]. The major hurdles in the use of platinum agents are the development of chemoresistance and severe side effects [5, 6]. The most common side effects include ototoxicity, neuropathy, nephrotoxicity, and myelosuppression. These effects are thought to be caused by increased production of reactive oxygen species and apoptosis in sensitive tissues. Several factors are known to influence a patient's response to therapy, including age, ethnicity, stage of disease, performance status, and co-morbidities. However, a patient's genetic background may also play an important role in modulating response to therapy. Therefore, one strategy to enhance the effectiveness of platinum-based treatment of NSCLC while avoiding adverse events is to gain a better understanding of the influence of genetic variations on the clinical outcome of patients.
Platinum-containing agents are cytotoxic through the creation of platinum-DNA crosslinks and the induction of cell cycle arrest and ultimately apoptosis if not properly repaired [7]. Several pathways are involved in this process, including the PI3K/PTEN/AKT/mTOR pathway that is responsible for balancing cell survival and apoptosis [8, 9]. This pathway is activated in various cancer types and plays a role in the development of chemoresistance to platinum-based chemotherapy [10-14]. This pathway is complex; however, the core components include PI3K (phosphoinositide-3-kinase), PTEN (phosphatase and tensin homolog), AKT (v-akt murine thymoma viral oncogene homolog) and mTOR (mammalian target of rapamycin). Genetic variations in the genes encoding these important molecules may modulate signaling through this pathway and result in variation in the development of toxicity or clinical outcomes following platinum-based therapy.
Genetic variations within the PI3K/PTEN/AKT/mTOR have recently been reported to modulate clinical outcomes in esophageal cancer [15]. Because of the variation in response to platinum-based chemotherapy in NSCLC patients, there is a need for efficient biomarkers to predict who will benefit from the chemotherapy while avoiding the development of unnecessary adverse events. In the current study, we set out to determine the association between genetic variations in AKT1, AKT2, PIK3CA (catalytic subunit of PI3K), PTEN, and FRAP1 (encoding for mTOR) with development of toxicity and disease progression in NSCLC patients treated with platinum-compounds.
Materials and Methods
Patient Population
All of the patients were selected from an ongoing epidemiology lung cancer study. The patients included in this analysis were enrolled from 1995 to 2004 and were newly diagnosed, histological confirmed NSCLC cases treated with primary platinum-based (carboplatin or cisplatin) combination chemotherapy at the University of Texas M. D. Anderson Cancer Center. We further restricted the analysis to non-Hispanic Caucasian patients with stage IIIB (wet) or IV NSCLC. All the subjects signed a consent form and the study was approved by the Institutional Review Board of The University of Texas M. D. Anderson Cancer Center. Peripheral blood specimens for genetic analysis were collected from each patient at the time of diagnosis prior to chemotherapy or radiotherapy treatment.
Epidemiological and Clinical Data Collection
Epidemiological data was collected using a structured questionnaire including demographic characteristics, family history of cancer, smoking history, and alcohol consumption. We defined an individual who had never smoked or had smoked no more than 100 cigarettes in his or her lifetime as never smoker; an individual who had quit smoking at least one year before diagnosis was defined as former smoker; a person who currently smoking or had quit smoking less than one year prior to diagnosis was defined as current & recent quitter. Clinical and follow-up information were abstracted from medical records. Performance status was determined based on the ECOG scale prior to treatment [16]. Complete blood counts were performed prior to each treatment based on M. D. Anderson's practice guidelines. Toxicities included in this study were neutropenia, neutropenic fever, anemia, thromobocytopenia, leukocytopenia, and nephrotoxicity that occurred during any of the primary chemotherapy treatment cycles [17]. Time to progression was measured from date of first treatment to date of progression of disease, last follow-up or death. Local progression was limited to primary tumor site and regional lymph nodes while distant progression was defined as a metastasis located outside of the thoracic cavity or in the other lung.
SNP Selection and Genotyping
Genomic DNA was extracted from peripheral blood lymphocytes using the Human Whole Blood Genomic DNA Extraction Kit (Qiagen, Valencia, CA). We selected tagging SNPs from 5-kb flanking and within the gene regions of five genes: AKT1, AKT2, PIK3CA, PTEN and FRAP1 (mTOR). Sixteen tagging SNPs were identified by the tagger algorithm with a cut-off value of r2 = 0.8 and a MAF (minor allele frequency) = 0.1-0.35, based on the allele frequencies from CEPH samples that were genotyped by the International HapMap Project. For each SNP, genotyping was performed using the TaqMan Pre-Designed SNP Genotyping Assays (Applied Biosystems, Foster City, CA) following manufacturer's instructions. End-point fluorescence was read by ABI Prism 7900HT Sequence Detection System with genotype calls being made with SDS software (SDS 2.1, Applied Biosystems, Foster City, CA).
Statistical Analysis
For toxicity risk, unconditional multivariate logistic regression analysis was performed to estimate adjusted odds ratios (ORs) along with the corresponding 95% confident intervals (95% CIs) for each SNP. The Cox proportional hazard model was used to assess the effect of individual SNPs on progression (local and distant)-free survival. Hazard ratios (HRs) and 95% CIs were estimated by fitting the Cox model while adjusting for age, gender, clinical stage, performance status, and smoking status. Kaplan-Meier curves and log-rank tests were used to assess progression-free survival time. All statistical analyses were performed using STATA software (version 10, STATA Corporation, College Station, TX) with P < 0.05 being considered statistically significant. The Benjamini-Hochberg method was used to correct for multiple comparisons based on an false discovery rate (FDR) of 10% [18].
Results
Patient Characteristics
Our patient population consisted of 168 non-Hispanic Caucasian patients with advanced stage NSCLC who received primary platinum-based chemotherapy (Table 1). A majority were treated with carboplatin-based treatment (88.7%) compared to cisplatin-based (11.3%) with an average number of treatment cycles of 4.5. Seventeen (10.1%) presented with stage IIIB (wet) and 151 (89.9%) with stage IV disease. The mean age was 58.1 years (SD: 11.08, range: 28-81 years). There were 94 men (56%) and 74 women (44%). There were 48 (28.6%) never smokers, 58 (34.5%) former smokers, and 62 (36.9%) current smokers or recent quitters. The median time enrolled in the study was 10.94 months with an overall median survival time of 10.92 months.
Table 1. Patient Characteristics.
| Characteristic | # of Patients | % |
|---|---|---|
| Total | 168 | |
| Age | ||
| Mean | 58.1 | |
| SD | 11.08 | |
| Range | 28-81 | |
| Sex | ||
| Male | 94 | 56 |
| Female | 74 | 44 |
| Clinical Stage | ||
| Stage IIIB (wet) | 17 | 10.1 |
| Stage IV | 151 | 89.9 |
| Smoking Status | ||
| Never | 48 | 28.6 |
| Former | 58 | 34.5 |
| Current & Recent Quitter | 62 | 36.9 |
| Performance Status | ||
| 0 | 38 | 22.6 |
| 1 | 109 | 64.9 |
| 2-4 | 21 | 12.5 |
| Treatment Regimen | ||
| Carboplatin-based | 149 | 88.7 |
| Cisplatin-based | 19 | 11.3 |
| Local Progression | ||
| No | 102 | 60.7 |
| Yes | 66 | 39.3 |
| Distant Progression | ||
| No | 51 | 30.4 |
| Yes | 117 | 69.6+ |
| Toxicity | ||
| No | 100 | 59.5 |
| Yes | 68 | 40.5 |
| Histology | ||
| Adenocarcinoma | 102 | 60.7 |
| Non-small cell carcinoma | 23 | 13.7 |
| Squamous cell cacinoma | 24 | 14.3 |
| Other NSCLC | 19 | 11.3 |
Associations between SNPs and Risk of Toxicity
We analyzed the 16 SNPs for associations with toxicity due to platinum-based chemotherapy. Two SNPs were found to be significantly associated with toxicity (Table 2). PTEN:rs2299939 showed a negative association with patients carrying at least one variant allele having a 56% reduced risk of developing a severe side effect (OR: 0.44, 95% CI: 0.20 - 0.95, P = 0.036). In contrast, patients who were homozygous for the PIK3CA:rs2699887 variant exhibited a significantly increased risk of toxicity (OR: 3.86, 95% CI: 1.08 - 13.82, P = 0.038). Both of these associations remained significant after correct for multiple comparisons at an FDR of 10%. No other SNPs were significantly associated with toxicity risk. Because cisplatin and carboplatin-based treatment regimens differ slightly in toxicity profiles, we stratified our analysis by platinum agent. The results in the carboplatin treatment group were similar to the full population (data not shown). Due to small sample size, we were not able to perform stratified analysis in the cisplatin group.
Table 2. PI3K/PTEN/AKT/mTOR pathway genotypes and toxicity.
| SNP and Genotype | Toxicity | ||||
|---|---|---|---|---|---|
| Yes | No | OR* | 95% CI | P value | |
| AKT1:rs3803304 | 62 | 93 | |||
| CC | 26 | 49 | 1.00 | ||
| CG | 30 | 37 | 1.43 | 0.71 to 2.86 | 0.317 |
| GG | 6 | 7 | 1.67 | 0.50 to 5.60 | 0.408 |
| CG + GG | 1.46 | 0.75 to 2.84 | 0.261 | ||
| AKT1:rs2494738 | 65 | 99 | |||
| AA | 53 | 81 | 1.00 | ||
| AG | 11 | 18 | |||
| GG | 1 | 0 | |||
| AG+GG | 1.04 | 0.45 to 2.42 | 0.921 | ||
| AKT1:rs2498804 | 66 | 97 | |||
| GG | 21 | 41 | 1.00 | ||
| GT | 37 | 46 | 1.45 | 0.71 to 2.95 | 0.305 |
| TT | 8 | 10 | 1.65 | 0.55 to 4.96 | 0.373 |
| GT + TT | 1.48 | 0.75 to 2.94 | 0.257 | ||
| AKT1:rs1130214 | 66 | 97 | |||
| GG | 30 | 54 | 1.00 | ||
| GT | 29 | 38 | 1.38 | 0.69 to 2.77 | 0.364 |
| TT | 7 | 5 | 2.92 | 0.80 to 10.67 | 0.105 |
| GT + TT | 1.55 | 0.79 to 3.01 | 0.200 | ||
| AKT2:rs892119 | 65 | 97 | |||
| AA | 40 | 72 | 1.00 | ||
| AG | 24 | 23 | 2.06 | 1.00 to 4.27 | 0.051 |
| GG | 1 | 2 | 1.00 | 0.08 to 12.26 | 1.000 |
| AG + GG | 1.97 | 0.97 to 4.02 | 0.061 | ||
| AKT2:rs8100018 | 63 | 95 | |||
| CC | 39 | 46 | 1.00 | ||
| CG | 20 | 39 | 0.54 | 0.26 to 1.12 | 0.096 |
| GG | 4 | 10 | 0.42 | 0.12 to 1.52 | 0.186 |
| CG + GG | 0.51 | 0.26 to 1.02 | 0.056 | ||
| FRAP1:rs11121704 | 66 | 97 | |||
| CC | 32 | 56 | 1.00 | ||
| CT | 28 | 36 | 1.31 | 0.65 to 2.62 | 0.448 |
| TT | 6 | 5 | 2.79 | 0.73 to 10.61 | 0.133 |
| CT + TT | 1.47 | 0.76 to 2.84 | 0.253 | ||
| FRAP1:rs2295080 | 66 | 98 | |||
| GG | 27 | 49 | 1.00 | ||
| GT | 32 | 42 | 1.40 | 0.71 to 2.76 | 0.333 |
| TT | 7 | 7 | 2.18 | 0.66 to 7.20 | 0.202 |
| GT + TT | 1.50 | 0.78 to 2.88 | 0.224 | ||
| PIK3CA:rs7651265 | 66 | 98 | |||
| AA | 58 | 76 | 1.00 | ||
| AG | 8 | 21 | |||
| GG | 0 | 1 | |||
| AG + GG | 0.43 | 0.17 to 1.06 | 0.065 | ||
| PIK3CA:rs7640662 | 65 | 99 | |||
| CC | 45 | 74 | 1.00 | ||
| CG | 17 | 24 | 1.05 | 0.49 to 2.25 | 0.909 |
| GG | 3 | 1 | 4.52 | 0.44 to 46.59 | 0.205 |
| CG + GG | 1.19 | 0.57 to 2.48 | 0.649 | ||
| PIK3CA:rs7621329 | 65 | 99 | |||
| CC | 47 | 63 | 1.00 | ||
| CT | 18 | 30 | |||
| TT | 0 | 6 | |||
| CT+TT | 0.63 | 0.31 to 1.28 | 0.202 | ||
| PIK3CA:rs6443624 | 66 | 99 | |||
| AA | 42 | 55 | 1.00 | ||
| AC | 24 | 36 | |||
| CC | 0 | 8 | |||
| AC + CC | 0.71 | 0.37 to 1.37 | 0.310 | ||
| PIK3CA:r52699887 | 65 | 98 | |||
| AA | 34 | 56 | 1.00 | ||
| AG | 21 | 38 | 0.93 | 0.46 to 1.89 | 0.845 |
| GG | 10 | 4 | 3.86 | 1.08 to 13.82 | 0.038 |
| AG + GG | 1.21 | 0.62 to 2.34 | 0.576 | ||
| PTEN:rs2299939 | 64 | 93 | |||
| AA | 51 | 58 | 1.00 | ||
| AC | 13 | 29 | |||
| CC | 0 | 6 | |||
| AC + CC | 0.44 | 0.20 to 0.95 | 0.036 | ||
| PTEN:rs12569998 | 66 | 99 | |||
| GG | 46 | 75 | 1.00 | ||
| GT | 19 | 23 | 1.40 | 0.66 to 2.97 | 0.386 |
| TT | 1 | 1 | 4.48 | 0.20 to 100.80 | 0.346 |
| GT + TT | 1.45 | 0.69 to 3.06 | 0.323 | ||
| PTEN:rs12357281 | 66 | 99 | |||
| CC | 61 | 86 | 1.00 | ||
| CG | 5 | 12 | |||
| GG | 0 | 1 | |||
| CG + GG | 0.54 | 0.17 to 1.66 | 0.281 | ||
adjusted for age, gender, clinical stage, performance status, and smoking status
Associations between SNPs and Progression Risk and Progression-free Survival
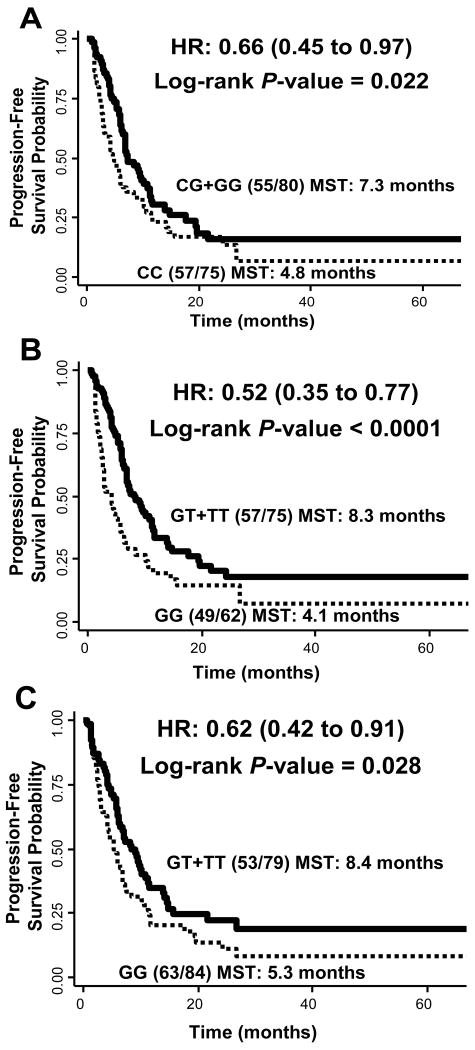
We next analyzed the association between SNPs and distant progression. None of the variants were associated with local progression (Table 3), but three of the 16 SNPs were associated with distant progression risk. These SNPs (rs3803304, rs2498804, rs1130214) all tagged genetic variation in AKT1. Patients carrying at least one variant allele exhibited similarly reduced risks with HRs of 0.66 (95% CI: 0.45 - 0.97, P = 0.035), 0.52 (95% CI: 0.35 - 0.77, P = 0.001) and 0.62 (95% CI: 0.42 - 0.91, P = 0.016), respectively. Although only AKT1:rs2498804 remained at an FDR of 10%, as shown in Figure 1, all three of these variants conferred a nearly two-fold prolonged progression-free time, from 4.84 to 7.30 months for rs3803304 (P = 0.022), 4.11 to 8.29 months for rs2498804 (P = 0.0005), and 5.3 to 8.42 months for rs1130214 (P = 0.028). Table 4 shows the linkage disequilibrium (LD) between the four AKT1 SNPs included in this analysis. Of the three identified as significantly associated with risk, only rs3803304 and rs2498804 exhibited modest LD (r2=0.75), with rs1130214 not sharing any LD with these two SNPs (r2=0.00 and 0.16, respectively). Similar to the toxicity analysis, the results in the carboplatin treatment group were comparable to the full population (data not shown).
Table 3. PI3K/PTEN/AK17mTOR pathway genotypes and disease progression.
| SNP and Genotype | Local Progression | Distant Progression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Yes | No | HR* | 95% CI | P value | Yes | No | HR* | 95% CI | P value | |
| AKT1:rs3803304 | 58 | 97 | 112 | 43 | ||||||
| CC | 28 | 47 | 1.00 | 57 | 18 | 1.00 | ||||
| CG | 22 | 45 | 0.80 | 0.45 to 1.42 | 0.448 | 44 | 23 | 0.61 | 0.41 to 0.92 | 0.017 |
| GG | 8 | 5 | 1.59 | 0.71 to 3.54 | 0.261 | 11 | 2 | 0.97 | 0.50 to 1.86 | 0.921 |
| CG + GG | 0.93 | 0.55 to 1.57 | 0.790 | 0.66 | 045 to 0.97 | 0.035 | ||||
| AKT1:rs2494738 | 65 | 99 | 117 | 47 | ||||||
| AA | 52 | 82 | 1.00 | 96 | 38 | 1.00 | ||||
| AG | 12 | 17 | 0.94 | 0.49 to 1.79 | 0.847 | 20 | 9 | |||
| GG | 1 | 0 | 1 | 0 | ||||||
| AG+GG | 0.99 | 0.53 to 1.86 | 0.985 | 0.80 | 0.49 to 1.29 | 0.357 | ||||
| AKT1:rs2498804 | 64 | 99 | 116 | 47 | ||||||
| GG | 24 | 38 | 1.00 | 49 | 13 | 1.00 | ||||
| GT | 30 | 53 | 0.80 | 0.46 to 1.40 | 0.432 | 52 | 31 | 0.47 | 0.31 to 0.72 | < 0.001 |
| TT | 10 | 8 | 1.26 | 0.60 to 2.68 | 0.540 | 15 | 3 | 0.76 | 0.42 to 1.37 | 0.357 |
| GT + TT | 0.89 | 0.53 to 1.50 | 0.661 | 0.52 | 0.35 to 0.77 | 0.001 | ||||
| AKT1:rs1130214 | 64 | 99 | 116 | 47 | ||||||
| GG | 35 | 49 | 1.00 | 63 | 21 | 1.00 | ||||
| GT | 24 | 43 | 0.74 | 0.43 to 1.27 | 0.275 | 45 | 22 | 0.60 | 0.40 to 0.90 | 0.013 |
| TT | 5 | 7 | 0.91 | 0.35 to 2.36 | 0.840 | 8 | 4 | 0.77 | 0.36 to 1.63 | 0.492 |
| GT+TT | 0.76 | 0.45 to 1.28 | 0.305 | 0.62 | 0.42 to 0.91 | 0.016 | ||||
| AKT2:rs892119 | 65 | 97 | 114 | 48 | ||||||
| AA | 46 | 66 | 1.00 | 80 | 32 | 1.00 | ||||
| AG | 18 | 29 | 0.93 | 0.53 to 1.63 | 0.788 | 32 | 15 | 0.83 | 0.54 to 1.25 | 0.370 |
| GG | 1 | 2 | 0.97 | 0.13 to 7.26 | 0.973 | 2 | 1 | 0.68 | 0.16 to 2.84 | 0.595 |
| AG + GG | 0.93 | 0.53 to 1.61 | 0.790 | 0.82 | 0.54 to 1.23 | 0.328 | ||||
| AKT2:rs8100018 | 60 | 98 | 110 | 48 | ||||||
| CC | 30 | 55 | 1.00 | 57 | 28 | 1.00 | ||||
| CG | 24 | 35 | 1.17 | 0.67 to 2.02 | 0.588 | 43 | 16 | 1.31 | 0.86 to 1.93 | 0.208 |
| GG | 6 | 8 | 1.09 | 0.43 to 2.76 | 0.856 | 10 | 4 | 1.30 | 0.66 to 2.56 | 0.456 |
| CG + GG | 1.15 | 0.68 to 1.93 | 0.599 | 1.30 | 0.88 to 1.92 | 0.181 | ||||
| FRAP1:rs11121704 | 65 | 98 | 115 | 48 | ||||||
| CC | 34 | 54 | 1.00 | 64 | 24 | 1.00 | ||||
| CT | 26 | 38 | 1.14 | 0.67 to 1.94 | 0.622 | 43 | 21 | 0.73 | 0.49 to 1.10 | 0.130 |
| TT | 5 | 6 | 1.34 | 0.51 to 3.57 | 0.553 | 8 | 3 | 0.81 | 0.38 to 1.74 | 0.593 |
| CT + TT | 1.17 | 0.71 to 1.94 | 0.528 | 0.75 | 0.51 to 1.09 | 0.129 | ||||
| FRAP1:rs2295080 | 63 | 101 | 117 | 47 | ||||||
| GG | 26 | 50 | 1.00 | 55 | 21 | 1.00 | ||||
| GT | 30 | 44 | 1.21 | 0.71 to 2.06 | 0.486 | 51 | 23 | 0.77 | 0.52 to 1.13 | 0.186 |
| TT | 7 | 7 | 1.70 | 0.72 to 4.03 | 0.224 | 11 | 3 | 0.91 | 0.47 to 1.77 | 0.784 |
| GT + TT | 1.28 | 0.77 to 2.12 | 0.343 | 0.79 | 0.55 to1.14 | 0.215 | ||||
| PIK3CA:rs7651265 | 65 | 99 | 116 | 48 | ||||||
| AA | 53 | 81 | 1.00 | 94 | 40 | 1.00 | ||||
| AG | 11 | 18 | 0.96 | 0.50 to 1.85 | 0.901 | 21 | 8 | 0.98 | 0.60 to 1.58 | 0.919 |
| GG | 1 | 0 | 1 | 0 | ||||||
| AG + GG | 1.04 | 0.55 to 1.96 | 0.902 | 1.02 | 0.64 to 1.64 | 0.921 | ||||
| PIK3CA:rs7640662 | 65 | 99 | 116 | 48 | ||||||
| CC | 46 | 73 | 1.00 | 83 | 36 | 1.00 | ||||
| CG | 16 | 25 | 1.1 | 0.60 to 2.02 | 0.753 | 31 | 10 | 1.06 | 0.69 to 1.64 | 0.791 |
| GG | 3 | 1 | 2.29 | 0.67 to 7.81 | 0.186 | 2 | 2 | 0.48 | 0.11 to 2.01 | 0.312 |
| CG + GG | 1.20 | 0.68 to 2.12 | 0.538 | 0.99 | 0.65 to 1.52 | 0.974 | ||||
| PIK3CA:rs7621329 | 65 | 99 | 117 | 47 | ||||||
| CC | 44 | 66 | 1.00 | 79 | 31 | 1.00 | ||||
| CT | 17 | 31 | 0.71 | 0.40 to 1.26 | 0.238 | 34 | 14 | 0.76 | 0.51 to 1.15 | 0.197 |
| TT | 4 | 2 | 1.95 | 0.68 to 5.61 | 0.213 | 4 | 2 | 0.77 | 0.27 to 2.21 | 0.629 |
| CT + TT | 0.82 | 0.48 to 1.39 | 0.460 | 0.76 | 0.51 to 1.14 | 0.183 | ||||
| PIK3CA:rs6443624 | 65 | 100 | 117 | 48 | ||||||
| AA | 38 | 59 | 1.00 | 69 | 28 | 1.00 | ||||
| AC | 22 | 38 | 0.81 | 0.47 to 1.39 | 0.439 | 44 | 16 | 0.88 | 0.59 to 1.30 | 0.516 |
| CC | 5 | 3 | 1.66 | 0.63 to 4.39 | 0.307 | 4 | 4 | 0.58 | 0.20 to 1.65 | 0.307 |
| AC + CC | 0.91 | 0.55 to 1.51 | 0.705 | 0.84 | 0.58 to 1.22 | 0.360 | ||||
| PIK3CA:rs2699887 | 65 | 98 | 115 | 48 | ||||||
| AA | 34 | 56 | 1.00 | 67 | 23 | 1.00 | ||||
| AG | 25 | 34 | 1.28 | 0.76 to 2.18 | 0.353 | 37 | 22 | 0.77 | 0.51 to 1.17 | 0.225 |
| GG | 6 | 8 | 1.17 | 0.47 to 2.89 | 0.733 | 11 | 3 | 0.92 | 0.47 to 1.80 | 0.800 |
| AG + GG | 1.26 | 0.77 to 2.08 | 0.362 | 0.80 | 0.54 to 1.18 | 0.258 | ||||
| PTEN:rs2299939 | 61 | 96 | 110 | 47 | ||||||
| AA | 45 | 64 | 1.00 | 77 | 32 | 1.00 | ||||
| AC | 15 | 27 | 0.78 | 0.42 to 1.43 | 0.416 | 31 | 11 | 0.94 | 0.61 to 1.44 | 0.770 |
| CC | 1 | 5 | 1.54 | 0.20 to 11.82 | 0.681 | 2 | 4 | 1.16 | 0.28 to 4.80 | 0.839 |
| AC + CC | 0.80 | 0.44 to 1.45 | 0.466 | 0.95 | 0.62 to 1.45 | 0.806 | ||||
| PTEN:rs12569998 | 65 | 100 | 117 | 48 | ||||||
| GG | 48 | 73 | 1.00 | 83 | 38 | 1.00 | ||||
| GT | 15 | 27 | 1.10 | 0.60 to 2.02 | 0.761 | 32 | 10 | 1.34 | 0.86 to 2.07 | 0.195 |
| TT | 2 | 0 | 2 | 0 | ||||||
| GT + TT | 1.23 | 0.69 to 2.19 | 0.492 | 1.40 | 0.91 to 2.14 | 0.127 | ||||
| PTEN:rs12357281 | 65 | 100 | 117 | 48 | ||||||
| CC | 56 | 91 | 1.00 | 105 | 42 | 1.00 | ||||
| CG | 8 | 9 | 0.94 | 0.42 to 2.07 | 0.876 | 11 | 6 | 0.98 | 0.51 to 1.88 | 0.945 |
| GG | 1 | 0 | 1 | 0 | 1.10 | 0.14 to 8.41 | 0.929 | |||
| CG + GG | 1.08 | 0.51 to 2.29 | 0.848 | 0.99 | 0.53 to 1.84 | 0.969 | ||||
adjusted for age, gender, clinical stage, performance status, and smoking status
Fig 1.
Kaplan-Meier curves of distant progression-free survival times in lung cancer patients by AKT1 SNPs A) rs3803304, B) rs2498804, and C) rs1130214. The numbers in parentheses are the numbers of patients with progression over the total number of patients by genotype. MST = median time to progression in months.
Table 4. Linkage disequilibrium (r2) between AKT1 SNPs.
| AKT1:rs3803304 | AKT1:rs2498804 | AKT1:rs1130214 | |
|---|---|---|---|
| AKT1:rs2498804 | 0.75 | ||
| AKT1:rs1130214 | 0.00 | 0.16 | |
| AKT1: rs2494738 | 0.11 | 0.12 | 0.00 |
Discussion
Lung cancer has remained the leading cause for cancer-related mortality in the United States [1]. A growing body of evidence suggests that lung tumors activate certain cellular signaling pathways to become invasive and resistant to platinum-based chemotherapy [19]. The deregulation of the PI3K/PTEN/AKT/mTOR pathway in human cancers has been extensively studied over the past few years [20-23]. Furthermore, this pathway has been reported to be associated with response to platinum-based chemotherapy treatment in lung cancer cell lines [13, 24]. In this study, we determined whether common variations in genes in this pathway (PIK3CA, PTEN, AKT1, AKT2, and FRAP1) were able to modulate the development of toxicity and clinical outcomes of NSCLC patients receiving platinum-based chemotherapy.
Although platinum-based agents are successful in treating several types of cancer, treatment is often associated with adverse side effects, including myleosuppression, ototoxicity, nephrotoxicity, and peripheral neurotoxicity due to increased apoptosis in cells with platinum-related DNA damage [5, 6]. Cisplatin and carboplatin are the most commonly used platinum-containing chemotherapeutic agents in NSCLC. Cisplatin-based therapy has been found to provide a better survival benefit for NSCLC patients, but it is associated with more severe toxicity compared to carboplatin-based therapy. However, treatment with either of these agents is hindered by development of severe toxicities and chemoresistance [25].
Since the PI3K/PTEN/AKT/mTOR pathway is involved in the balance between cell survival and death, genetic variation in the core components of this pathway may shift this balance, resulting in altered toxicity risk. In the current study, a genetic variation (rs2299939) in the negative regulator of this pathway, PTEN, was associated with a 54% decreased risk of toxicity. PTEN protein expression is often lost in NSCLC, but this loss is rarely due to inactivating mutations, loss of heterozygosity, or hypermethlyation of the gene [26-30]. Our results suggest that genetic variations in PTEN may modulate PTEN activity. Specifically, since rs2299939 is associated with a decrease in toxicity, we speculate that this SNP, or another functional SNP that is tagged by this variant, may decrease the expression of PTEN and hence the inhibitory effect of PTEN on signaling through this pathway. Further investigation will be needed to understand the effect of this SNP on PTEN function. In contrast, patients carrying at least one variant rs2699887 allele in PIK3CA, the catalytic domain for PI3K, had a nearly 4-fold increased risk of toxicity. PIK3CA is a known oncogene and is responsible for initiating signaling through this pathway activating cell survival signals [31]. Decreased PI3K activity would result increased apoptosis in sensitive, non-cancer cells causing an increase in toxic side-effects. Therefore, the genetic variation tagged by the PIK3CA:rs2699887 SNP would likely cause an decrease in PI3K signaling. The contrasting results of PTEN and PIK3CA genetic variation have biological plausibility based on their function in regulating signaling through this pathway.
The serine-threonine kinase AKT is a central node in cell signaling that regulates several processes, including cell survival, proliferation, and protein synthesis [8]. AKT activation is a common molecular alteration during carcinogenesis, and it has been reported that AKT is constitutively activated in NSCLC resulting in cell survival by blocking induction of apoptosis [32]. In addition, forced expression of AKT1 was found to be sufficient to regulate cisplatin resistance in cultured lung cancer cells [13]. We found that three AKT1 tagging SNPs decreased risk for distant disease progression. These three SNPs do not share a high degree of linkage disequilibrium, suggesting the presence of at least two independent causal variants. The directionality of the effect indicates that the functional variants all diminish AKT1 activity causing decreased signaling through this pathway, and thus a reduction in cell survival signals. However, since the variants genotyped in this study were tagging SNPs, we are unable to identify the causative SNP and mechanism responsible. Future studies are clearly warranted in this regard.
Platinum-based chemotherapy is still the core treatment for NSCLC patients. Although knowledge and chemotherapeutic methods for treating NSCLC keep evolving, the survival rate has not improved notably with chemotherapy. New biologic insight and biomarkers are desired to find new approaches for treating patients with advanced disease. In the current study, although based on a small sample size, the homogenous nature of the treatment regimens the patients received allowed us to identify genetic variations within the PI3K/PTEN/AKT/mTOR signaling pathway that are associated with variation in development of toxicity and clinical outcomes for NSCLC patients. With validation, our findings may provide additional biomarkers for individualized treatment in order to enhance the efficiency and reduce toxicity during chemotherapy with platinum-based agents for NSCLC.
Acknowledgments
This research was supported in part, by National Cancer Institute (NCI) grants R01 CA111646 P50 CA070907, and R01 CA055769. MATH is supported by an NCI Cancer Prevention Research Fellowship training grant R25T CA57730. The study sponsors have no involvement in any aspects of the preparation of this manuscript.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Ries LAG, Melbert D, Krapcho M, Stinchcomb DG, Howlader N, Horner MJ, Mariotto A, Miller BA, Feuer EJ, Altekruse SF, Lewis DR, Clegg L, Eisner MP, Reichman M, Edwards BK. SEER Cancer Statistics Review, 1975-2005. National Cancer Institute; Bethesda, MD: 2008. [Google Scholar]
- 3.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 4.Burdett S, Stephens R, Stewart L, Tierney J, Auperin A, Le Chevalier T, Le Pechoux C, Pignon J, Arriagada R, Higgins J, Johnson D, van Meerbeeck J, Parmar M, Souhami R, Bell D, Cartei G, Cormier Y, Cullen M, Ganz P, Gridelli C, Kaasa S, Quoix E, Rapp E, Seymour L, Spiro S, Thatcher N, Tummarello D, Williams C, Williamson I. Chemotherapy in addition to supportive care improves survival in advanced non-small-cell lung cancer: a systematic review and meta-analysis of individual patient data from 16 randomized controlled trials. J Clin Oncol. 2008;26:4617–4625. doi: 10.1200/JCO.2008.17.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res. 2001;478:23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 6.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 8.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal. 2002;14:381–395. doi: 10.1016/s0898-6568(01)00271-6. [DOI] [PubMed] [Google Scholar]
- 10.Gagnon V, Van Themsche C, Turner S, Leblanc V, Asselin E. Akt and XIAP regulate the sensitivity of human uterine cancer cells to cisplatin, doxorubicin and taxol. Apoptosis. 2008;13:259–271. doi: 10.1007/s10495-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 11.Kim SH, Juhnn YS, Song YS. Akt involvement in paclitaxel chemoresistance of human ovarian cancer cells. Ann N Y Acad Sci. 2007;1095:82–89. doi: 10.1196/annals.1397.012. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Choi EJ, Jin C, Kim DH. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol. 2005;97:26–34. doi: 10.1016/j.ygyno.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 13.Liu LZ, Zhou XD, Qian G, Shi X, Fang J, Jiang BH. AKT1 amplification regulates cisplatin resistance in human lung cancer cells through the mammalian target of rapamycin/p70S6K1 pathway. Cancer Res. 2007;67:6325–6332. doi: 10.1158/0008-5472.CAN-06-4261. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Fraser M, Moll UM, Basak A, Tsang BK. Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006;66:3126–3136. doi: 10.1158/0008-5472.CAN-05-0425. [DOI] [PubMed] [Google Scholar]
- 15.Hildebrandt MA, Yang H, Hung MC, Izzo JG, Huang M, Lin J, Ajani JA, Wu X. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol. 2009;27:857–871. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oken MM, Creech RH, Tormey DC, Horton J, Davis TE, McFadden ET, Carbone PP. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5:649–655. [PubMed] [Google Scholar]
- 17.Trotti A, Colevas AD, Setser A, Rusch V, Jaques D, Budach V, Langer C, Murphy B, Cumberlin R, Coleman CN, Rubin P. CTCAE v3.0: development of a comprehensive grading system for the adverse effects of cancer treatment. Semin Radiat Oncol. 2003;13:176–181. doi: 10.1016/S1053-4296(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 19.Pisick E, Jagadeesh S, Salgia R. Receptor tyrosine kinases and inhibitors in lung cancer. Scientific World Journal. 2004;4:589–604. doi: 10.1100/tsw.2004.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 21.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 22.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 23.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MW, Kim DS, Min NY, Kim HT. Akt1 inhibition by RNA interference sensitizes human non-small cell lung cancer cells to cisplatin. Int J Cancer. 2008;122:2380–2384. doi: 10.1002/ijc.23371. [DOI] [PubMed] [Google Scholar]
- 25.Sanborn RE. Cisplatin versus carboplatin in NSCLC: is there one “best” answer? Curr Treat Options Oncol. 2008;9:326–342. doi: 10.1007/s11864-009-0085-5. [DOI] [PubMed] [Google Scholar]
- 26.Forgacs E, Biesterveld EJ, Sekido Y, Fong K, Muneer S, Wistuba II, Milchgrub S, Brezinschek R, Virmani A, Gazdar AF, Minna JD. Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene. 1998;17:1557–1565. doi: 10.1038/sj.onc.1202070. [DOI] [PubMed] [Google Scholar]
- 27.Marsit CJ, Zheng S, Aldape K, Hinds PW, Nelson HH, Wiencke JK, Kelsey KT. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol. 2005;36:768–776. doi: 10.1016/j.humpath.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Petersen S, Rudolf J, Bockmuhl U, Gellert K, Wolf G, Dietel M, Petersen I. Distinct regions of allelic imbalance on chromosome 10q22-q26 in squamous cell carcinomas of the lung. Oncogene. 1998;17:449–454. doi: 10.1038/sj.onc.1201949. [DOI] [PubMed] [Google Scholar]
- 29.Soria JC, Lee HY, Lee JI, Wang L, Issa JP, Kemp BL, Liu DD, Kurie JM, Mao L, Khuri FR. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2002;8:1178–1184. [PubMed] [Google Scholar]
- 30.Yokomizo A, Tindall DJ, Drabkin H, Gemmill R, Franklin W, Yang P, Sugio K, Smith DI, Liu W. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene. 1998;17:475–479. doi: 10.1038/sj.onc.1201956. [DOI] [PubMed] [Google Scholar]
- 31.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 32.Brognard J, Clark AS, Ni Y, Dennis PA. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001;61:3986–3997. [PubMed] [Google Scholar]