Abstract
Secreted Frizzled-Related Proteins (sFRP) are involved in embryonic development as well as pathological conditions including bone and myocardial disorders and cancer. Because of their sequence homology with the Wnt-binding domain of Frizzled, they have generally been considered antagonists of canonical Wnt signaling. However, additional activities of various sFRPs including both synergism and mimicry of Wnt signaling as well as functions other than modulation of Wnt signaling have been reported. Using human embryonic kidney cells (HEK293A), we found that sFRP2 enhanced Wnt3a-dependent phosphorylation of LRP6 as well as both cytosolic β-catenin levels and its nuclear translocation. While addition of recombinant sFRP2 had no activity by itself, Top/Fop luciferase reporter assays showed a dose-dependent increase of Wnt3a-mediated transcriptional activity. sFRP2 enhancement of Wnt3a signaling was abolished by treatment with the Wnt antagonist, Dickkopf-1 (DKK1). Wnt-signaling pathway qPCR arrays showed that sFRP2 enhanced the Wnt3a-mediated transcriptional up-regulation of several genes regulated by Wnt3a including its antagonists, DKK1 and Naked cuticle-1 homolog (NKD1). These results support sFRP2’s role as an enhancer of Wnt/β-catenin signaling, a result with biological impact for both normal development and diverse pathologies such as tumorigenesis.
Keywords: Wnt3a, sFRP2, Wnt signaling, LRP6/5, β-catenin
Introduction
Canonical Wnt/β-catenin signaling is a highly conserved pathway that is essential for cell-fate, patterning during development, and is often involved in human diseases such as cancer [1; 2]. The stabilization and nuclear translocation of β-catenin are two hallmarks of canonical Wnt signaling [1; 2]. In the absence of Wnt, β-catenin binds to scaffold proteins, Axin and adenomatosis polyposis coli (APC), and undergoes phosphorylation thereby triggering its proteasome-dependent degradation. Signaling starts with Wnt proteins binding to two cell surface receptors, a member of the Frizzled (Fz) serpentine receptor family [3; 4] and a single-pass transmembrane receptor, LRP6 (low density lipoprotein receptor-related protein-6), or closely the related, LRP5 [5; 6]. When Wnt binds to its receptors, β-catenin phosphorylation is suppressed leading to its accumulation in the cytoplasm and translocation to the nucleus where it interacts with members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors, consequently initiating target gene transcription [7]. Wnt/β-catenin signaling is modulated by an array of secreted molecules, including Wnt inhibitory signaling factor-1 (WIF1), Cerebrus, Sclerostin, Dickkopf-1 (DKK1), and secreted Frizzled-related proteins (sFRP). Sclerostin and DKK1 antagonize canonical signaling by binding to LRP5/6, whereas WIF1, Cerebrus, and sFRP2 are reported to interact directly with Wnt proteins [8; 9].
The sFRPs constitute a family of 5 proteins in mammals: Frzb (sFRP3), sFRP1, SFRP2, sFRP4, and sFRP5 [10]. They are expressed in adults and during embryonic development in dynamic as well as spatially restricted manners. Furthermore, their expression is altered in various bone pathologies [11], retinal degradation [12], hypophosphatemic diseases [13], and myocardial disorders [14; 15] as well as in different types of cancer [9]. Because of their sequence homology with the Wnt-binding domain of the Fz receptors, sFRPs have been considered antagonists of canonical Wnt signaling by binding to Wnt proteins and preventing signal transduction [16; 17]. However, sFRPs have recently been reported to synergize or mimic Wnt activities by direct interaction with Fz receptors [18; 19; 20], by antagonizing each other’s action [21], enhancing extracellular transport of Wnt proteins [22], or by playing roles other than directly controlling Wnt signaling pathways [23].
In this study, we used two cell lines of epithelial tissue origin (HEK293 and human salivary gland intercalated duct cell line, HSG) to show that sFRP2 enhances Wnt3a’s ability to induce: 1) phosphorylation of LRP5/6; 2) β-catenin stabilization and its nuclear translocation; and 3) changes in expression of known Wnt-mediated genes. Furthermore, we show that a specific inhibitor of Wnt canonical signaling, DKK1, negates all sFRP2 effects on Wnt3a signaling.
Materials and Methods
Cell culture
The murine L-cells M(TK−) (ATCC, Manassas, VA) and the human HEK293A cells (Invitrogen, Carlsbad, CA) were propagated in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco-BRL, Gaithersburg, MD) containing 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA). HSG cells (established from an irradiated human salivary gland as described [24]) were grown in DMEM/F12 medium (Gibco-BRL) containing 10% FBS. Media were supplemented with L-glutamine (22 mM), penicillin (100 IU/mL), and streptomycin (100 μg/mL). For conditioned media (CM), L-cells were grown to subconfluence, replenished with fresh complete culture medium that was then collected 48 h later, centrifuged at 700 × g for 10 minutes, and stored in aliquots at −80 °C.
Antibodies and recombinant proteins
Mouse monoclonal anti-β-catenin antibody was from BD Transduction Laboratories (Cat. #610154, Franklin Lakes, NJ). Rabbit anti-phospho-LRP6 (Ser1490) antibody was from Cell Signaling (Cat. #2568, Beverly, NJ). Mouse monoclonal anti-β-actin antibody was from Sigma-Aldrich (Cat. #A1978, St. Louis, MO) and mouse monoclonal anti-TATA-binding protein (TBP) was from Abcam (Cat. #ab61411, Cambridge, UK), IRDye 800 goat anti-mouse IgG and IRDye 680 goat anti-rabbit IgG second antibodies were from LI-COR Biosciences (Lincoln, NE). Carrier-free recombinant mouse sFRP2, recombinant human Wnt3a, and recombinant human DKK1 were purchased from R & D Systems (Minneapolis, MN).
Western blot
1×106 cells/well were seeded into 6-well plates. After overnight culture, cells were treated with Wnt3a alone or in combinations of sFRP2 and DKK1 for 2 h at 37 °C. For total cellular protein, cells were lysed with 100 μl 2x Laemmli Sample Buffer. Alternatively, cytosolic and nuclear fractions were prepared using the NE-PER Nuclear and Cytoplasmatic Extraction Kit (Pierce, Rockford, IL). Equal volumes of extracts were electrophoresed on 4–12 % Bis-Tris Nu-PAGE gels (Invitrogen) and transferred to Immobilon-FL membrane (Millipore, Billerica, MA). Blots were blocked for 1h in Odyssey Blocking Buffer (LI-COR Biosciences), followed by incubation with the first antibody overnight at 4°C and then with IRDye 680 goat anti-rabbit IgG or IRDye 800 goat anti-mouse for 1h at room temperature (RT). Blots were analyzed using the LI-COR Odyssey infrared imaging system (LI-COR Biosciences).
Transfection and reporter assays
1.5×104 HEK293A cells/well were plated into 96-well plates. After overnight culture, the cells were transfected with 50 ng of the Super 8x TopFlash plasmid or the control plasmid Super 8x FopFlash (Addgene Inc., Cambridge, MA) and 1 ng of the pRL-SV40 vector (Promega, Madison, WI) in Opti-MEM medium using 0.25 μl Lipofectamine 2000 per well according to manufacturer’s protocol (Invitrogen). After 6 h, cells were switched to serum-containing medium. Eighteen hours later, cells were treated with Wnt3a and/or sFRP2. For inhibition experiments, cells were incubated with DKK1 for 30 min prior to treatment with Wnt3a and sFRP2. After 24-h treatment, cells were lysed and the luciferase activity was determined using the Dual Luciferase kit from Promega. All reporter assays were performed in triplicate and each experiment was repeated at least three times. The relative firefly luciferase activity was normalized with its respective Renilla luciferase activity.
RNA Extraction, cDNA synthesis, and PCR array analysis
Using a Trizol/RNeasy hybrid protocol, total RNA was isolated from cells treated for 2 h with Wnt3a alone or in combination with sFRP2. This protocol takes the RNA extracted with Trizol (Invitrogen) through an additional column-based clean-up RNeasy step (Qiagen, Germantown, MD) following instructions for both kits. The first-strand cDNA was synthesized from 1 μg of total RNA (quantified by NanoDrop, Wilmington, DE) using the RT2 First Strand Kit (SABiociences, Frederick, MD) (including a DNase pretreatment to remove any genomic DNA). cDNA reactions were mixed with the SYBR Green qPCR Master Mix and transferred into 96-well RT2 Profiler PCR Array for human or mouse WNT-Signaling Pathway (SABiosciences), containing primers for 84 genes related to Wnt-mediated signal transduction as well as housekeeping genes. The two-step real-time PCR reaction was performed using the MyiQ5 instrument (Bio-Rad, Hercules, CA) according to SABioscience’s MyiQ5-specific instructions.
Statistical analysis
Threshold cycle values obtained from RT2 Profiler PCR Arrays (three experiments) were analyzed using the PCR Array Data Analysis Web Portal from SABioscienecs. All other statistical analyses were performed using GraphPad software (San Diego, CA). Multiple comparisons were performed with a one-way analysis of variance (ANOVA) followed by Newman-Keuls test comparing all pairs of columns. Results are expressed as mean ± SEM.
Results
sFRP2 enhances the Wnt3a-induced β-catenin stabilization and its nuclear translocation
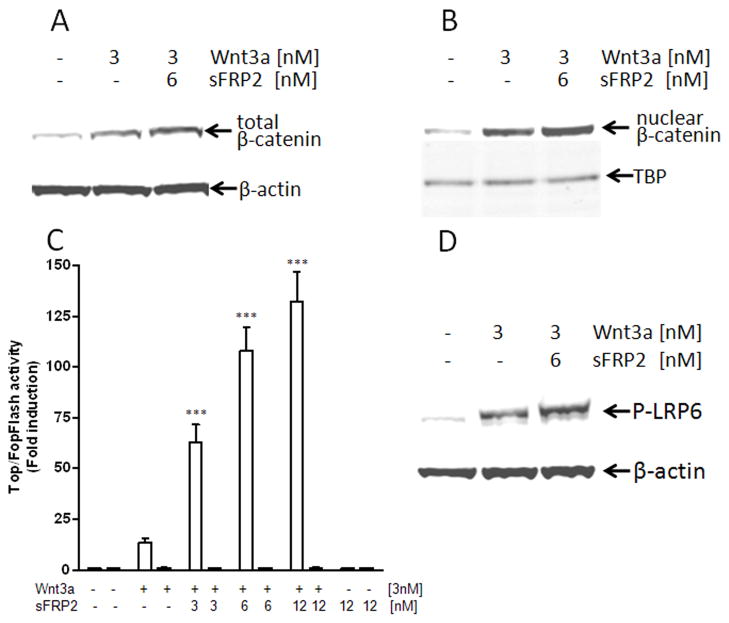
The stabilization of β-catenin and its nuclear translocation are two hallmarks of canonical Wnt-signaling. HEK293A cells were treated for 2 h with purified recombinant Wnt3a plus/minus recombinant sFRP2 and the total β-catenin levels in cell lysates were visualized by immunodetection on Western blots. As shown in Fig. 1A, incubation with 3 nM Wnt3a alone increased the total β-catenin level in HEK293A cells, indicating that Wnt3a was able to activate the canonical pathway in these cells. The addition of 6 nM recombinant sFRP2 with the 3 nM Wnt3a increased the total β-catenin accumulated. Furthermore, using nuclear protein-enriched cell fractions, the induction of overall β-catenin protein level upon Wnt3a/sFRP2 treatment was also associated with greater β-catenin nuclear translocation (Fig. 1B). Treatment with 6 nM sFRP2 alone did not effect either β-catenin accumulation or its relative nuclear translocation (data not shown).
Fig. 1.
sFRP2 enhances: (A)Wnt3a-mediated β-catenin accumulation, (B) β-catenin nuclear translocation, (C) TCF/LEF transcriptional activity, and (D) LRP6 phosphorylation in HEK293A. Cells were treated with Wnt3a ± indicated sFRP2 for 2 h. The levels of total cellular and nuclear β-catenin as well as phosphorylated LRP6 were detected on Western blots. Antibodies against β-actin and TATA-binding protein (TBS) were used as loading controls. Shown are representative results of three independent experiments. For luciferase assays, cells were co-transfected with Renilla transfection-control plasmids plus either TopFlash (□) or FopFlash (■) plasmids. After transfection, cells were treated with Wnt3a ± sFRP2 for 24 h before extraction for luciferase assays. Shown are means ± SEM from at least 3 experiments each performed in triplicate wells. ***P<0.0001 compared with control by using of ANOVA and the Newman-Keuls test.
sFRP2 enhances Wnt3a/β-catenin transcriptional reporter activity
Nuclear β-catenin associates with transcription factors of the T-cell factor/lymphoid enhancing factor family (TCF/LEF), leading to transcriptional activation of Wnt-targeted genes. Therefore, to further characterize the sFRP2 effects on the Wnt3a/β-catenin signaling, HEK293A cells were transfected with either the reporter plasmid, Super 8x TopFlash (containing 8 TCF/LEF-binding sites driving the transcription of the luciferase enzyme), or the control plasmid, Super 8x FopFlash (containing inactive TCF/LEF-binding sites). After a recovery period, the TCF/LEF transcriptional activity was measured upon treatment with 3 nM Wnt3a alone or in combination with increasing doses of sFRP2 (3 nM, 6 nM, and 12 nM). As expected, Wnt3a alone increased the transcription of the luciferase reporter gene (Fig. 1C) by 13.7-fold (±1.9, n=5). As seen in the same figure, addition of 3–12 nM sFRP2 further enhanced this transcriptional activity an additional 6 to 10-fold in a dose-dependent manner (e.g., 132-fold ±14.6 over control for 3 nM Wnt3a and 12 nM sFRP2; n=5). Neither treatment with Wnt3a alone nor together with sFRP2 changed the luciferase activity of the control, FopFlash plasmid (Fig. 1C).
sFRP2 enhances LRP6 phosphorylation
Wnt3a mediates its effects through engagement with two distinct cell surface receptors, Frizzled (Fz) and LRP6/5, resulting in the phosphorylation of the latter [5]. Therefore, phosphorylation-dependent LRP6 antibody was used on Western blots to visualize its phosphorylation levels upon treatment with 3 nM Wnt3a ± 6 nM sFRP2. As shown in Fig. 1D, treatment with Wnt3a induced the expected phosphorylation of LRP6 and simultaneous treatment with both Wnt3a and sFRP2 further augmented the LRP6 phosphorylation in HEK293A cells.
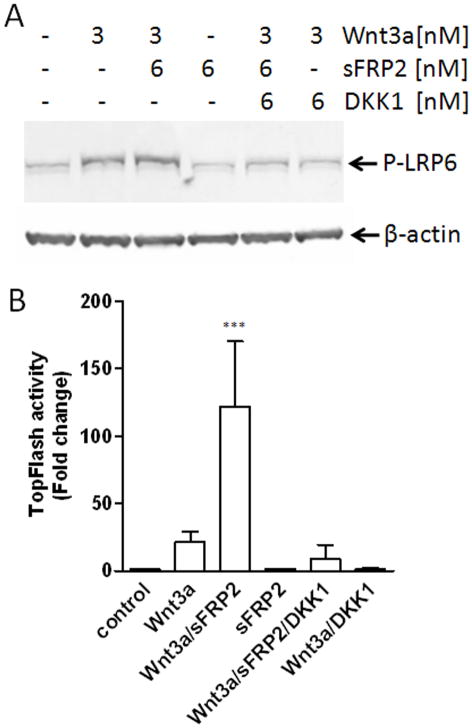
Dickkopf-1 (DKK1) antagonizes both Wnt3a/sFRP2-induced LRP6 phosphorylation and transcriptional activity
DKK1 is known to antagonize the Wnt signaling by binding LRP6 thereby preventing the Fz/LRP6 complex formation and subsequent LRP6 phosphorylation [8]. To gain more insight into the mechanisms of the sFRP2 action on the Wnt3a/β-catenin signaling pathway, HEK293 cells were first incubated with DKK1 and then Wnt3a plus/minus sFRP2. Next, the levels of LRP6 phosphorylation and TCF/LEF transcriptional activities were determined. Recombinant DKK1 blocked both the Wnt3a and the Wnt3a/sFRP2-enhanced phosphorylation of LRP6 (Fig. 2A). DKK1 pretreatment also abolished TCF/LEF transcriptional activity induced by both Wnt3a and Wnt3a/sFRP2 (Fig. 2B). Together these data indicate that sFRP2 augments canonical Wnt3a/β-catenin signaling at the level of its cell surface receptor.
Fig. 2.
DKK1 blocks sFRP2-enhanced Wnt3a LRP6 phosphorylation (A) and TCF/LEF transcriptional activity (B). HEK293A cells were treated with Wnt3a alone or with indicated sFRP2 for 2 h. For inhibition studies, cells were pre-treated with 6 nM DKK1 for 30 min. The levels of LRP6 phosphorylation were analyzed by Western blot with equal loading verified by detection of cellular β-actin. For luciferase assay, cells were co-transfected with TOPflash and Renilla plasmids for 6 h and then treated with Wnt3a with/without sFRP2 at concentrations indicated for 24 h. Shown are mean ± SEM of at least 3 independent experiments, each performed in triplicate wells. ***P<0.0001compared with control by using of ANOVA and the Newman-Keuls test.
sFRP2 enhances expression of four genes known to be regulated by Wnt3a signaling
To show that the sFRP2 enhancement of Wnt3a signaling had downstream biological effects, the expression of genes known to be related to Wnt signaling was studied using Wnt-signaling Pathway qPCR arrays. Treatment with Wnt3a plus sFRP2 enhanced mRNA expression of human WNT11 (4.0-fold; p= 0.03) and T gene (4.2-fold; p=0.002) over control. Interestingly, mRNA expression of two Wnt signaling inhibitors, DKK1 and NKD1, was also significantly elevated by treatment with the Wnt3a/sFRP2 combination (4.5-fold; p=0.003 and 5.7-fold; p=0.008, respectively) (Fig. 3).
Fig. 3.
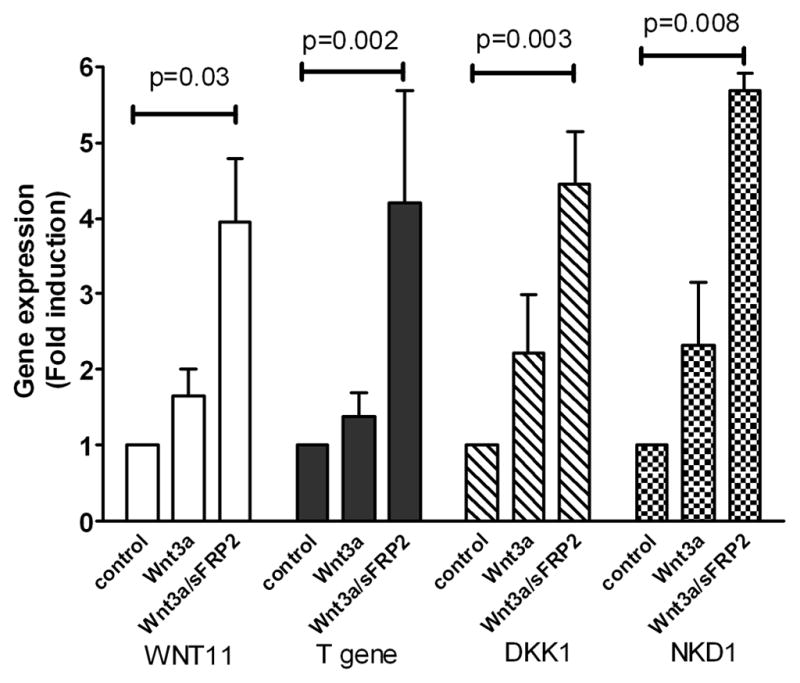
sFRP2 augments Wnt3a-induced gene expression in HEK293A cells. Cells were treated with 3 nM Wnt3a alone or in combination with 6 nM sFRP2 for 6 h. RNA was isolated and analyzed using Wnt Signaling Pathway RT2 Profile PCR Arrays. Shown are mean ± SEM of 3 independent experiments.
sFRP2 enhances Wnt3a-transcriptional activity in HSG cells but not in L-cells
Next, we asked whether sFRP2 had the same Wnt3a signal-enhancing effect on cells of a different origin. Since HEK293 cell line has recently been suggested to be of neuronal lineage [25], we tested a human salivary gland cell line, HSG, of epithelial origin and a murine L-cells of fibroblastic origin (commonly used in studies of Wnt signaling). As shown in Fig. 4A, Wnt3a alone increased the TopFlash luciferase activity by 2.4-fold (±0.4; n=6) in HSG cells and by 11.9-fold (±1.9; n=3) fold in L-cells. The luciferase activity increased significantly in a dose-dependent manner up to 13.0-fold (±2.7; n=6) fold in HSG cells with increasing concentrations of sFRP2. The addition of two molar excess of sFRP2 to Wnt3a-treated HSG cells also increased phosphorylation of the LRP6 receptor as well as the β-catenin accumulation suggesting the same pathways are enhanced in these epithelial cells (data not shown). Surprisingly, co-treatment with Wnt3a and recombinant sFRP2 protein did not significantly change the luciferase activity in L-cells (Fig. 4B).
Fig. 4.
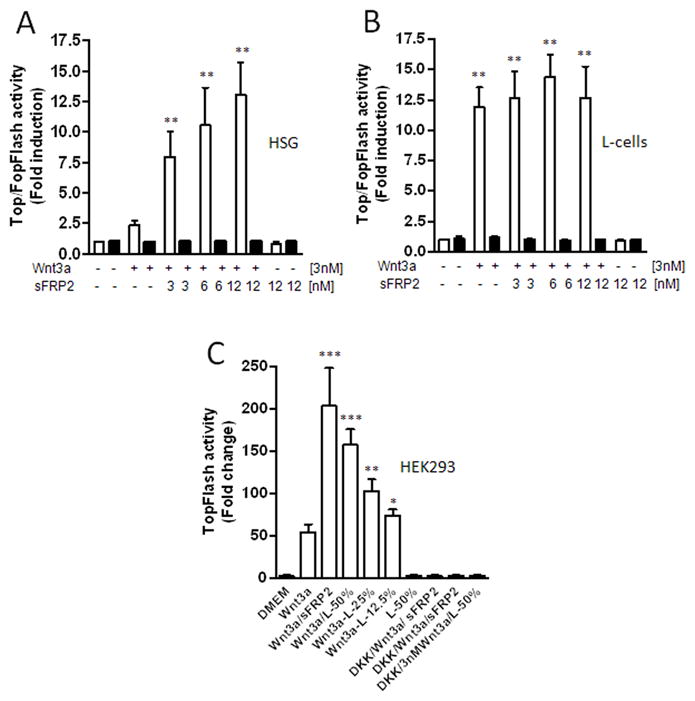
sFRP2 enhances Wnt3a-mediated transcriptional activation in HSG cells (A) but not in L-cells (B). Cells were co-transfected with TopFlash (□) or FopFlash (■) construct and Renilla (transfection control) plasmids and 24 h later treated with Wnt3a alone or together with indicated sFRP2 for 24h. (C) After transfection with TopFlash and Renilla plasmids, HEK293A cells were treated with L-cell-conditioned medium plus/minus 3 nM Wnt3a. For inhibition experiments, cells were pretreated with 6 nM DKK1 for 30 minutes. For controls, cells were treated with Wnt3a and sFRP2 or with 50 % CM alone. After 24 h, cell lysates were analyzed for luciferase activity. Shown are mean ± SEM of at least three experiments, each performed in triplicate wells. ***P<0.0001; **P<0.001; *P<0.01 compared with control by using of ANOVA and the Newman-Keuls test.
Analysis of mouse Wnt-signaling Pathway qPCR arrays revealed a low expression of sFRP2 in L-cells (data not shown), suggesting that the lack of enhancement by the exogenously added sFRP2 in L-cells may have been due to endogenous expression of sFRP2 or other enhancing factor by the L-cells themselves. To test this hypothesis, HEK293A cells were transfected with the reporter construct, Super 8x TopFlash, and later exposed to increasing concentrations of L-cell-conditioned media (12.5%, 25%, and 50%; v/v) with or without the presence of 3 nM Wnt3a. As shown in Fig. 4C, co-treatment with Wnt3a and L-cell CM resulted in a dose-dependent increase of luciferase reporter gene activity when compared with Wnt3a treatment alone. Treatment with 50% CM alone did not increase luciferase activity (remaining at the level observed for DMEM medium control) showing that L-cells did not produce significant amounts of Wnt3a. The increased transcriptional activity upon treatment with 50% CM plus Wnt3a was comparable with that observed upon treatment with Wnt3a plus recombinant sFRP2 and was completely inhibited by DKK1 (Fig. 4C).
Discussion
Currently, sFRP proteins appear to represent the largest family of Wnt modulators. These proteins were first predicted to be antagonists of canonical Wnt signaling based on their sequence homology with the ligand-binding domain of Frizzled receptors and were expected to exert their inhibitory effects by sequestering Wnt proteins from their receptors [16; 17]. However, some recent reports suggest that the function of sFRPs is more complex [23]. In this study, we provide several lines of evidence that sFRP2 enhances the Wnt3a-mediated canonical signaling in the context of HEK293A and HSG cells. Moreover, our results indicate that sFRP2 acts at the ligand/receptor level because recombinant sFRP2 substantially enhanced the Wnt3a-mediated phosphorylation of the LRP6 co-receptor and was efficiently blocked by the antagonist, DKK1. Furthermore, exogenously added DKK1 blocked the activation of the TCF/LEF reporter gene. Finally, co-treatment with Wnt3a and sFRP2 significantly increased expression of several known downstream target genes of β-catenin/TCF/LEF, including two Wnt antagonists DKK1 [26] and NKD1 [27], the transcription factor T [28], as well as WNT11 [29]. Our data support a recently published report showing that different sFRPs, including sFRP2, may co-function with Wnt3a to enhance osteoblastic differentiation and its transcriptional activity [30].
The precise molecular mechanism responsible for the sFRP2-mediated augmentation of the Wnt3a-mediated canonical signaling, however, remains unclear. At this time, sFRPs are proposed to act not only by directly binding to Wnt proteins but also directly with Frizzled proteins [18; 19]. More recently, it was shown that sFRP2 stabilizes β-catenin via interaction with Frizzled receptors without functionally interfering with Wnt3a ligand in a model of intestinal epithelium [20]. Although, we did not observe any effects on β-catenin signaling by sFRP2 alone, our results do not enable us to conclude whether sFRP2 binds Wnt3a or a cell/tissue specific Frizzled receptor. The latter model provides the possibility that the ability of sFRP2 to modulate Wnt3a signaling may depend on its ability to bind a specific Frizzled receptor. This suggests that the ability of sFRP2 to modulate Wnt3a may vary with the expression of the Frizzled receptors and, thus, depend on molecular and cellular context. For example, GeneChip assays performed on NIH3T3 cells and the rat pheochromocytoma cell line, PC12, revealed changes in expression in 355 and 129 genes, respectively, after Wnt3a-stimulation. However, only two Wnt-regulated genes were shared by both cell lines [31]. Consistent with this notion, that sFRP2 function might occur in a tissue-specific manner, we did not observe any enhancing effects of sFRP2 on Wnt3a signaling in L-cells of fibroblastic origin, which is agreement with the study by Lee et al. [32] showing that sFRP2-transfected fibroblasts do not counteract the dermomyotome-inducing activity of Wnt3a in presometic mesoderm explants. However, other studies have observed that sFRP2 is capable of antagonizing Wnt3a-activity in L-cells [33; 34]. One possible explanation for this difference might be that many earlier studies relied on the use of conditioned media that may have included bioactive proteins of unknown type and concentrations, whereas recent efforts, including ours, are based on the use of currently available purified proteins.
Exogenously added sFRP2 did not enhance the Wnt3a-mediated signaling in L-cells, perhaps reflecting the cell specific expression of different Frizzled receptors. However, since HEK293A cells treated with L-cell-conditioned media (which according to the Wnt-Signaling Pathway qPCR Array analyses express sFRP2 message) showed an enhanced transcriptional activity, one can speculate that cultured L-cells produce endogenous sFRP2 (or other factor) which induced the Wnt3a signaling in these cells, thereby making them insensitive to exogenous sFRP2.
In conclusion, our results further establish sFRP2 as a potential enhancer of canonical Wnt signaling and therefore also support its possible role as a paracrine/autocrine positive modulator of this key cellular signal transduction pathway and, therefore, may have biological relevance for both development and various diseases.
Acknowledgments
This work was supported by the Division of Intramural Research, NIDCR, of the Intramural Research Program, NIH, DHHS.
Abbreviations
- APC
adenomatosis polyposis coli
- LRP6/5
low density lipoprotein receptor-related protein-6/5
- TCF/LEF
T-cell factor/lymphoid enhancer factor
- Fz
Frizzled
- WIF1
Wnt inhibitory signaling factor-1
- DKK1
Dickkopf-1
- sFRP
Frizzled-related protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 3.Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–30. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 4.Schulte G, Bryja V. The Frizzled family of unconventional G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:518–25. doi: 10.1016/j.tips.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 5.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–77. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 6.Holmen SL, Salic A, Zylstra CR, Kirschner MW, Williams BO. A novel set of Wnt-Frizzled fusion proteins identifies receptor components that activate beta -catenin-dependent signaling. J Biol Chem. 2002;277:34727–35. doi: 10.1074/jbc.M204989200. [DOI] [PubMed] [Google Scholar]
- 7.Cong F, Schweizer L, Chamorro M, Varmus H. Requirement for a nuclear function of beta-catenin in Wnt signaling. Mol Cell Biol. 2003;23:8462–70. doi: 10.1128/MCB.23.23.8462-8470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–34. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 9.Rubin JS, Barshishat-Kupper M, Feroze-Merzoug F, Xi ZF. Secreted WNT antagonists as tumor suppressors: pro and con. Front Biosci. 2006;11:2093–105. doi: 10.2741/1952. [DOI] [PubMed] [Google Scholar]
- 10.Jones SE, Jomary C. Secreted Frizzled-related proteins: searching for relationships and patterns. Bioessays. 2002;24:811–20. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 11.Bodine PV, Komm BS. Wnt signaling and osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:33–9. doi: 10.1007/s11154-006-9002-4. [DOI] [PubMed] [Google Scholar]
- 12.Jones SE, Jomary C, Grist J, Stewart HJ, Neal MJ. Altered expression of secreted frizzled-related protein-2 in retinitis pigmentosa retinas. Invest Ophthalmol Vis Sci. 2000;41:1297–301. [PubMed] [Google Scholar]
- 13.Shaikh A, Berndt T, Kumar R. Regulation of phosphate homeostasis by the phosphatonins and other novel mediators. Pediatr Nephrol. 2008;23:1203–10. doi: 10.1007/s00467-008-0751-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteve P, Bovolenta P. The advantages and disadvantages of sfrp1 and sfrp2 expression in pathological events. Tohoku J Exp Med. 221:11–7. doi: 10.1620/tjem.221.11. [DOI] [PubMed] [Google Scholar]
- 15.Mangi AA, Noiseux N, Kong D, He H, Rezvani M, Ingwall JS, Dzau VJ. Mesenchymal stem cells modified with Akt prevent remodeling and restore performance of infarcted hearts. Nat Med. 2003;9:1195–201. doi: 10.1038/nm912. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Krinks M, Lin K, Luyten FP, Moos M., Jr Frzb, a secreted protein expressed in the Spemann organizer, binds and inhibits Wnt-8. Cell. 1997;88:757–66. doi: 10.1016/s0092-8674(00)81922-4. [DOI] [PubMed] [Google Scholar]
- 17.Lin K, Wang S, Julius MA, Kitajewski J, Moos M, Jr, Luyten FP. The cysteine-rich frizzled domain of Frzb-1 is required and sufficient for modulation of Wnt signaling. Proc Natl Acad Sci U S A. 1997;94:11196–200. doi: 10.1073/pnas.94.21.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bafico A, Gazit A, Pramila T, Finch PW, Yaniv A, Aaronson SA. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J Biol Chem. 1999;274:16180–7. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez J, Esteve P, Weinl C, Ruiz JM, Fermin Y, Trousse F, Dwivedy A, Holt C, Bovolenta P. SFRP1 regulates the growth of retinal ganglion cell axons through the Fz2 receptor. Nat Neurosci. 2005;8:1301–9. doi: 10.1038/nn1547. [DOI] [PubMed] [Google Scholar]
- 20.Kress E, Rezza A, Nadjar J, Samarut J, Plateroti M. The frizzled-related sFRP2 gene is a target of thyroid hormone receptor alpha1 and activates beta-catenin signaling in mouse intestine. J Biol Chem. 2009;284:1234–41. doi: 10.1074/jbc.M806548200. [DOI] [PubMed] [Google Scholar]
- 21.Yoshino K, Rubin JS, Higinbotham KG, Uren A, Anest V, Plisov SY, Perantoni AO. Secreted Frizzled-related proteins can regulate metanephric development. Mech Dev. 2001;102:45–55. doi: 10.1016/s0925-4773(01)00282-9. [DOI] [PubMed] [Google Scholar]
- 22.Mii Y, Taira M. Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development. 2009;136:4083–8. doi: 10.1242/dev.032524. [DOI] [PubMed] [Google Scholar]
- 23.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121:737–46. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 24.Shirasuna K, Sato M, Miyazaki T. A neoplastic epithelial duct cell line established from an irradiated human salivary gland. Cancer. 1981;48:745–52. doi: 10.1002/1097-0142(19810801)48:3<745::aid-cncr2820480314>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 25.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. Faseb J. 2002;16:869–71. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Sancho JM, Aguilera O, Garcia JM, Pendas-Franco N, Pena C, Cal S, Garcia de Herreros A, Bonilla F, Munoz A. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098–103. doi: 10.1038/sj.onc.1208303. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt C, Otto A, Luke G, Valasek P, Otto WR, Patel K. Expression and regulation of Nkd-1, an intracellular component of Wnt signalling pathway in the chick embryo. Anat Embryol (Berl) 2006;211:525–34. doi: 10.1007/s00429-006-0102-4. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, McMahon AP. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–90. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh M, Katoh M. Integrative genomic analyses of WNT11: transcriptional mechanisms based on canonical WNT signals and GATA transcription factors signaling. Int J Mol Med. 2009;24:247–51. doi: 10.3892/ijmm_00000227. [DOI] [PubMed] [Google Scholar]
- 30.Cho SW, Her SJ, Sun HJ, Choi OK, Yang JY, Kim SW, Kim SY, Shin CS. Differential effects of secreted frizzled-related proteins (sFRPs) on osteoblastic differentiation of mouse mesenchymal cells and apoptosis of osteoblasts. Biochem Biophys Res Commun. 2008;367:399–405. doi: 10.1016/j.bbrc.2007.12.128. [DOI] [PubMed] [Google Scholar]
- 31.Railo A, Pajunen A, Itaranta P, Naillat F, Vuoristo J, Kilpelainen P, Vainio S. Genomic response to Wnt signalling is highly context-dependent--evidence from DNA microarray and chromatin immunoprecipitation screens of Wnt/TCF targets. Exp Cell Res. 2009;315:2690–704. doi: 10.1016/j.yexcr.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 32.Lee CS, Buttitta LA, May NR, Kispert A, Fan CM. SHH-N upregulates Sfrp2 to mediate its competitive interaction with WNT1 and WNT4 in the somitic mesoderm. Development. 2000;127:109–18. doi: 10.1242/dev.127.1.109. [DOI] [PubMed] [Google Scholar]
- 33.Galli LM, Barnes T, Cheng T, Acosta L, Anglade A, Willert K, Nusse R, Burrus LW. Differential inhibition of Wnt-3a by Sfrp-1, Sfrp-2, and Sfrp-3. Dev Dyn. 2006;235:681–90. doi: 10.1002/dvdy.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wawrzak D, Metioui M, Willems E, Hendrickx M, de Genst E, Leyns L. Wnt3a binds to several sFRPs in the nanomolar range. Biochem Biophys Res Commun. 2007;357:1119–23. doi: 10.1016/j.bbrc.2007.04.069. [DOI] [PubMed] [Google Scholar]