Abstract
The mammalian MRG15 gene encodes a chromodomain protein predicted to bind to chromatin via methylated histone tails. Human MORF4 encodes a related but truncated protein that is capable of promoting cellular senescence in a subset of human tumor cell lines. Drosophila contains a single homolog of human MRG15, called DmMRG15. Null mutation of MRG15 is embryonic lethal in mice and Drosophila, making study of MRG15 requirements in adults difficult. In these studies the DmMRG15 gene was over-expressed in Drosophila, during developmental stages and in adults, using a doxycycline-regulated system (Tet-on). In addition an inverted-repeated construct was designed to inactivate DmMRG15 via the RNAi pathway, and RNAi constructs were expressed using both the Tet-on system and Geneswitch system. The DmMRG15 protein was readily expressed in adult flies in a doxycycline-dependent manner. A truncated form of DmMRG15 (called DmMT1) was designed to mimic the structure of human MORF4, and expression of this mutant protein or the inverted repeat constructs inhibited fertility in females. Conditional expression of the DmMRG15 inverted-repeat constructs during larval development or in adults caused reductions in survival. These experiments indicate that Drosophila DmMRG15 gene function is required for female fertility, larval survival and adult life span, and provide reagents that should be useful for further dissecting the role of DmMRG15 in cell proliferation and aging.
Keywords: senescence, chromatin, epigenetics, stem cells, aging
Introduction
Normal human cells cultured in vitro undergo a limited number of population doublings before entering an irreversible cell cycle arrest referred to as cellular senescence, associated with characteristic changes in cell morphology and gene expression patterns (Pena and Pereira-Smith, 2007). Immortal cell lines lack this growth-restricting mechanism and will divide indefinitely. Cell fusion studies reveal that replicative senescence is a dominant phenotype, because the hybrid cells produced by fusion of normal cells and immortal cells will senesce. Fusions between numerous immortal human cell lines identified at least four different complementation groups that could induce senescence (Pereira-Smith and Smith, 1988). The evolutionarily conserved MORF4/MRG15 family of genes was discovered based on the ability of the MORF4 gene (Mortality factor, 4th chromosome) to induce replicative senescence in complementation group B human tumor cell lines (Bertram et al., 1999; Pena and Pereira-Smith, 2007). Additional members of this gene family were identified in humans based on sequence homology, including the MRG15 gene (MORF4-related gene on chromosome 15) and the gene MRGX (MORF4-related gene on chromosome X), as well as several pseudo-genes (Bertram et al., 1999). MRG15 appears to represent the evolutionary ancestor of the gene family based on sequence conservation with genes in mice, Drosophila, C. elegans, yeast, and plants, while MRGX is unique to mammals, and MORF4 is unique to humans. Based on the presence of a conserved chromodomain, MRG15 proteins are predicted to bind to chromatin via interactions with methylated histone tails. Consistent with this idea, studies with yeast and cultured human and Drosophila cells implicate MRG15 in chromatin remodeling (Kusch et al., 2004; Pardo et al., 2002). Studies in yeast suggest that the MRG15-related protein Eaf3 may direct histone acetyltransferase complexes to the promoters of target genes and direct deacetylase complexes to gene coding regions through interaction of its chromodomain with methylated lysine 36 in histone 3 protein in nucleosomes (Carrozza et al., 2005; Joshi and Struhl, 2005; Keogh et al., 2005). A knock-out of the MRG15 in mice produces phenotypes of reduced cell proliferation and embryonic lethality (Tominaga et al., 2005). Taken together, the data suggest that MRG15 plays a conserved role essential for normal cell proliferation, and that the related but truncated human MORF4 gene might induce cellular senescence by antagonizing the function(s) of MRG15.
Histone modifications are implicated in modulating aging phenotypes across multiple species (Dimauro and David, 2009; Willis-Martinez et al., 2009). Tip60 (HIV tat interacting protein 60) is a member of the conserved MYST-family of histone acetyltransferases (HATs) that regulate diverse cellular processes (Thomas and Voss, 2007; Tyteca et al., 2006), and has recently been re-named KAT5 in humans (Allis et al., 2007). In humans Tip60/KAT5 is required for double-strand DNA break (DSB) repair and apoptotic responses to DSBs (Ikura et al., 2000; Ikura et al., 2007; Squatrito et al., 2006), and is a haplo-insufficient tumor suppressor (Gorrini et al., 2007). Drosophila contains a gene related to the MORF4/MRG15 family, hereafter referred to as DmMRG15 (CG6363, FBgn0027378). In Drosophila the DmMRG15 protein is present in a complex with Tip60/KAT5 and is implicated in repair of double-strand DNA breaks (DSBs) (Kusch et al., 2004). Specifically, in cultured Drosophila cells depleted for either Tip60/KAT5 or DmMRG15, and in Drosophila DmMRG15 null mutant embryos, chromatin remodeling failed to occur normally after DSBs were generated with gamma-irradiation. Drosophila DmMRG15 is also implicated in epigenetic gene silencing mediated by the Tip60/KAT5 complex, since a P element mutation in DmMRG15 was found to suppress position-effect-variegation (PEV) and to interact genetically with polycomb-group (PcG) genes (Qi et al., 2006). Histone acetylation and consequent changes in gene expression are implicated in Drosophila life span regulation. Feeding adult Drosophila the drug 4-phenylbutyrate (PBA) is reported to increase global levels of histone acetylation and to increase life span (Kang et al., 2002), suggesting generally positive effects of histone acetylation on life span. Consistent with this idea, reduced activity of the histone deacetylase rpd3 is reported to increase fly life span (Rogina et al., 2002). In contrast, increased activity of the Drosophila deacetylase Sir2 is reported to increase life span through a mechanism related to dietary restriction (Rogina and Helfand, 2004). Taken together these results support the conclusion that different targets for acetylation may produce opposing effects on life span (Frankel and Rogina, 2005). Consistent with an important role for chromatin modifications in modulating aging phenotypes, heterozygous mutations in two core subunits of the Drosophila Polycomb Repressive Complex 2 (the histone H3 lysine 27-specific methyltransferase E(Z) and the H3 binding protein ESC) were recently found to reduce adult levels of trimethylated H3K27 and to increase life span, suggesting negative effects of histone methylation on life span (Siebold et al. 2010).
The Drosophila gene Mof (males absent on first) is another conserved member of the MYST-family of HATs, called MYST1 or KAT8 in humans. Mof is part of a protein complex required for X chromosome dosage compensation in male Drosophila (Hilfiker et al., 1997), along with the protein MSL3 that contains an MRG-related domain (Morales et al., 2005). The C. elegans MRG15 homolog, MRG-1, is associated with autosomes and silences X-linked genes. Genetic analysis in C. elegans suggests a role for MRG-1 in maintaining the immortality of the germ-line cells and influencing the germ-line/soma distinction (Fujita et al., 2002; Olgun et al., 2005; Takasaki et al., 2007).
Each of these processes in which MRG15 is implicated, DSB repair (Li et al., 2008), dosage compensation (Hartman and Ishii, 2007) and maintenance of germ-line/soma distinction (Curran et al., 2009), are ones that may be directly relevant to aging phenotypes such as cellular senescence and life span. In particular, sexual differentiation is implicated in aging across species, but the mechanism(s) are not yet clear (Hartman and Ishii, 2007; Tower, 2006; Yoshida et al., 2006).
Analyzing the role of MRG15 family genes in development and aging is impeded by the embryonic-lethal phenotype of null mutations. To begin to better understand the function of the evolutionarily conserved MORF4/MRG15 gene family, and in particular its relevance to aging and senescence phenotypes, the function of this gene family was investigated in Drosophila using a conditional transgenic system. The generation of conditional mutant phenotypes allowed for assay of effects on survival both during development and specifically in adult animals.
Materials and methods
DNA constructs
Constructs were generated using standard methods, and details of cloning steps and oligonucleotide sequences used for PCR are presented in Supplementary Materials.
Generation and other sources of transgenic lines
Transgenic strains were generated by P element-mediated germline transformation (Rubin and Spradling, 1982), using the w1118 recipient strain. All inserts were made homozygous by crosses to appropriate balancer stocks, and single copy insertions were confirmed by genomic Southern blots (data not shown).
The Tet-on system driver strain w[1118]; rtTA(3)E2/TM3 and the Geneswitch system driver strain w[1118]; Act-GS-255B have been previously described and characterized (Ford et al., 2007; Shen et al., 2009), and two UAS-RNAi strains directed against DmMrg15, w[1118];P{UAS-RNAi-CG6363}[45A2]/TM3 and w[1118];P{UAS-RNAi-CG6363}[189E] were obtained from the VDRC. Two strains containing a Tet-on system target construct that causes RNAi inhibition of the phosphogluconate mutase (PGM) gene were used as controls for possible effects of RNAi pathway activation, w[1118];p{PGMinvrpt}(2)[12A1] and w[1118];p{PGMinvrpt}(3)[23B1] (Allikian et al., 2002).
Western blot assay
Male flies of the indicated transgenic lines were crossed with virgin females of the rtTA(3)E2 driver strain. Age-synchronized progeny containing both constructs were collected and maintained in single-sex vials at ~25 flies per vial, by every-other-day passage on food supplemented +drug or −drug for seven days. The drug treatments were doxycycline for rtTA(3)E2 crosses and Tet-on system constructs, as previously described (Ford et al., 2007). Control flies were generated by crossing the driver strains to Oregon-R wild-type, to produce progeny containing the driver but no target transgene. Protein extracts were generated from whole flies, and were separated by SDS-PAGE. Proteins were transferred to nitrocellulose membrane and antibody staining was performed using standard methods. The anti-MRG15 antibody (Kusch et al., 2004) was a gift from Jerry Workman and was used at 1:3000 dilution. The anti-rabbit secondary antibody (Applied Bioscience) was used at 1:5000 dilution. The ECL kit (Kodak) was used to detect the protein bands.
Northern blot Assay
Male flies of the indicated transgenic lines were crossed with virgin females of the rtTA(3)E2 driver strain or the Act-GS-255B driver strain. Age-synchronized progeny containing both constructs were collected and maintained in single-sex vials at ~25 flies per vial, by every-other-day passage on food supplemented +drug or −drug for seven days. The drug treatments were doxycycline for rtTA(3)E2 crosses and Tet-on system constructs, or RU486/Mifepristone for Act-GS-255B crosses and Geneswitch system constructs, as previously described (Ford et al., 2007). Control flies were generated by crossing the driver strains to Oregon-R wild-type, to produce progeny containing the driver but no target transgene. Total RNA was extracted using Trizol reagent (invitrogen), fractionated on 1% agarose gels and transferred to GeneScreen membrane (Dupont/NEN). RNA loaded 10 μg per lane. The blot was hybridized with 32P-labelled DNA probe corresponding to DmMRG15 gene sequences from −53 to +303 (generated with primer SET9), which will recognize the endogenous DmMRG15 mRNA but not the inverted repeat species. A probe specific for the ribosomal protein 49 (Rp49) gene was used as a loading control.
Life span assay
Life span assays were performed essentially as previously described (Ford et al., 2007). Male and female progeny were collected from the cross of the indicated transgenic lines and the rtTA(3)E2 driver strain or the Act-GS-255B driver strain. Control flies were generated by crossing the respective driver strains to Oregon-R wild-type strain or to w[1118] strain, to produce progeny containing the driver but no target transgene. Approximately 150 flies for each sex were cultured at ~25 flies per vial with +drug or −drug food (doxycycline or RU486, respectively). The flies were transferred to new vials every other day, and the number of dead flies was recorded. A survival curve was generated based on the survival data, and the median life span was calculated. The significance of changes in median life span between −drug and +drug flies for each genotype was determined using log-rank test in R 2.6.2. Life span assays were performed in parallel in four experiments (Exp 1, Exp 2, Exp 3, Exp 4), and data is summarized in Table 2.
Female fertility assay
Approximately 30 age-synchronized female progeny from the cross of the indicated transgenic lines and the rtTA(3)E2 driver strain or Act-GS-255B driver strain were collected and maintained on +drug or −drug food. Control flies were generated by crossing the driver strains to Oregon-R wild-type. The flies were transferred to new vials every other day. On the 40th day (Exp 1 and Exp 2) or on the 60th day (Exp 3) these flies were distributed into four vials with four females each, and combined with an equal number of young Oregon R male flies. Four days later, egg production assay was initiated. The flies were transferred to new vials every other day for a total of 12 time points (24 days total), and the old vials were kept and scored for total progeny production. The total number of pupa were counted in each of the 12 vials. The average number of progeny per female and SD were calculated, and +drug and −drug groups were compared for each genotype using unpaired, two-sided t-tests.
Male fertile period assay
The indicated transgenic strains were each crossed to the rtTA(3)E2 driver strain or the Act-GS-255B driver strain, and male progeny were collected within 48 hours of eclosion. Half of the flies (the experimental group) were cultured on +drug food while the other half (the control group) were cultured on −drug food, until the flies were 30 days old. Each male fly was then cultured individually in a vial with four young wild-type (Oregon R) virgins. The male was transferred to a new vial with new virgins every four days. The date of the last vial to produce progeny was scored as the fertile life span for each individual male fly, and the median fertile period of +drug and −drug flies for each genotype was compared using log-rank tests.
Results
Generation and characterization of transgenic strains
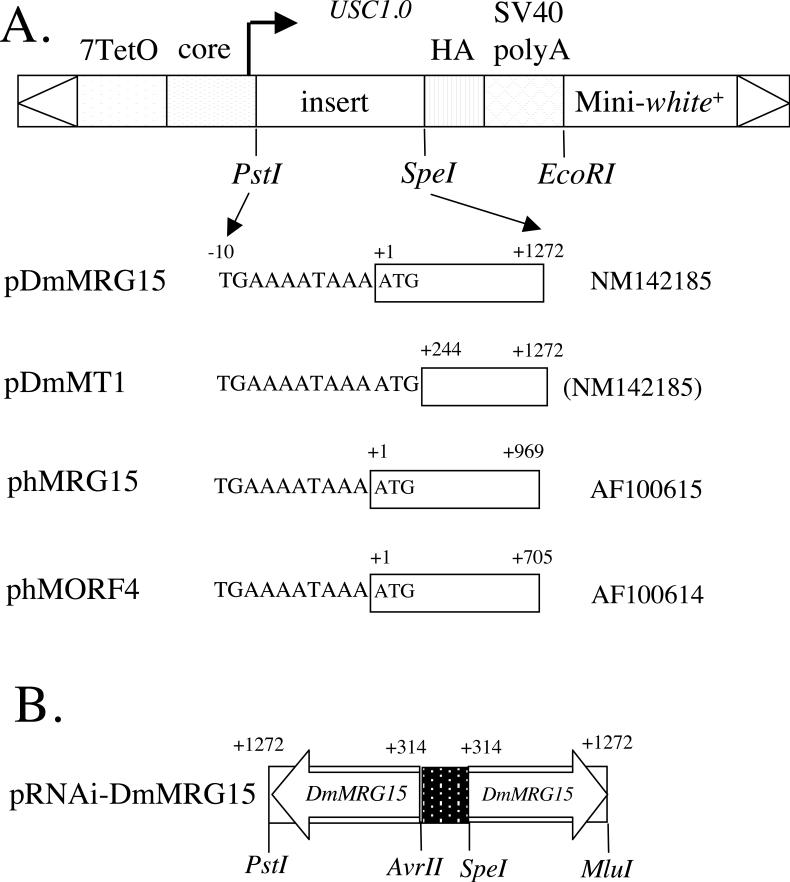
In mice loss of MRG15 gene function is associated with embryonic lethality (Tominaga et al., 2005), making analysis of MRG15 function in adults difficult. Similarly, in Drosophila, loss-of-function alleles of DmMRG15 are recessive embryonic-lethal (Kusch et al., 2004; Qi et al., 2006; Spradling et al., 1999), and possible effects on adult fertility and life span have not been reported. To enable studies of MRG15-family gene functions during aging, a conditional gene expression system was chosen so that over-expression and RNAi phenotypes could be generated specifically in adult Drosophila, as well as during development. Constructs were generated where the human and Drosophila MRG-family gene coding regions would be expressed under control of the doxycycline-regulated promoter in the USC1.0 vector (the Tet-on system)(Allikian et al., 2002; Bieschke et al., 1998)(Figure 1). For the protein-encoding constructs the corresponding cDNA fragments were amplified using PCR, and a triple HA epitope tag was added to each protein (Figure 1A). The phMRG15 and phMORF4 constructs encode the corresponding human proteins, while the pDmMRG15 construct encodes the Drosophila DmMRG15 gene. The pDmMT1 construct is a version of the Drosophila DmMRG15 gene truncated at nucleotide residue +243 in order to mimic the structure of the human MORF4 protein. Finally, a fragment extending from nucleotide residues +314 to +1272 of the Drosophila DmMRG15 cDNA was cloned as an inverted-repeat downstream of the doxycycline-regulated promoter (Figure 1B). This construct is expected to produce expression of a RNA hairpin product and cause conditional RNAi (Allikian et al., 2002). For each construct multiple independent transgenic strains were generated and the chromosome of insertion was identified (Supplemental Table S1). For the Tet-on system the rtTA(3)E2 driver strain was employed, which will drive doxycycline-dependent expression of the target constructs throughout the somatic tissues of larvae or adult flies (Bieschke et al., 1998; Ford et al., 2007). For certain experiments the Geneswitch system was also used, where the cytoplasmic Actin5C gene promoter is used to drive expression of the transcription factor Geneswitch in line Act-GS-255B. Act-GS-255B drives RU486-dependent expression of target constructs throughout the somatic tissues of larvae or adult flies (Ford et al., 2007; Shen et al., 2009).
Figure 1. Diagram of transgenic constructs.
A. Protein encoding constructs. The indicated insert fragments derived from Drosophila and human cDNAs were cloned into USC1.0 vector downstream of the doxycycline-regulated promoter, with the addition of a 3×HA tag to each protein. Genbank accession numbers are given to the right. B. Inverted-repeat construct pRNAi-DmMRG15.
Western blot analysis of conditional transgene expression
To confirm conditional expression of the MRG proteins using the Tet-on system, young adult flies were cultured in the presence and absence of doxycycline for one week, and whole-fly extracts were assayed by Western analysis using an antibody specific for Drosophila DmMRG15 protein (Kusch et al., 2004). In addition to the endogenous Drosophila DmMRG15 protein, expression of the slightly larger HA-tagged DmMRG15 transgenic protein was observed only in the presence of doxycycline (Figure 2A), thereby demonstrating conditional expression. Western analysis using an antibody specific for the HA tag confirmed the conditional expression of the DmMRG15 protein, as well as conditional expression of the truncated DmMT1 protein (Figure 2B). Interestingly, expression of the human MRG15 and MORF4 proteins could not be detected in adult flies (data not shown).
Figure 2. Western analysis of conditional transgene expression in adult flies.
A. Adult flies from four independent transgenic strains containing the DmMRG15 over-expression construct were assayed for doxycycline-dependent protein expression, using an antibody specific for DmMRG15 protein. The transgenic protein is slightly larger due to the presence of the triple HA epitope tag. B. The DmMRG15–26 line and several lines expressing the truncated MT mutant form of DmMRG15 were assayed for expression of protein in adult flies, in the presence and absence of doxycycline, using an antibody specific for the HA epitope tag. As expected the truncated MT protein migrates at a smaller apparent MW than the full length DmMRG15 protein.
Northern blot analysis of conditional transgene expression and RNAi
Using Northern blot analysis the endogenous Drosophila DmMRG15 RNA was readily detected, and as expected the amount of this RNA species showed no consistent changes upon doxycycline treatment or RU486 treatment in control flies (Figure 3). Independent transgenic lines of an RNAi construct typically vary in their activity. Each of three independent transgenic lines containing the Tet-on system RNAi-DmMRG15 construct were assayed for the ability to cause conditional knock-down of endogenous DmMRG15 RNA levels in adult males and females (lines RNAi-DmMRG15-4-1, RNAi-DmMRG15–53, and RNAi-DmMRG15-4-3). In males a conditional reduction in DmMRG15 message levels of ~−45% was observed with each line, whereas in females a smaller reduction was observed, and only with lines RNAi-DmMRG15-4-3 and RNAi-DmMRG15–53. With the Geneswitch system, the two RNAi lines (line 189E and line 45A2) produced a conditional knock-down of DmMRG15 message levels of −36 % and −20 % in males, and −14 % and −10 % in females, respectively. Therefore the independent transgenic lines for the RNAi constructs varied in their activity, and were in generally more effective in males than in females.
Figure 3. Northern analysis of conditional transgene expression in adult flies.
The indicated transgenic strains containing either the RNAi-DmMRG15 construct, the DmMRG15 wild type construct, or the MT1 construct were crossed to the rtTA(3)E2 driver strain or to the Act-GS-255B driver strain, as indicated. Progeny containing both constructs were cultured in the presence and absence of drug for one week. The drug was doxycycline for rtTA(3)E2 crosses and RU486 for Act-GS-255B crosses. Control flies were the progeny of a cross of the drivers to Oregon-R wild-type flies or to strain bearing a PGM-RNAi construct, as indicated. Total RNA was analyzed by Northern blot, and the membrane was hybridized sequentially to probes that will recognize the endogenous DmMRG15 mRNA and the Rp49 loading control, as indicated. Data for males is presented in the upper two panels and data for females in the lower two panels. Band intensities were quantified, and DmMRG15 expression was normalized to the Rp49 loading control. The percent change in DmMRG15 message levels between minus-drug and plus-drug samples is indicated below each pair of lanes. Values in parentheses indicates uncertainty in measurements due to interference from larger transcripts associated with line DmMRG-WT8.
Effect of conditional transgene expression on adult life span
To assay for effects on life span, the various transgenic strains were crossed to the rtTA(3)E2 driver strain or the Act-GS-255B driver strain, and survival of male and female progeny was assayed in the presence and absence of drug in replicated experiments. Representative survival curves are presented in Figure 4 for the Tet-on system and in Figure 5 for the Geneswitch system, and median life spans and statistical analyses for replicated experiments are presented in Supplemental Table S2. To control for any possible effects of the drugs, the driver strains were crossed to Oregon-R wild type flies, and to w[1118] control strain flies, and the progeny assayed for life span in the presence and absence of drug. In these control flies the effect of doxycycline on life span was either not significant and/or caused increases of +0% to +7% (Supplemental Table S2), consistent with our previous observations that doxycycline generally has neutral or small positive effects on adult fly life span (Bieschke et al., 1998; Landis et al., 2003). Over-expression of DmMRG15 was sometimes associated with small positive effects on life span, however these were generally within the +0% to +7% range of effects associated with doxycycline itself (Supplemental Table S2). In contrast, the RNAi lines produced negative effects on adult life span (Figure 4).
Figure 4. Life span analyses using the Tet-on doxycycline-regulated system.
The indicated transgenic strains were crossed to the rtTA(3)E2 driver strain, and the adult progeny containing both constructs were assayed for survival when cultured on food plus and minus doxycycline. Control flies were generated by a cross of Oregon-R wild type strain to rtTA(3)E2. The percent change in life span and p value from log-rank test are indicated in each panel. Additional statistical details and results of replicate experiments are summarized in Table 2. The plotted data are from replicate experiment number 2 (Table 2). A. Control flies. B. Line RNAi-DmMRG15-4-1. C. Line RNAi-DmMRG15–53. D. Line RNAi-MRG15-4-3.
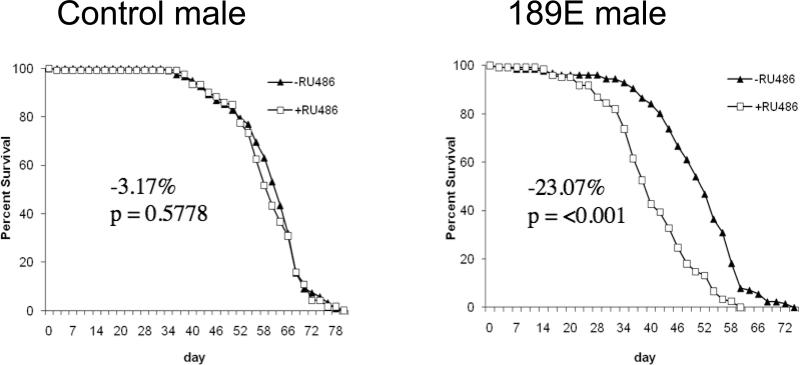
Figure 5. Life span analyses using the Geneswitch RU486-regulated system.
The indicated RNAi-Mrg15-189E transgenic strain was crossed to the Act-GS-255B driver strain, and the adult progeny containing both constructs were assayed for survival when cultured on food plus and minus RU486. Control flies were generated by a cross of Oregon-R wild type strain to Act-GS-255B. The percent change in life span and p value from log-rank test are indicated in each panel. Additional statistical details and results are summarized in Table 2. A. Control flies. B. Line 189E.
Each of the Tet-on system RNAi lines caused decreased life span in adult males: Line RNAi-DmMRG15-4-1 decreased male life span by −7% (p = 0.00143; Figure 4B), line RNAi-DmMRG15–53 decreased male life span ranging from −10% (p = 0.046; Supplemental Table S2, Exp 1) to −15.9% (p < 0.001; Figure 4C), and line RNAi-DmMRG15-4-3 decreased male life span by −7.32% (p = 0.0177; Figure 4D). In contrast, effects on female life span were generally smaller and less consistently observed: Line RNAi-DmMRG15-4-1 decreased female life span by −8% in experiment 1 (p = 0.0191; Supplemental Table S2), but had no significant effect on female life span in experiment 2 (−2%; p = 0.754). Line RNAi-DmMRG15–53 caused a small decrease in female life span in experiment 1 (−3.88%, p < 0.001; Supplemental Table S2) but had no effect on female life span in experiment 2 (−1.89%, p = 0.147; Figure 4C), whereas line RNAi-DmMRG15-4-3 decreased female life span by −16.7% (p = 0.0029; Figure 4D). The fact that line RNAi-DmMRG15-4-3 had the greatest negative effect on female life span may be related to the fact that it was the most effective of the Tet-on system lines at reducing DmMRG15 RNA levels in females (Figure 3).
Two Tet-on system lines that cause efficient RNAi knock-down of the PGM gene (Allikian et al., 2002) were used as controls to demonstrate that RNAi pathway activation per se does not decrease life span (Supplemental Table S2). Line PGM3–23 produced no significant change in life span in males (−1.32%, p = 0.71), and a small increase in females (+4.88%, p = 0.0011), whereas line PGM2–12 produced a small increase in males (+7.06%, p < 0.001) and no significant change in females (+4.94%, p = 0.246). As these changes are within the background of the assay, this result is consistent with our previous observations that activation of the RNAi pathway alone does not cause life span decrease.
With the two Geneswitch system MRG15-RNAi lines, line 189E was the most effective at RNA knock-down (Figure 3), and this line caused decreased life span in males (−23%, p < 0.001; Figure 4) and no change in life span in females (Supplemental Table S2). In contrast, line 45A2 was less effective at RNA knock-down, and caused no significant change in either males (−3.57%, p = 0.86) or females (−1.64%, p = 0.13; Supplemental Table S2). Therefore, with both Tet-on and Geneswitch systems, the decrease in life span produced by the RNAi lines generally corresponded with the amount of RNA knock-down achieved, with greater effects observed in males than in females. Taken together, the data suggest that DmMRG15 expression is required for normal longevity, and the larger and more consistent effect on life span observed in males relative to females may indicate a relatively greater requirement for MRG15 for normal life span in males, or may reflect the generally greater efficacy of RNAi achieved in males relative to females.
Effect of conditional transgene expression on maintenance of reproduction during aging
One of the defining characteristics of aging and senescence across species is a decline in reproductive fitness. To investigate if MRG-family genes function in the maintenance of reproductive capacity in Drosophila, experiments were designed to test the effect of various MRG-family transgenes on the length of the reproductive period in female and male flies. Using the Tet-on system, the effect of various transgenes on female fertility (progeny per female per day) was assayed across a period of 24 days in adult flies starting at age 40 days (Exp 1 and Exp 2), or at age 60 days (Exp 3). The flies were cultured in the presence or absence of drug throughout their adult life span, so any tendency of the transgenes to shorten or lengthen the female fertile period should be apparent as a difference in the average fertility between plus and minus drug groups at these age ranges. In control flies doxycycline treatment caused no statistically significant effect on female fertility in two out of three experiments (Experiments 1 and 3; p > 0.05) and a small but significant negative effect on female fertility in the other experiment Experiment 2, −26%, p = 0.0034) (Supplemental Table S3). These results are consistent with our previous observations that doxycycline itself has neutral or small negative effects on female fertility (Li and Tower, 2009). The RNAi-DmMRG15 lines caused conditional negative effects on female fertility in each experiment that were considerably greater in magnitude than the effects observed in control flies, consistent with a role for DmMRG15 in maintaining female fertility during aging. For example, line RNAi-DmMRG15-4-1 decreased female fertility by −50%, line RNAi-DmMRG15–53 decreased female fertility by −72%, and line RNAi-DmMRG15-4-3 decreased female fertility by −45% (p < 0.001 in each case; Supplemental Table S3). In addition, the multiple independent transgenic lines for the MT1 construct caused negative effects on female fertility ranging from −30% (line MT-106; p = 0.0166) to −60% (line MT1-3; p < 0.001), which could also be interpreted to suggest a disruption of normal DmMRG15 function required to maintain female fertility. Taken together the data suggest that DmMRG15 gene function is required for a normal reproductive life span in female flies. Notably, at the concentrations used here, the drug RU486 had a significant negative effect on female fertility (−66%, p < 0.001; Supplemental Table S3) that precluded use of the Geneswitch system. No consistent effect of the transgenes on the male fertile period was detected with the assay employed (Supplemental Table S4).
Effect of transgenes on larval survival
In both mice and flies, null mutation of the MRG15 gene is embryonic lethal. To investigate the possible role of DmMRG15 function in later stages of Drosophila development, transgenes were expressed during larval development by crossing the indicated transgenic strains to the rtTA(3)E3 driver strain, and growing the resultant larvae in media supplemented with doxycycline or in control media, in three replicate bottles for each cross. In the parents of the cross the rtTA(3)E2 chromosome was balanced over the 3rd chromosome balancer TM3, which is marked with the dominant mutation Stubble. In this way the survival of progeny containing the transgene and the rtTA(3)E2 driver, in which conditional transgene expression will occur, could be compared to the survival of sibling progeny, containing the transgene and the balancer marked with Stubble, where transgene expression will not occur. The control cross of the rtTA(3)E2 driver to Oregon-R wild-type flies controls for any possible effects of doxycycline that might alter the expected 50:50 ratio, and no significant effect of doxycycline on larval survival was observed (Supplemental Table S5). Over-expression of wild-type DmMRG15 or the truncated mutant form MT1 also had no significant effect on larval survival. In contrast, a doxycycline-dependent reduction in survival was observed with the RNAi lines. Line RNAi-DmMRG15-4-1 decreased survival of males (−9%, p < 0.001) and females (−13.5%, p = 0.029), and similarly line RNAi-DmMRG15-4-3 decreased survival of males (−4.4%, p = 0.018) and females (−10.2%, p = 0.033), whereas line RNAi-DmMRG15–53 decreased male survival (−24.5%, p < 0.001) but the decrease in females was not statistically significant (−6.05%, p = 0.133). Taken together these results are consistent with a requirement for normal DmMRG15 expression to support larval viability.
Discussion
Constructs were designed to over-express a variety of MRG15/MORF4 family proteins from human and Drosophila in transgenic flies. Interestingly, expression of the human proteins, hMRG15 and MORF4 could not be detected in adult flies, despite the assay of multiple independent transgenic lines. In contrast, doxycycline-dependent expression of protein was readily detected for the Drosophila DmMRG15 transgene and for a truncated version of DmMRG15 (called MT1) that was designed to resemble the structure of human MORF4. This suggests that either the human proteins are particularly unstable in Drosophila, or that there is some other barrier to their expression in flies.
The RNAi construct pRNAi-DmMRG15 proved difficult to transform, and one possibility is that the inverted repeat sequences may have made the construct unstable in Drosophila thereby reducing transformation efficiency. Alternatively, the low yield of transformation might reflect negative effects of expression of this construct on viability, such as was observed upon conditional expression of this construct during larval development. Conceivably this low frequency of transformation could have selected for strains where the efficiency of RNAi was relatively modest, such as the ~50% decreases observed here. Despite the modest efficiency of RNAi knock-down, the RNAi-DmMRG15 strains yielded consistent negative effects on adult life span, particularly in males. In addition the RNAi-DmMRG15 lines caused a conditional reduction in the female fertile period, as well as reductions in larval survival. The data suggest that we have successfully generated a conditional phenotype caused by a partial loss of DmMRG15 activity, and suggest that DmMRG15 normally plays a role in maintaining Drosophila viability both during development and in adults. The fact that a partial reduction in adult DmMRG15 expression was sufficient to reduce life span may indicate that function of this pathway is one limiting factor in longevity. Consistent with this conclusion, we note that the DmMRG15 null mutation strain is sickly and difficult to maintain, even as a heterozygous stock. Over-expression of wild-type DmMRG15 protein alone was not sufficient to cause a life span increase, however this may be due to the fact that DmMRG15 normally functions in a complex, where an appropriate balance of subunits may be required for optimal function.
The mammalian MRG15 gene has been found to be required for cell proliferation and embryonic viability (Tominaga et al., 2005). The phenotypes observed here for altered DmMRG15 gene expression in Drosophila are consistent with those observations, for example, expression of RNAi directed against DmMRG15 during larval development was associated with decreased survival. The fertile period of adult flies depends upon continued division of both germ-line and somatic stem cells in the gonads (Boyle et al., 2007; Pan et al., 2007; Wallenfang et al., 2006; Waskar et al., 2005), and alterations in DmMRG15 gene expression specifically in adult flies was associated with reduced female fertility. The lack of phenotype observed for male fertile period may indicate a relatively smaller requirement for DmMRG15 function in male reproduction, or may indicate less efficient expression of the transgenes in male reproductive tissues than in female reproductive tissues.
The MT1 construct encodes a truncated form of Drosophila MRG15 designed to mimic the structure of human MORF4. Conditional expression of MT1 in adult flies reduced female fertility similar to DmMRG15 RNAi, but MT1 did not have a detectable effect on adult life span or larval survival like DmMRG15 RNAi. We interpret these results to suggest that fertility has a more stringent requirement for the level of DmMRG15 activity than does life span, and/or that there is a mechanistic difference in the requirements for DmMRG15 activity in fertility versus life span. Relevant to this, DmMRG15 RNAi reduces the expression of endogenous DmMRG15, whereas the MT1 truncated protein is expected to act as a dominant negative and to compete with DmMRG15 protein in complex formation and/or other activities. Therefore these two different ways of inhibiting DmMRG15 function may have different outcomes. For example, oogenesis may involve the initiation of epigenetic states, whereas larval and adult survival might involve mostly maintenance of epigenetic states, and therefore these processes may respond differently to inhibition of DmMRG15 activity.
It will be important in the future to determine what are the critical tissues in which Drosophila DmMRG15 must function to ensure normal fertility and survival during development and aging. Mouse embryos that are null for MRG15 exhibit defects in the proliferation and differentiation of neural stem/progenitor cells, suggesting a particular requirement for MRG15 gene function in stem cells (Chen et al., 2009). In Drosophila, stem cells populations have recently been identified that continue to divide in adult flies in the gut (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006) and in the malpighian tubule (the fly kidney homolog)(Singh et al., 2007). It remains to be determined if the continued proliferation of these somatic stem cell populations are required for normal adult Drosophila life span, and one interesting question to ask in the future is if DmMRG15 inactivation in adult flies causes decreased life span by inhibiting the proliferation of stem cells, or through some other mechanism.
Supplementary Material
Acknowledgements
We thank J. Workman for providing antibody directed against DmMRG15, and thank Gary Landis for help with experiments. This work was supported by grants from the Department of Health and Human Services to JT (AG011833) and OMPS (AG032134), and grants from the Ellison Medical Foundation (OMPS) and the American Federation for Aging Research (KT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allikian MJ, Deckert-Cruz D, Rose MR, Landis GN, Tower J. Doxycycline-induced expression of sense and inverted-repeat constructs modulates phosphogluconate mutase (Pgm) gene expression in adult Drosophila melanogaster. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-research0021. research0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–6. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Bertram MJ, Berube NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, Clark LJ, Parkinson EK, Barrett JC, Smith JR, Pereira-Smith OM. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol. 1999;19:1479–85. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieschke ET, Wheeler JC, Tower J. Doxycycline-induced transgene expression during Drosophila development and aging. Mol Gen Genet. 1998;258:571–9. doi: 10.1007/s004380050770. [DOI] [PubMed] [Google Scholar]
- Boyle M, Wong C, Rocha M, Jones DL. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1:470–8. doi: 10.1016/j.stem.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Chen M, Takano-Maruyama M, Pereira-Smith OM, Gaufo GO, Tominaga K. MRG15, a component of HAT and HDAC complexes, is essential for proliferation and differentiation of neural precursor cells. J Neurosci Res. 2009;87:1522–31. doi: 10.1002/jnr.21976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–84. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimauro T, David G. Chromatin modifications: the driving force of senescence and aging? Aging (Albany NY) 2009;1:182–90. doi: 10.18632/aging.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford D, Hoe N, Landis GN, Tozer K, Luu A, Bhole D, Badrinath A, Tower J. Alteration of Drosophila life span using conditional, tissue-specific expression of transgenes triggered by doxycycline or RU486/Mifepristone. Exp Gerontol. 2007;42:483–97. doi: 10.1016/j.exger.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel S, Rogina B. Drosophila longevity is not affected by heterochromatin-mediated gene silencing. Aging Cell. 2005;4:53–6. doi: 10.1111/j.1474-9726.2005.00143.x. [DOI] [PubMed] [Google Scholar]
- Fujita M, Takasaki T, Nakajima N, Kawano T, Shimura Y, Sakamoto H. MRG-1, a mortality factor-related chromodomain protein, is required maternally for primordial germ cells to initiate mitotic proliferation in C. elegans. Mech Dev. 2002;114:61–9. doi: 10.1016/s0925-4773(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, Confalonieri S, Cesaroni M, Marchesi F, Gasco M, Scanziani E, Capra M, Mai S, Nuciforo P, Crook T, Lough J, Amati B. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–7. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Ishii N. Chromosome dosage as a life span determinant in Caenorhabiditis elegans. Mech Ageing Dev. 2007;128:437–43. doi: 10.1016/j.mad.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. Embo J. 1997;16:2054–60. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–40. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–8. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kang HL, Benzer S, Min KT. Life extension in Drosophila by feeding a drug. Proc Natl Acad Sci U S A. 2002;99:838–43. doi: 10.1073/pnas.022631999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Landis GN, Bhole D, Tower J. A search for doxycycline-dependent mutations that increase Drosophila melanogaster life span identifies the VhaSFD, Sugar baby, filamin, fwd and Cctl genes. Genome Biol. 2003;4:R8. doi: 10.1186/gb-2003-4-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–9. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Li H, Mitchell JR, Hasty P. DNA double-strand breaks: a potential causative factor for mammalian aging? Mech Ageing Dev. 2008;129:416–24. doi: 10.1016/j.mad.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tower J. Adult-specific over-expression of the Drosophila genes magu and hebe increases life span and modulates late-age female fecundity. Mol Genet Genomics. 2009;281:147–62. doi: 10.1007/s00438-008-0400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–9. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Morales V, Regnard C, Izzo A, Vetter I, Becker PB. The MRG domain mediates the functional integration of MSL3 into the dosage compensation complex. Mol Cell Biol. 2005;25:5947–54. doi: 10.1128/MCB.25.14.5947-5954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–4. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Olgun A, Aleksenko T, Pereira-Smith OM, Vassilatis DK. Functional analysis of MRG-1: the ortholog of human MRG15 in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 2005;60:543–8. doi: 10.1093/gerona/60.5.543. [DOI] [PubMed] [Google Scholar]
- Pan L, Chen S, Weng C, Call G, Zhu D, Tang H, Zhang N, Xie T. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell. 2007;1:458–69. doi: 10.1016/j.stem.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem. 2002;277:50860–6. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- Pena AN, Pereira-Smith OM. The role of the MORF/MRG family of genes in cell growth, differentiation, DNA repair, and thereby aging. Ann N Y Acad Sci. 2007;1100:299–305. doi: 10.1196/annals.1395.031. [DOI] [PubMed] [Google Scholar]
- Pereira-Smith OM, Smith JR. Genetic analysis of indefinite division in human cells: identification of four complementation groups. Proc Natl Acad Sci U S A. 1988;85:6042–6. doi: 10.1073/pnas.85.16.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Jin H, Lilja T, Mannervik M. Drosophila Reptin and other TIP60 complex components promote generation of silent chromatin. Genetics. 2006;174:241–51. doi: 10.1534/genetics.106.059980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- Shen J, Curtis C, Tavaré S, Tower J. A screen of apoptosis and senescence regulatory genes for life span effects when over-expressed in Drosophila. Impact Aging. 2009;1:191–211. doi: 10.18632/aging.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci U S A. 2010;107:169–74. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SR, Liu W, Hou SX. The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem Cell. 2007;1:191–203. doi: 10.1016/j.stem.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–77. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–42. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Takasaki T, Liu Z, Habara Y, Nishiwaki K, Nakayama J, Inoue K, Sakamoto H, Strome S. MRG-1, an autosome-associated protein, silences X-linked genes and protects germline immortality in Caenorhabditis elegans. Development. 2007;134:757–67. doi: 10.1242/dev.02771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Voss AK. The diverse biological roles of MYST histone acetyltransferase family proteins. Cell Cycle. 2007;6:696–704. doi: 10.4161/cc.6.6.4013. [DOI] [PubMed] [Google Scholar]
- Tominaga K, Kirtane B, Jackson JG, Ikeno Y, Ikeda T, Hawks C, Smith JR, Matzuk MM, Pereira-Smith OM. MRG15 regulates embryonic development and cell proliferation. Mol Cell Biol. 2005;25:2924–37. doi: 10.1128/MCB.25.8.2924-2937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J. Sex-specific regulation of aging and apoptosis. Mech Ageing Dev. 2006;127:705–18. doi: 10.1016/j.mad.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Tyteca S, Legube G, Trouche D. To die or not to die: a HAT trick. Mol Cell. 2006;24:807–8. doi: 10.1016/j.molcel.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Wallenfang MR, Nayak R, DiNardo S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell. 2006;5:297–304. doi: 10.1111/j.1474-9726.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- Waskar M, Li Y, Tower J. Stem Cell Aging in the Drosophila Ovary. AGE. 2005;27 doi: 10.1007/s11357-005-2914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45 doi: 10.1016/j.exger.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Fujisawa T, Hwang JS, Ikeo K, Gojobori T. Degeneration after sexual differentiation in hydra and its relevance to the evolution of aging. Gene. 2006;385:64–70. doi: 10.1016/j.gene.2006.06.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.