Summary
Left-right (LR) patterning is an intriguing but poorly understood process aspect of bilaterian embryogenesis. We report a mechanism for LR patterning in C. elegans in which the embryo uncouples its midline from the anteroposterior (AP) axis. Specifically, the eight-cell embryo establishes a midline that is tilted rightward from the AP axis and positions more cells on the left, allowing subsequent differential LR fate inductions. To establish the tilted midline, cells exhibit LR asymmetric protrusions and a handed collective movement. This process, termed chiral morphogenesis, involves differential regulation of cortical contractility between a pair of sister cells that are bilateral counterparts fate-wise, and is activated by non-canonical Wnt signaling. Chiral morphogenesis is timed by the cytokinetic furrow of a neighbor of the sister pair, providing a developmental clock and an unexpected signaling interaction between the contractile ring and adjacent cells.
Introduction
Many bilateral organisms exhibit stereotypic left-right (LR) asymmetries in e.g. the position and shape of internal organs. However, we are only beginning to understand how this asymmetry arises, propagates and integrates with the overall superficial bilateral symmetry during embryogenesis (Brown and Wolpert, 1990; Wood, 1997). In vertebrates, the early events that initiate LR asymmetries are unclear and appear to vary among species (Levin, 2005; Tabin, 2005). For example, in the mouse it requires the mechanical asymmetry of directional cilia rotation and the resulting leftward nodal flow (Hirokawa et al., 2006; Shiratori and Hamada, 2006). In the chick, it involves actomyosin based asymmetric cell movements to create a LR asymmetric node and to rearrange the initially symmetric gene expression pattern (Cui et al., 2009; Gros et al., 2009). Nonetheless, the early events in different species converge to the asymmetric expression of Nodal on the left side of the midline around gastrulation in an embryo with gross morphological LR symmetry. The asymmetric Nodal expression eventually leads to visceral asymmetry (Levin, 2005; Tabin, 2005; Shiratori and Hamada, 2006).
In contrast to vertebrates, gross morphological asymmetry arises early during Caenorhabditis elegans embryogenesis and without any obvious differential fate induction between the left and right sides. The morphological symmetry is broken during the four- to six-cell transition via a skew of the spindle in two blastomeres, ABa and ABp (Sulston et al., 1983; Wood, 1997; Figure S1A). From an initial orientation orthogonal to the anteroposterior (AP) axis, the spindles skew during elongation so that the left daughters of ABa and ABp are localized more anteriorly than the right daughters. At this stage, each pair of LR sisters (ABal-ABar and ABpl-ABpr) is equivalent in fate potential, but this fate symmetry is subsequently broken via a series of Notch signaling events (Hutter and Schnabel, 1995; Priess, 2005). Intriguingly, reversing the direction of the ABa/ABp spindle skew through micromanipulations generates organisms with reversed handedness (Wood, 1991). Given the mechanical nature of the manipulation, this result suggests that asymmetry in cytoskeletal and spindle mechanics may instruct handedness choice upstream of differential gene expression. However, it is currently not clear how initial symmetry breaking translates into later LR asymmetries, especially whether there are other morphogenetic events downstream that ensure maintenance of the initial LR bias, and if they exist, when and how they are triggered.
By systematically tracking cells through C. elegans embryogenesis (Bao et al., 2006), we analyzed the cellular events in the establishment and elaboration of the bilateral body plan. Our results suggest a strategy for embryonic LR patterning that involves uncoupling the midline, which bisects the bilateral structures, from the AP axis. At the eight-cell stage, the C. elegans embryo establishes a midline that is tilted from the AP axis to the right to break morphological symmetry and to allow differential Notch inductions between the two sides. We further show that starting with the spindle skew of ABa/ABp, the embryo undergoes a complex morphogenesis to establish the tilted midline, involving LR asymmetric protrusions and a chiral rearrangement of cells. We term this process chiral morphogenesis and demonstrate that it is based on differential regulation of cortical actomyosin contractility in ABpl and ABpr, a pair of otherwise bilaterally equivalent cells. In addition, we present identification and analyses on the cellular and molecular signals regulating chiral morphogenesis, including a non-canonical Wnt signal that is required to activate actomyosin dynamics in ABpl/ABpr; a novel signal from the cytokinetic furrow of a neighboring cell (EMS) that times the protrusion formation in ABpl/ABpr, whose molecular identity is yet to be identified; and the direction of the ABa/ABp spindle skew that likely sets the handedness of chiral morphogenesis.
Results
An Asymmetric Bilateral Body Plan at the Eight-Cell Stage
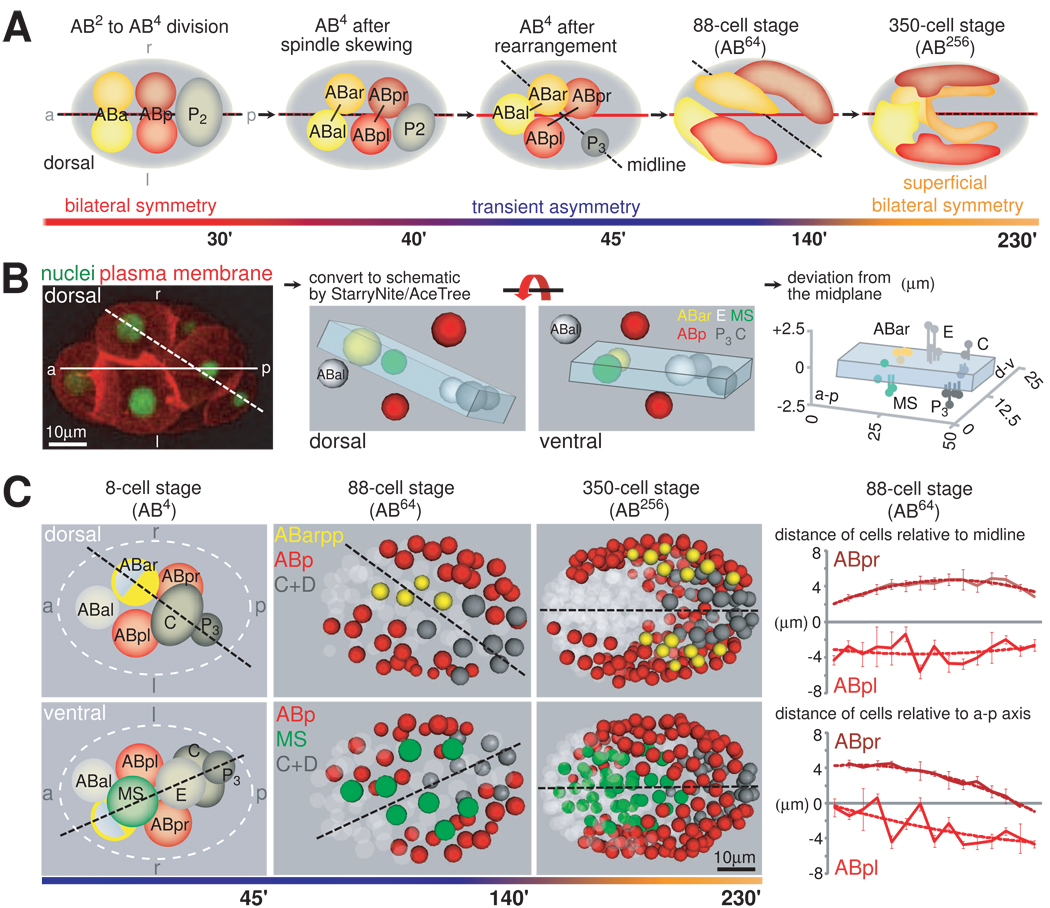
We find that after the ABa/ABp spindle skew that breaks the morphological LR symmetry of the embryo (Wood, 1991), cells are further rearranged to assemble a stereotypical configuration at the 8-cell stage (Figure 1A). Specifically, 5 of the embryo’s 8 cells (ABar, MS, E, C, and P3) are positioned on a plane that tilts from the embryo’s AP axis to the right side by 22±2° degrees (n=10) (Figure 1B; Movie S1). The formation of the plane is highly reproducible as on average cells deviate from the plane by less than a quarter of nuclear diameter (Figure 1B, Figure S1B). Two other cells, namely ABpl and ABpr, are positioned symmetrically on two sides of the tilted plane (Figure 1B, red nuclei). The eighth cell, ABal, is on the left side of the plane.
Figure 1. Formation of a Midline through Blastomere Rearrangements.
A. Positioning of AB blastomeres from the 6- to 350-cell Stage. Schematic dorsal views rendered from lineaging data, which show the two repositioning events of the midline (dashed line) relative to the AP-axis (solid red line) at the AB8- and the AB256 cell stage. B. Generation of a midplane at the 8-cell stage. Left panel: Illustration of the AP-axis (solid line) and the midplane (dashed line) in a dorsally oriented embryo with fluorescently labeled nuclei and plasma membrane (HIS-72::GFP and PH-domain of PLC1δ1 fused to mCherry, respectively). Middle panel: Schematic rendering of the midplane. The midplane comprises ABar, MS, E, C and P3. Note that the illustrations represent data obtained from lineaging and thus real embryos. Right panel: Deviation of cells from the midplane. Values for each cell from 4 embryos are shown. Note the different scale of the axes. Orientation and colors of cells match those in the middle panel showing a ventral view. (For details see also Figure S1 and Movie S1). C. A tilted midline is maintained until late gastrulation. Left panel: Schematic dorsal and ventral views of 8-cell stage embryos. Cells are colored as in A. The cell of which only the posterior half is colored represents ABar, where only one posterior granddaughter generates bilateral symmetric structures (see text). Middle panel: Representation of embryos lineaged with StarryNite/AceTree. Dashed lines represent midlines. Right panels: Quantification of distances of ABpl/ABpr derived blastomeres in 4 individual 88-cell embryos (mean +/− SD). The upper panel indicates the distances relative to the tilted midline and the lower panels relative to the AP-axis. See also Movie S2.
This stereotypical configuration provides the blueprint for the bilateral body plan of C. elegans. Specifically, the tilted plane serves as the center of bilateral symmetry. According to the invariant cell lineage and fates, the five cells on the plane each generates a bilaterally symmetric structure and essentially contributes equally to the left and right sides of the organism (Figure 1C, see also below), with the minor exception of ABar, where one of its granddaughters (ABarpp) generates a bilateral structure. ABpl and ABpr, the two cells positioned symmetrically on the two sides of the plane, are equivalent fate-wise and give rise to symmetric structures on the left and right side, respectively. Thus, the tilted plane, bisecting the bilaterally symmetric founder cells and hence future bilateral structures, qualifies as the midplane, or the midline as it is commonly referred to. Notably, with the rightward tilt, the midline is uncoupled from the AP axis (i.e. the long axis of the ellipsoidal egg), which has not been seen in other bilaterian organisms.
Furthermore, the eighth cell, ABal, which does not have a bilateral counterpart in the cell lineage, is segregated to the left side of the tilted midline with no cell mirroring it on the right. Thus, the eight cells are organized into an inherently asymmetric bilateral body plan, with more cells on the left side of the midline. This asymmetric bilateral body plan provides the anatomical basis for how the invariant Notch inductions can generate LR fate asymmetry, which was not fully understood before (see Discussion; Hutter and Schnabel, 1995; Hermann et al., 2000).
The tilted midline configuration is maintained until the 88-cell stage (the AB64 stage; Figure 1C, middle panel), as the bilaterally symmetric founder cells divide symmetrically with respect to the tilted midline (Figure 2A). Cell movements then start to gradually adjust the midline over the next two cell cycles, so that by the 350-cell stage (AB256; Figure 1C, right panel), it aligns with the AP axis (Movie S2 and see Sulston et al., 1983; Schnabel et al., 1997; Zhao et al., 2008). The re-alignment of the midline with the AP axis restores the spatial symmetry of the bilateral structures of the worm. Thus, the dynamic positioning of the midline relative to the AP axis organizes both the LR asymmetric structures and the integration of these asymmetries into a superficially symmetric body plan (Figure 1A).
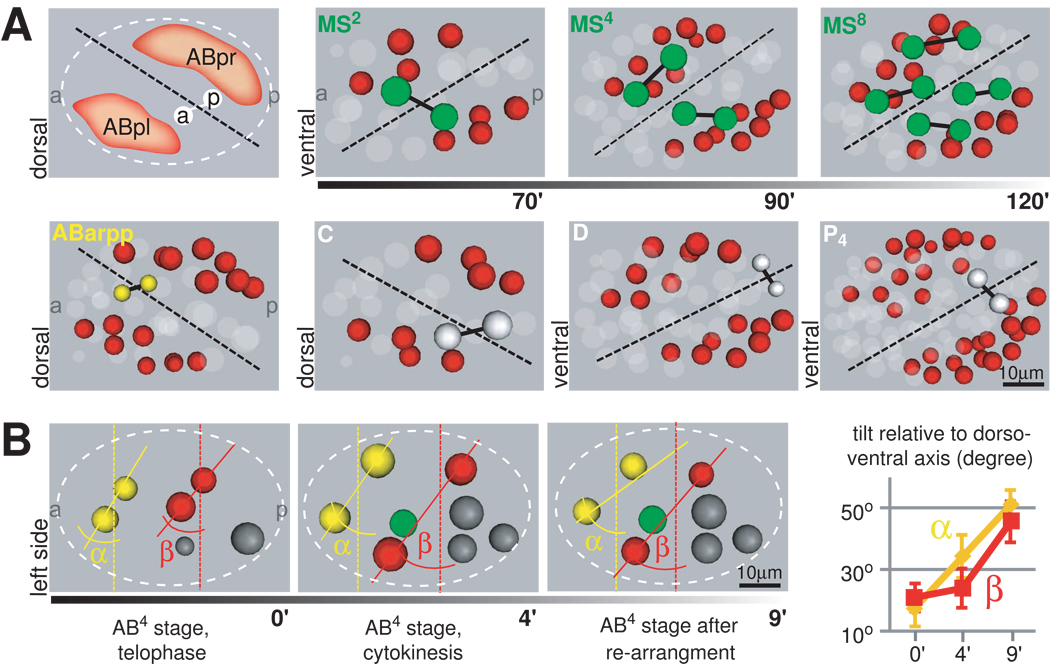
Figure 2. The Tilted Midline Organizes Body Plan LR Asymmetry and LR Lineage Symmetry.
A. Cells of different lineages are positioned symmetrically to the midline. Schematic and representations of embryos lineaged with StarryNite/AceTree displayed from the dorsal side. Sister pairs from an LR symmetric division of the respective founder cells are shown. Subsequent divisions are shown for MS (upper right panel). Color code for cells is depicted on the left, ABp cells are in red. Note that positioning of MS, D and P4 cells is depicted from the ventral side. B. Contributions of spindle skew vs. blastomere rearrangement to LR symmetry breaking. Left: Time-lapse data from lineaged embryos. Cells are color-coded as in Figure 1. α represents the angle between the connecting line of the ABar/al nucleus (solid yellow line) and the geometric LR axis (dashed yellow line), β the respective angle for ABpr/pl (red lines). Right: The change in α and β over time was measured (n=10, mean +/− SD).
Additionally, the rearrangement of cells during the assembly of the body plan further increases the morphological LR asymmetry caused by the ABa/ABp spindle skew, on top of creating the architectural framework for the bilateral body plan. Quantitatively, the skew increases from 19±5° at the end of the ABa/ABp division to 48±6° after the rearrangement (Figure 2B).
Chiral Morphogenesis Assembles the Asymmetric Bilateral Body Plan
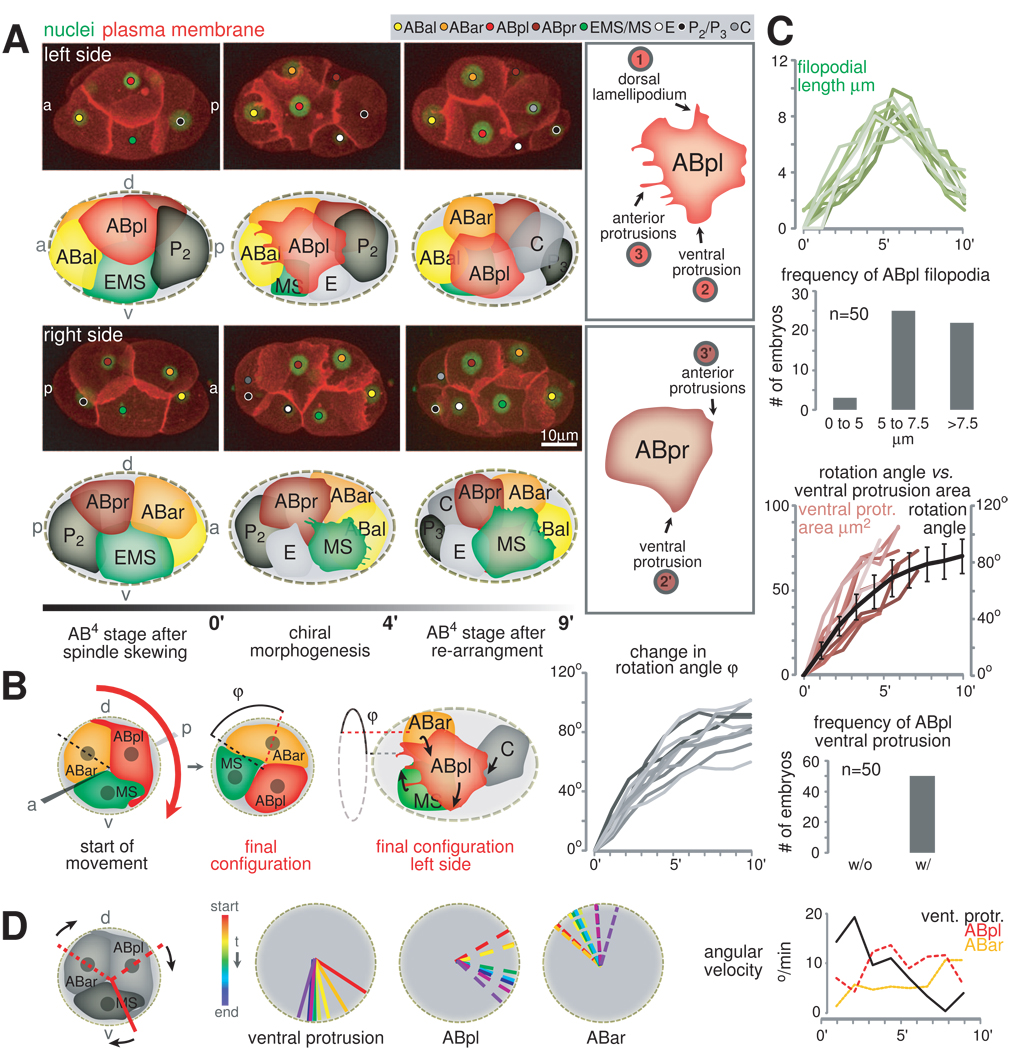
Analyzing time lapse recordings of embryos expressing a plasma membrane marker (PH-domain of PLC1δ1 fused to mCherry; Audhya et al., 2005), we find that the rearrangement of cells during the assembly of the bilateral body plan involves reproducible LR asymmetric cell behaviors and movements with a specific handedness (Figure 3A left panel). We term this process chiral morphogenesis. As the ABa/ABp spindle skew finishes and before the contractile ring fully closes, the LR equivalents, ABpl and ABpr, start to exhibit a set of asymmetric behaviors. ABpl undergoes dramatic shape changes: It forms a dorsal lamellipodium, a ventral protrusion as well as anterior filopodial extensions (Figure 3A, upper right panel, marked 1, 2 and 3, respectively), and migrates anteroventrally. Besides these protrusions, ABpl also forms an apparently adhesive contact with C, as the two cells form an hourglass shape (Figure 3A, upper left panel, middle frame and Movie S3). In contrast, ABpr shows only a rudimentary ventral protrusion and ruffling on its anterior front instead of filopodial extensions (Figure 3A, lower right panel, marked 2’ and 3’, respectively and Movie S3) and does not move significantly. Another cell with significant shape changes is MS, which forms an elaborate lamellipodial protrusion at its anterior front (Figure 3A, lower left panel).
Figure 3. Chiral Morphogenesis Mediates the Formation of the Asymmetric Bilateral Body Plan.
A. Chiral morphogenesis and its underlying cellular behaviors. Left panel: 3D-projection stills, left side and right side views with matched schematics. The color code of this schematic matches that of the small circles in 3D-projections throughout the paper aiming to help the reader identifying individual blastomeres. Plasma membrane is shown in red (PH-domain of PLC1δ1 fused to mCherry) and nuclei in green (HIS-72::GFP, HIS-72 is a H3 variant histone). Right panel: Features specific to ABpl and ABpr are indicated with circled numbers in the boxes outlined in gray (see text and Movie S3 for details). B. Chiral morphogenesis is a rotational cellular rearrangement. Left panels: Cartoon depicting a view from the anterior onto a transverse section and a side view that detail the rotation. The anterior view shows two points in time, first, the starting configuration and, second, the configuration after completion of the collective rotation. Right panel: Robustness of the rotational rearrangement. The graphs display the dynamic change in the rotation angle φ of the ABar/ABpl/MS-group during chiral morphogenesis as measured in 10 different wild-type embryos (details on the measurement can be found in Supplemental Data). 0’ represents the timepoint just prior to the start of noticeable changes in nuclear positions. For details on nuclear movements see also Figure S2 and Movie S4. C. Quantification of protrusion formation. Time scales as in B, right panel. Upper panel: Growth of the longest filopodium is shown on top of a graph showing the frequency of the appearance of filopodia on ABpl. Lower panel: Correlation of growth of the ABpl’s ventral protrusion with cell rotation is shown on top of a graph showing the frequency of the appearance of ABpl’s ventral protrusion. D. ABpl’s ventral protrusion leads the rotational rearrangement. Left: The radial position of the ventral protrusion and of ABpl’s and ABar’s nuclei were analyzed in a representative embryo. Right: Angular velocity for the three elements depicted on the left (average; n=3).
With the LR asymmetric protrusive activities, cells show a handed movement and rearrange accordingly. The first major aspect of the cell rearrangement involves three cells, namely ABpl, MS, and ABar (Figure S2A). Their movements can best be described as a collective rotation around the AP axis (Figure 3B and Movie S4). Looking from the anterior end of the embryo, the rotation is clockwise by 83±7° (n=10), and brings ABar and MS onto the midplane (Figure 3B and S2B). As other cells do not show circumferential movements (Movie S4; Figure S2B), this is not a whole-embryo rotation as previously indicated (Sulston et al., 1983; Schnabel et al., 1997). The second major aspect of the cell rearrangement is the juxtaposition of ABar and C. As ABar shifts posteriorly during the three-cell rotation, C shifts anteriorly to meet ABar, likely pulled by ABpl given the hourglass shape (see above). The juxtaposition makes the five-cell midplane contiguous (Figure 1B) and separates ABpl and ABpr (red in Figure 1B), which are sisters and born next to each other, to two sides of the midplane. Besides this, the ABar-C contact is also required for ABar to receive a Wnt signal from C to orient its spindle (Walston et al., 2004).
The protrusion formation and the collective cell movement both occur reproducibly and are well coordinated temporally (Figure 3C).
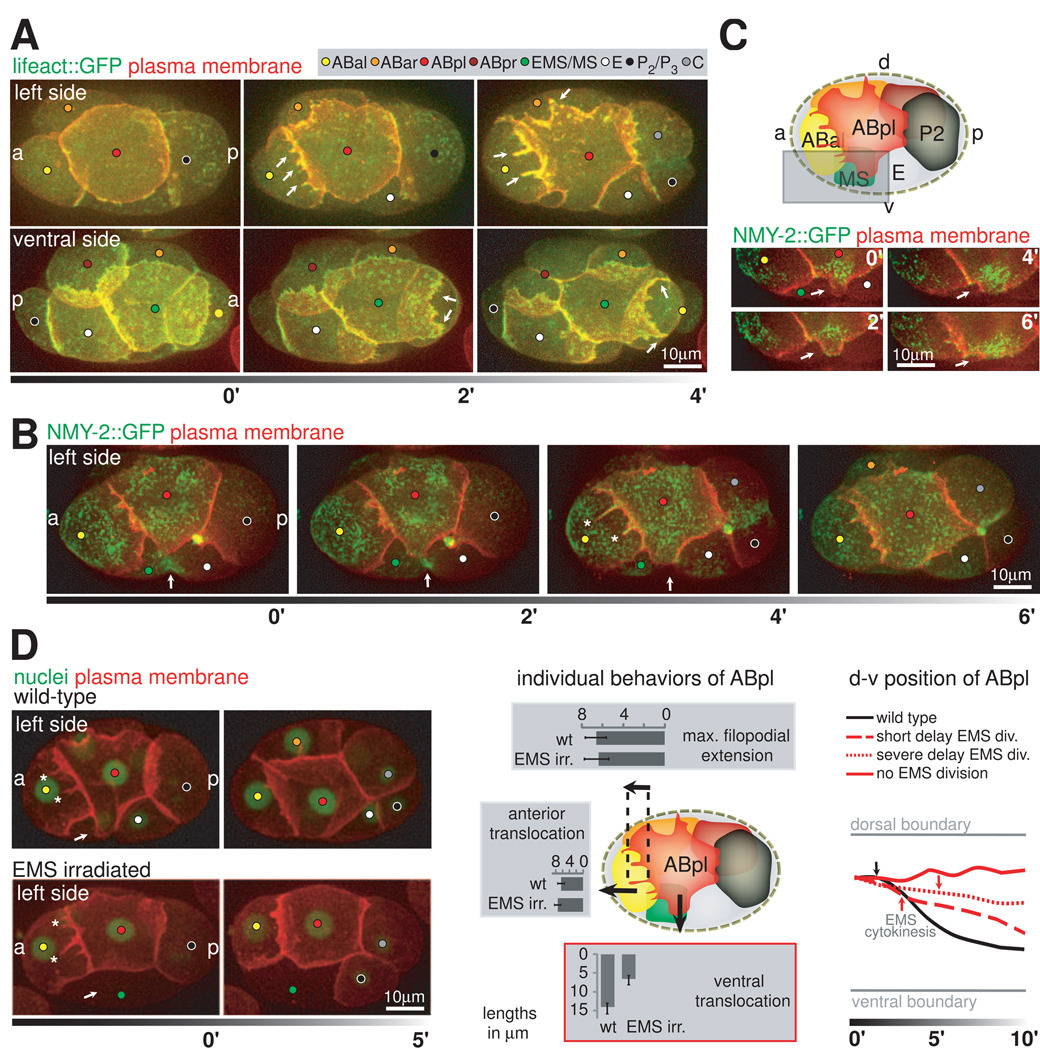
Coordination and Timing of Chiral Morphogenesis
In order to understand how the dynamic protrusions described above mediate the cell movements, we analyzed actomyosin dynamics during chiral morphogenesis. To this end we imaged F-actin by using Lifeact::GFP (Riedl et al., 2008), and non-muscle myosin II heavy chain (NMY-2::GFP; Munro et al., 2004). F-actin strongly accumulates in the anterior and dorsal protrusions of ABpl (Figure 4A, upper panel; Movie S5) and the lamellipodium of MS (Figure 4A, lower panel). Furthermore, as actin polymerization driving protrusion formation requires the Rho GTPase Cdc42 (Pollard et al., 2000), we observed a loss of protrusions when we deplete embryos for CDC-42 by RNAi (Figure S3A and data not shown). These findings show that actin polymerization mediates the formation of protrusions and the directional spreading of ABpl and MS. Furthermore, NMY-2 accumulates at the anterior front of ABpl and fills the filopodial protrusions as they grow (Figure 4B, asterisks), indicative for a role of NMY-2 in cell-cell contact formation in these protrusions (Conti et al., 2004).
Figure 4. Actomyosin Dynamics and the Coordination of Protrusion Formation.
A. F-actin dynamics during protrusion formation. Top panel: 3-D projection stills, left side views. Plasma membrane is shown in red F-actin in green (Lifeact::GFP). Arrows point at the anterior and dorsal protrusions of ABpl. Lower panel: As above, however an embryo imaged ventrally. Arrows indicate the lamellipodium of MS during maximal extension (2’) and during disassembly (4’). For details and a schematic see Movie S5 and Figures S3A and B. B. Non-muscle myosin II dynamics during cell movement. 3-D projection stills, left side views. Plasma membrane is shown in red, cortical non-muscle myosin in green (NMY-2::GFP). Arrows point to the EMS furrow. Asterisks (timepoint 4’) indicate NMY-2 accumulation in the growing anterior filopodia of ABpl. C. ABpl forms a ventral protrusion onto MS. Upper panel: Cartoon depicting the embryo’s left side at the time when ABpl has formed its protrusions. Lower panel: 3-D projection stills, left side views showing only the ventral protrusion of ABpl as indicated by the boxed area in the cartoon. Arrows point to the tip of the protrusion. D. ABpl’s ventral protrusion is triggered by EMS cytokinesis. Left panel: 3-D projection stills, left side views with plasma membrane (red) and nuclei (green) fluorescently labeled. In the lower panel, the EMS nucleus in the embryo was irradiated to delay division. Arrows point to the location where ABpl’s ventral protrusion forms. Asterisks point to ABpl’s anterior filopodia. Middle panel: Quantification of ABpl’s displacement in both anterior and ventral directions and of its longest filopodium in wt embryos and embryos where EMS nucleus was irradiated (n=6, respectively, mean +/− SD). Right panel: Impairment of ABpl’s ventral displacement correlates with the degree of EMS division delay. Gray lines indicate the dorsal and ventral boundary of an embryo. Black and red lines represent the position of ABpl’s nucleus over time. Arrows indicate the timepoint when the furrow appears in EMS. Note that when EMS does not divide during the lifetime of ABpl, ABpl essentially does not move ventrally (solid red line). For details on cell-cell contacts see Figure S4.
We further investigated the ventral protrusion of ABpl. Temporally and spatially, this protrusion leads the ventral movement of ABpl and the translocation of ABar (Figure 3D): it forms in all embryos examined (n=50) and its size is proportional to the degree of the ventral movement (n=10, Figure 3C). More specifically, the ABpl ventral protrusion starts to form as the EMS cell forms its cytokinetic contractile ring (Figure 4B, arrow). A small protrusion first forms onto the future MS part of the cell. It then turns into rapid and directional growth along the contact with MS, thus does not seem to simply fill the open space of the EMS furrow (Figure S4). Interestingly, formation of the ventral protrusion is not associated with an enrichment of F-actin (Figure 4A), but with NMY-2 (Figure 4C). It is therefore possible that it constitutes a ‘cryptic’ lamellipodium (Farooqui and Fenteany, 2004) which has been shown to mediate mechanical coupling during collective migration.
To test if ABpl’s ventral protrusion is triggered by EMS cytokinesis, or if the timing of the two events is coincidental, we delayed EMS division by UV irradiation. We observe that belated EMS divisions delay the formation of the ABpl protrusion, and the delayed protrusion forms when the EMS starts to form the cytokinetic furrow (Figure 4D). In the more severe cases where EMS division is delayed until after ABpl divides, ABpl does not form the protrusion (Figure 4D, left panel). These results suggest that the ABpl ventral protrusion is indeed triggered by EMS cytokinesis. Furthermore, delaying ABpl protrusion delays and reduces ventral movement of ABpl and the ABpl-MS-ABar rotation (Figure 4D, middle and right panels). When the protrusion does not form, there is no ventral movement or collective rotation (Figure 4D, right panel). The irradiation does not affect other protrusions of ABpl (Figure 4D, middle panel, see also below), suggesting that the effect is specific to the ventral protrusion. Furthermore, irradiating other neighbors of ABpl, such as ABal, does not affect the ABpl ventral protrusion or movement (data not shown), suggesting that the irradiation effect is EMS specific. Thus, the results suggest that the ABpl ventral protrusion is triggered by EMS cytokinesis and is required to mediate ABpl’s ventral movement and chiral morphogenesis.
Abolishing the ventral protrusion also reveals the nature of the other behaviors of ABpl: The anterior and dorsal protrusions are not affected, suggesting that their formation does not depend on the interaction with the EMS furrow (Figure 4D, middle panel). Furthermore, as ABpl does not move ventrally in this case, the result suggests that the dorsal protrusion is an active process, rather than a passive adhesion patch being stretched by ABpl’s movement. In contrast, the stretching of C by ABpl disappears, suggesting that it is the result of traction force as ABpl’s movement pulls on C.
Handedness of Chiral Morphogenesis
As described above, ABpr, the bilateral equivalent of ABpl fate-wise, has a much more reduced ventral protrusion in terms of size and duration compared to that of ABpl. Given that the ABpl ventral protrusion is required for chiral morphogenesis, this difference may determine the handedness of chiral morphogenesis, that is, the clockwise direction of the cell rearrangement. We therefore investigated how the asymmetry and handedness are brought about.
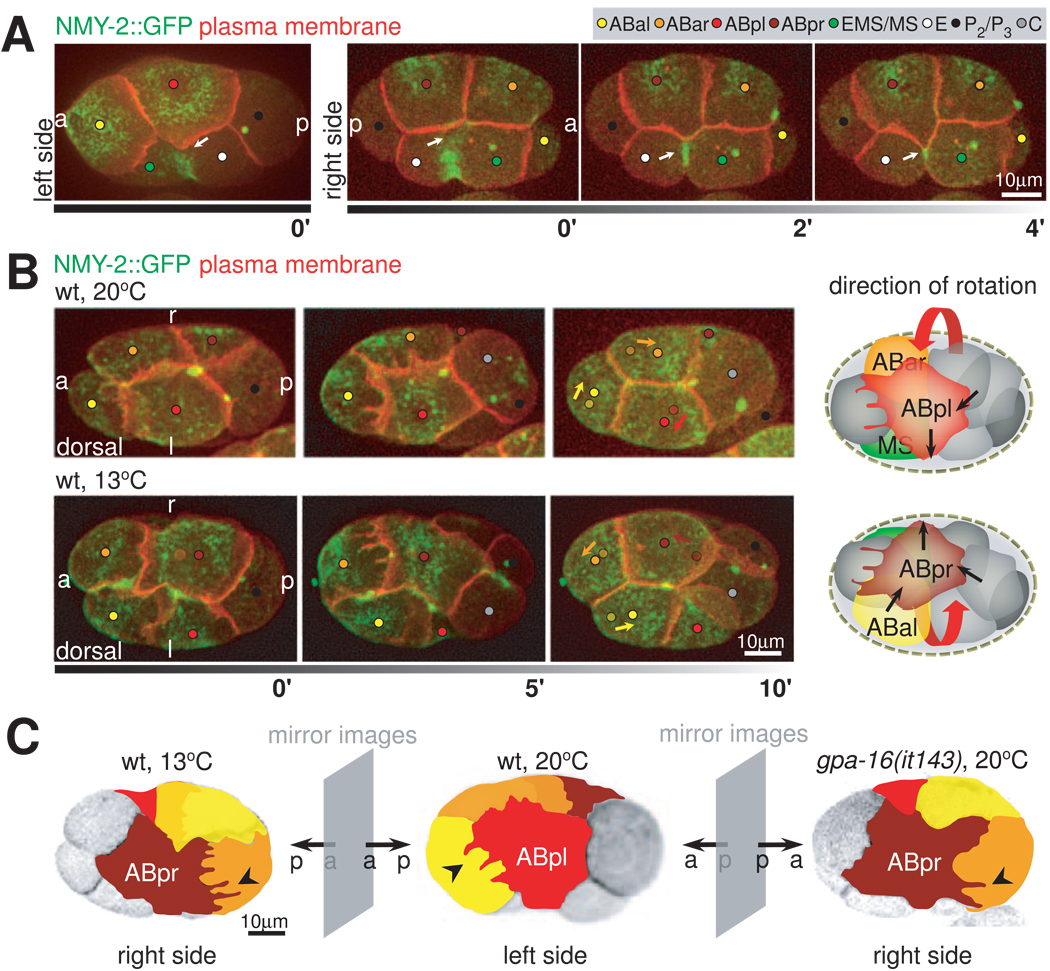
Given that EMS cytokinesis triggers the ventral protrusion, we first focused on the EMS furrow. EMS undergoes an asymmetric abscission with the contractile elements coalescing faster on the left side and the midbody forming on the right (Figure 5A). Thus, we tested if the asymmetric furrow, with faster contraction on the left, may direct ABpl to grow its ventral protrusion faster and/or more significantly than ABpr. Specifically, we perturbed the asymmetric furrow by depleting embryos of the septins UNC-59/61 with RNAi, which are required for asymmetric abscission (Maddox et al., 2007). The perturbation led to cases where the EMS midbody appears on the left instead of the right side (Movie S6). The result suggests that there is a novel mechanism that aligns the intrinsically asymmetric contractile ring with the LR body axis and that septins are required for the alignment. However, the asymmetry between ABpl and ABpr ventral protrusions is not affected and chiral morphogenesis occurs as in the wild type. This result further confirms that the ABpl ventral protrusion is not a passive response to fill the open space of the EMS furrow as the space on the left side is much smaller than the wild type. More importantly, the result suggests that while the EMS furrow triggers the ventral protrusion formation, its LR asymmetry does not regulate the handedness of chiral morphogenesis.
Figure 5. Handedness of Chiral Morphogenesis.
A. LR asymmetry of EMS cytokinesis correlates with asymmetry in ABpl/pr ventral protrusions. 3-D projection stills, left and right side views, respectively, with plasma membrane (red) and NMY-2 (green) fluorescently labeled. 0’ frames of left and right sides show the same point in developmental time in two carefully staged embryos. Arrows point at the contacts between the coalescing furrow of EMS and the ventral protrusions of ABpl and ABpr (0’) and at the developing midbody on the right side (2’ and 4’). B. Reversal of chiral morphogenesis by raising embryos at low temperature. Left panel: 3D- projection stills, dorsal views, right (r) and left (l) sides are indicated. Fluorescent markers are plasma membrane (red) and NMY-2 (green). Arrows in the last timepoint indicate the translocation of the corresponding cells, where the dots with lighter shade indicate the starting positions of the corresponding cells as shown in the first timepoint. Right panel: Schematic representation of wild-type and mirror-symmetric chiral morphogenesis. The three cells that perform the collective rotation are highlighted, red arrows indicate the handedness of the rotation. Black arrows indicate directions of force as deduced from the directionality of protrusions and cell movement. See also Movie S7. C. Overview of the effect of cold-treatment and gpa-16(it143) on chiral morphogenesis. 3D-projection stills from embryos expressing fluorescently labeled plasma membranes. Individual blastomeres are highlighted by false color representation to highlight mirror symmetry to the wild type; right side views of an embryo raised at 13°C and a gpa-16(it143) embryo at restrictive temperature (20°C) and left side view of a wild-type embryo. See also Movie S8.
We then tested if the handedness of the ABa/ABp spindle skew sets the handedness of the ABpl-ABpr asymmetry and chiral morphogenesis. To do so, we reversed the handedness of the ABa/ABp spindle skew with two different conditions. First, we cultivated worms at low temperature, which induces reversal of the ABa/ABp spindle skew in a fraction of embryos by impairing an unknown process before fertilization of the oocyte (Wood et al., 1996). In the cases where we observed reversed spindle skew, mirror-imaged chiral morphogenesis occurs: ABpr instead of ABpl undergoes a dramatic shape change, forms protrusions and the cellular rearrangement is a counterclockwise rotation (Figure 5B, lower panel; Movie S7). As a result, the midplane is tilted to the left and C induces a spindle rotation in ABal instead of ABar. Second, we analyzed a temperature-sensitive mutant for a Gαi gene, gpa-16(it143). At non-permissive temperature, gpa-16(it143) randomizes ABa/ABp spindle orientation, thus gives rise to a fraction of embryos where the spindles lie in the reversed direction (Bergmann et al., 2003). We observed 4 such reversed cases out of a total of 65 embryos. Consistent with the cold-induced reversals, all 4 embryos showed mirror-image chiral morphogenesis (Figure 5C and Movie S8). These results suggest either that the ABa/ABp spindle skew is an upstream event that sets the handedness of chiral morphogenesis, or that the two events are in parallel under the assumption that the two experimental conditions both perturb an early symmetry breaking event to which the spindle skew and chiral morphogenesis react identically.
LR Asymmetry in Cortical Contractility
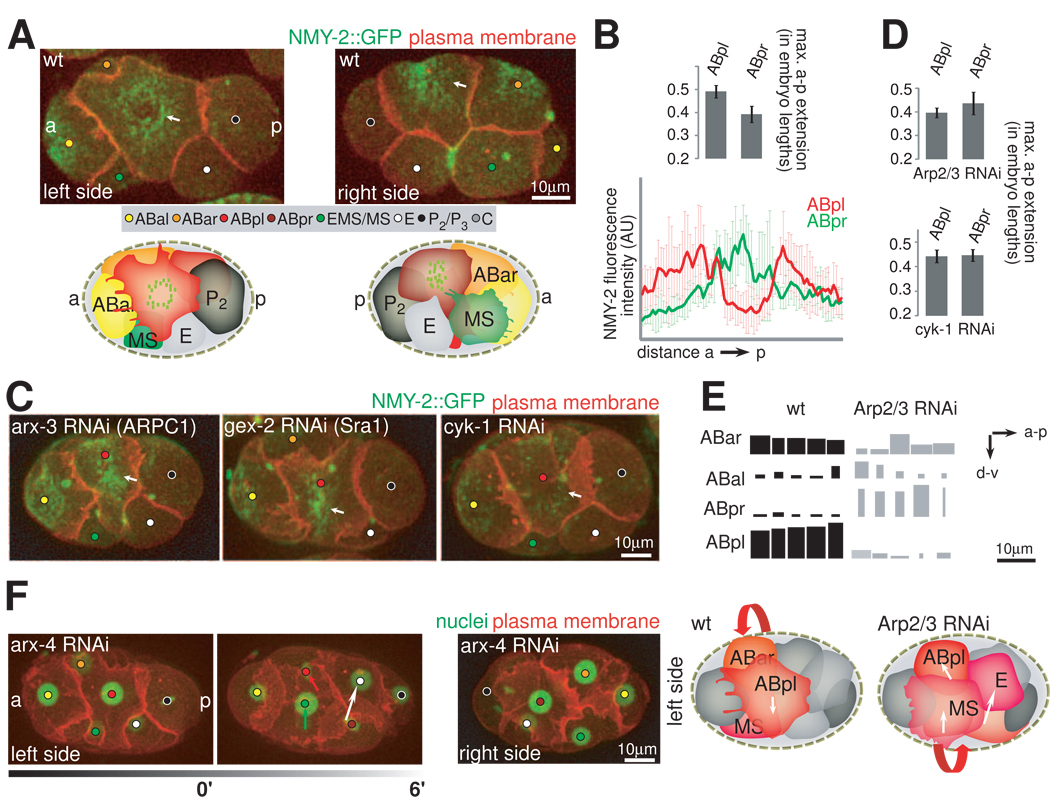
Given that ABpl and ABpr are bilateral equivalents fate-wise, we reason that their asymmetric behaviors might lie in mechanical LR asymmetries. Indeed, ABpl and ABpr show different cortical morphologies and actomyosin dynamics. Spreading of both cells is accompanied by cortical flow and the formation of an apical NMY-2 cap-like structure (Movie S9). Specifically, cortical NMY-2 flow starts as cells exit cytokinesis, from the periphery towards the center of the apical surface. When cortical flows reach a steady state, NMY-2 forms a torus-like structure in ABpl (Figure 6A, left panel). In ABpr, however, the torus collapses into a central patch (Figure 6A, right panel). As it has been shown that cortical flows originate from dynamic contractions of actomyosin (Munro et al., 2004), we reasoned that the observed cortical asymmetry might also reflect differential contractility of cells. We therefore quantified the spreading of the two cells by measuring the maximal apical extension of ABpl and ABpr along the AP axis. We find that ABpl occupies more space on the surface of the embryo than ABpr (Figure 6B). Thus, the molecular dynamics and the morphological differences suggest asymmetric contractility between ABpl and ABpr.
Figure 6. LR Asymmetries in ABpl/pr Cortical Contractility.
A. Asymmetry of the apical NMY-2 cap structure. 3D-projection stills, left side and right side views with matched schematics. Plasma membrane is shown in red and the actomyosin cortex (NMY-2::GFP) in green. Arrows point to the center of the cap structure on the respective sides. Green speckles in the cartoons indicate the distribution of cortical contractile elements on either side of the embryo. See also Movie S9. B. Quantification of the AP extension of ABpl and ABpr and of the cortical NMY-2 signature. Upper panel: AP extensions of the respective cells were measured from time-lapse microscopy data and normalized to the length of the respective embryo (n=10, mean +/− SD). Lower panel: Graphs displaying cortical NMY-2::GFP fluorescence intensity as measured on a line parallel to the AP axis that transects the center of ABpl (red) and ABpr (green), respectively (n=3, mean +/− SD). C. Perturbations of actomyosin and its consequences for asymmetric cortical dynamics. Representations as in A. Wt and embryos treated with RNAi targeting the indicated genes are shown. Arrows point to the center of the apical surface of ABpl. D. Loss of differential LR surface occupancy in Arp2/3 and cyk-1 RNAi embryos. Graph showing AP extensions as in B for the respective RNAi-treated embryos (n= 5, mean +/− SD). For details on protrusion formation see Figure S3C. E. Nuclear migration paths in wild-type and Arp2/3 RNAi embryos. Displacement of AB nuclei in AP and DV direction during chiral morphogenesis are shown as boxes for 5 wild-type and 5 Arp2/3 RNAi embryos with inverted handedness of collective rotation. Boxes are scaled with respect to the scale bar on the right. Displacements were measured from time-lapse microcopy images by taking nuclear positions at the start and end of chiral morphogenesis; see also Figure S3D. F. Perturbing asymmetric contractility can lead to inversion of the collective cell rotation. Left panel: 3D-projection stills, fluorescent markers are plasma membrane (red) and nuclei (green). The migration directions of cells are indicated by arrows, which is the opposite of the wild type (left side view). Note that ABpr forms a ventral protrusion in these embryos (right side view). Right panel: Cartoon of wild-type and Arp2/3 RNAi embryos with migrations paths as shown in E. Cells that are part of the collective rotation are highlighted in shades of red. The handedness of the rotation is indicated with red arrows. Note that in Arp2/3 RNAi embryos, the rotation comprises additional cells compared to wild-type. See also Movie S10.
To further test the hypothesis of mechanical LR asymmetry, we perturbed the cortical actin dynamics by depleting WAVE-Arp2/3 complex components with RNAi. The WAVE-Arp2/3 complex (Figure S3E) is required for polymerization of branched actin filaments and for strengthening of the cortex, hence reduction of WAVE-Arp2/3 should enhance contractility. Indeed, we find that RNAi of arx-2/-3/-4/-7 and gex-2/3 (summarized as WAVE-Arp2/3 RNAi) leads to large ruptures of the cortex and blebbing (Severson et al. 2002; Figure S3F and G). With RNAi of the WAVE-Arp2/3 complex, the ABpl cortex starts to resemble ABpr: The torus-like NMY-2 structure collapses into a solid patch (Figure 6C and S3B), and ABpl’s flattening is reduced (Figure 6D, upper panel).
According to our hypothesis, the reduction of actin polymerization and actomyosin contractility, should also abolish cortical asymmetries. To this end, we used RNAi to deplete CYK-1, which is a formin homolog responsible for linear actin polymerization and the Arp2/3-independent assembly of the actomyosin cortex (Severson et al, 2002). We find that ABpl and ABpr now resemble each other. In both cells, apical NMY-2 is greatly reduced, the apical surface is round (Figure 6C), and the two cells occupy comparable space on the surface of the embryo because ABpl now flattens less than the wild-type (Figure 6D, lower panel).
These results suggest that ABpl and ABpr regulate their cortical contractility differently and exert mechanical asymmetry during chiral morphogenesis. The observed direction of NMY-2 flow further suggests that cells do not roll freely into their new positions. Specifically, NMY-2 flows towards the apical center without a circumferential component in the flow, which would be expected if cells roll. Consistent with the notion that deformation-based forces are involved in cell movement, perturbation of actomyosin contractility in the above experiments greatly reduce cell movement and consequently the tilt of the midline decreases from 22±2° in wild-type embryos to 6±3° (WAVE-Arp2/3 RNAi) or 3±3° (cyk-1 RNAi) (Figure S3D).
More importantly, in 6 out of 21 embryos depleted for WAVE-Arp2/3, ABpr instead of ABpl undergoes significant ventral movement (Figure 6E) and the collective cell movement is counterclockwise, the opposite of normal chiral morphogenesis (Figure 6F; Movie S10). In contrast, the initial spindle skew of ABa/ABp in these cases is normal, suggesting that the chiral forces driving the handed collective cell movement is perturbed, rather than the upstream LR cue. Interestingly, in these cases ABpr forms a large ventral protrusion similar to that of wild-type ABpl. While the RNAi would affect actomyosin in all cells, a possible interpretation is that the enlarged ventral protrusion in ABpr competes with the ventral protrusion of ABpl, leading to a randomization in terms of the direction of the collective cell movement. Such an interpretation is consistent with the various results in the above sections, which all attest to an essential role of the ABpl ventral protrusion in chiral morphogenesis.
Non-Canonical Wnt Activates Cortical Dynamics and Chiral Morphogenesis
In the early embryo, the germline precursor P2 and its offspring serve as an organizing center through Wnt signaling (Bischoff and Schnabel, 2006), which induces fate differentiation (Rocheleau et al., 1997; Thorpe et al., 1997) and coordinates spindle orientations in EMS and ABar (schematically depicted in Figure 7A; Schlesinger et al., 1999; Walston et al., 2004). As P2 contacts ABpl and ABpr, we tested if Wnt signaling regulates chiral morphogenesis.
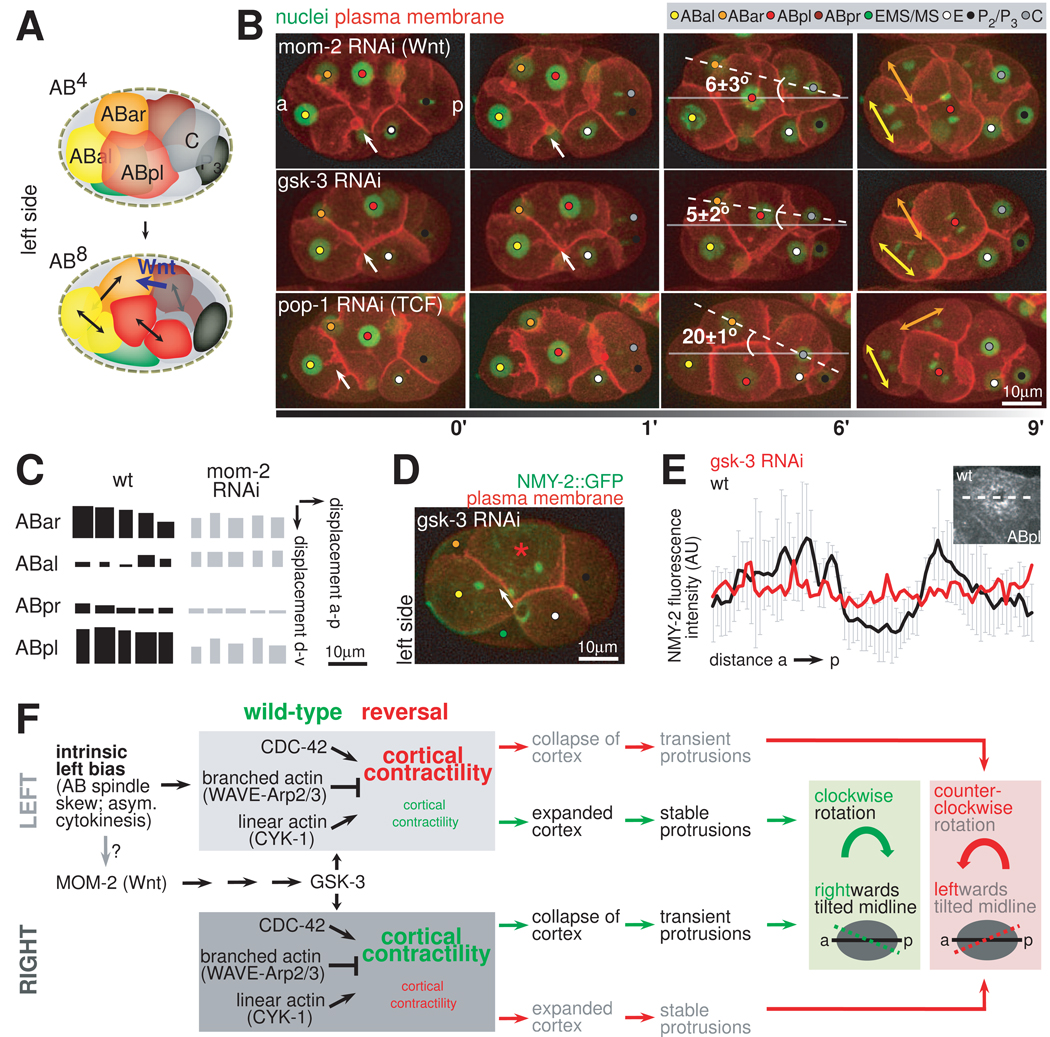
Figure 7. Non-Canonical Wnt Signaling is Required for the Formation of the Asymmetric Bilateral Body Plan.
A. Spindle orientations during AB4-AB8 division. Schematic representation of left side views. The Wnt signal from C that leads to spindle rotation in ABar is highlighted in blue. Note that all other AB spindles are parallel. B. RNAi of Wnt-signaling components and their effect on asymmetric midline placement. 3D-projection stills, left side views, fluorescent markers are plasma membrane (red) and nuclei (green). Dashed lines indicate the orientation of the midline, numbers indicate the angle between the midline and the AP axis (n=3, mean +/− SD). Large double arrows indicate spindle orientations of ABal and ABar, respectively. Small arrows indicate the interface of ABal/pl where filopodial protrusions are normally observed in wild-type embryos. C. Effects of mom-2 RNAi on individual cellular paths. Displacement of AB nuclei in AP and DV direction during chiral morphogenesis are shown as in Figure 6D for five wild-type and five mom-2 RNAi embryos. D. Severe impairment of apical NMY-2 accumulation in gsk-3 RNAi-treated embryos. 3D-projection still, left side view. Plasma membrane is shown in red and the actomyosin cortex (NMY-2) in green. Arrows indicate the interface of ABal/pl where filopodial protrusions are normally observed in wild-type embryos. The asterisk points to the center of the apex of ABpl. E. Quantification of the gsk-3 RNAi phenotype relative to wild-type. Graphs displaying cortical NMY-2::GFP fluorescence intensity as measured on a line parallel to the AP axis that transects the center of ABpl (as shown in the inset on the upper right) for a representative wild-type (gray) and gsk-3 RNAi embryo (red). F. Flowchart of the components and processes leading to LR mechanical differences, chiral morphogenesis and asymmetric midline placement. Wild-type behaviors are depicted in green. The behavior in mutants with inverted chiral morphogenesis is shown in red. Size of fonts depicts the degree of cortical contractility. Solid lines represent AP axes, dashed lines represent midlines.
We find that reduction of MOM-2 (Wnt) by RNAi greatly reduces the dynamic shape change of ABpl. The ventral protrusion as well as the dorsal protrusion connects ABpl to ABar in the clockwise rotation and the anterior filopodia onto ABal, are reduced or abolished in 30% of the mom-2 RNAi embryos (n=20). Meanwhile, the global translocation of the 4 AB cells is reduced by about 15% (n=5) (Figure 7B). Consequently, the tilt of the midplane is reduced from 22±2° to 6±3°. In 75% of the embryos (n=20), ABar and C fail to contact each other, which in turn leads to a failure of ABar spindle rotation (Figure 7C, upper panel). Other genes in the Wnt signaling pathway, namely gsk-3, mom-5 (Frizzled), dsh-2, and mig-5 (Disheveled) give similar phenotypes (Figure 7B, middle panel and data not shown). However, RNAi of pop-1, the sole homolog of the TCF/Lef transcription factor downstream of Wnt in C. elegans, does not affect the protrusions or chiral morphogenesis (Figure 7C, lower panel), even though the RNAi leads to the known phenotype of the MS to E fate transformation (Lin et al., 1995). Thus, our results suggest that it is the non-canonical Wnt pathway that regulates chiral morphogenesis without a transcriptional response. In mom-2 and gsk-3 RNAi embryos the apical flow of NMY-2 and the torus-like NMY-2 cap is impaired (Figure 7D), although NMY-2::GFP background levels in ABpl are similar to the wild-type (Figure 7E). As the spindle skew in ABa/ABp is not perturbed in these experiments, our results suggest that the non-canonical Wnt pathway acts as a permissive signal to activate the dynamics of the actomyosin cortex and chiral morphogenesis.
Discussion
A Strategy for Achieving Left-Right Patterning
Our results reveal a strategy for achieving LR patterning in bilateral animals based on positioning of the midline. Specifically the C. elegans embryo first establishes a midline tilted from the AP axis. The tilted midline establishes an inherently asymmetric bilateral body plan with more cells on the left side. The tilted midline is then re-positioned through cell movements to align with the AP axis to restore the superficial bilateral symmetry (Figure 1 and Movie S2). This is in contrast to vertebrate and Drosophila embryogenesis (Speder et al., 2007) where the midline appears static morphologically. In vertebrates, such a static midline may be essential to restrict symmetry breaking molecules from crossing over to the other side (Levin, 2005). The tilted bilateral body plan is also geometrically distinct from the gastropod where a spiral asymmetry rather than a bilateral asymmetry is established in early embryogenesis (Kuroda et al., 2009).
Our results also illustrate how the initial breaking of LR symmetry is propagated to the global anatomical asymmetry in an organism (Figure 7F). Following the initial skew of the ABa/p spindle that breaks the symmetry, the cellular pathway in C. elegans involves asymmetric actomyosin dynamics and protrusions (Figures 3 and 4) and a collective handed movement to rearrange cells into the tilted body plan (Movie S3), which we term chiral morphogenesis. Our perturbations of the ABa/p spindle skew (Figure 5) and the fact that that reversing the direction of the ABa/ABp spindle skew is sufficient to generate a mirror-image worm (Wood, 1991) suggest that the spindle skew is most likely the instructive step for chiral morphogenesis.
A Missing Link in C. elegans Morphogenesis
Chiral morphogenesis and the tilted bilateral body plan provide an important missing link in the LR patterning process in C. elegans. In particular, they illustrate the anatomical basis as to how the initial symmetry breaking by the ABa/p spindle skew leads to the subsequent Notch signaling to break fate symmetry. For example, a Notch signal first breaks the fate symmetry between ABal and ABar. This is achieved by ABar rotating its spindle so that the anterior daughter contacts the signaling cell (MS) while in ABal, it is the posterior daughter. As our results show, a major aspect of chiral morphogenesis is to juxtapose C to ABar, which allows C to signal and rotate the ABar spindle (Walston et al., 2004). Later on, additional Notch signaling, originating from the left side of the tilted midline, induces minor fate asymmetry within the LR symmetric part of the body plan (Hutter and Schnabel, 1995; Hermann et al., 2000). Interestingly, by the time the midline is re-aligned with the AP axis at the 350-cell stage, the known Notch inductions are complete.
In the light of the tilted bilateral body plan, various observations of the C. elegans LR patterning process that have been made as isolated and puzzling phenomena can now be readily viewed as sensible aspects of a coherent developmental strategy. In his seminal work tracing the C. elegans embryonic lineage, Sulston noted two different strategies to generate bilateral structures (Sulston et al., 1983). In the head, structures are assembled piecemeal by recruiting small sublineages that are not LR homologous by origin. However, in the posterior lineages, bilateral structures are generated from LR homologous lineages. It was however not clear how the embryo deploys and integrates the two different strategies. Our results show that blastomeres using the two strategies are organized differently in the bilateral body plan. The posterior lineages are organized into the tilted structure with a midplane, while the anterior lineages (ABal, ABara, ABarpa) are segregated from the structure where the overall LR fate symmetry between ABal and ABar descendants are first broken by Notch signaling and subsequently reassembled in small pieces.
The tilted bilateral body plan also explains other previously puzzling observations. For example, the first divisions of the bilaterally symmetric founder cells (ABarpp, MS, C and each daughter of P3, namely D and P4) give rise to daughters that are LR counterparts (Sulston et al., 1983). However, these divisions are bona fide AP divisions instead of LR based on their spindle orientation and molecular signatures (Lin et al., 1998). In light of the tilted midline, the apparent discrepancy can be reconciled: these AP divisions are symmetric with respect to the tilted midline, sending the anterior daughter to the left and the posterior to the right (Figure 2A). Another interesting case is the bi-directional signaling between EMS and P2 to orient each other’s spindle (Bei et al., 2002; Tsou et al., 2003). As a result, E and P3 are born at the future midplane (Figure S2C), suggesting that the E-P axis may help predefine the tilt of the midline.
Conservation of the LR Patterning Process
Previous studies have shown that the overall strategy of LR patterning is highly diverse among Bilateria. However, our studies of C. elegans suggest that the underlying cellular and molecular mechanisms may be conserved. In particular, C. elegans achieves morphological asymmetry before fate asymmetry, which involves LR asymmetric regulation of cortical contractility and handed cell movement. Our data show that cellular rearrangements in the early C. elegans embryo are dominated by cortical tension: (1) cells deform asymmetrically dependent on actomyosin forces and (2) perturbation of cortical tension prevents LR asymmetric cellular rearrangements. Moreover, we present evidence that a main aspect of the downstream LR asymmetry pathway is to establish differential cortical flows on the two sides of the embryo (Figure 7F). Interestingly, recent studies showed that in the chick embryo actomyosin-based and handed cell migration arise before the asymmetric expression of Nodal, a hallmark in vertebrate LR patterning, to achieve morphological asymmetry in the node and rearrange the initially symmetric gene expression pattern into an asymmetric (Cui et al., 2009; Gros et al., 2009). This is in contrast to the mouse embryo where asymmetric gene expression arises before noticeable morphological asymmetry. Given the complexity of veterbrate development, questions remain open as to whether the cells undergoing differential movement on the left and right sides in chick are equivalent fate-wise as assumed, that is, if handed movement and morphological asymmetry arise before fate differentiation in these cells. Previous studies in C. elegans have firmly established the equivalence of ABpl and ABpr (Wood, 1991; Hutter and Schnabel, 1995). Hence, our results suggest that asymmetric cellular behaviors and morphological asymmetry before fate asymmetry can be a viable hypothesis in vertebrates. Combined, these results suggest that a mechanism where morphological asymmetry determines fate asymmetry may constitute a more general mechanism for LR patterning.
Furthermore, our data identifies non-canonical Wnt signaling as a candidate pathway that might activate cortical dynamics by providing a permissive signal (Lee et al., 2006; Seifert and Mlodzik, 2007). Interestingly, it has been recently shown that non-canonical Wnt signaling is required to properly polarize cilia along the AP axis in mouse embryos to achieve leftward nodal flow (Hashimoto et al., 2010). Given the known role of Wnt signaling in defining the AP polarity in C. elegans cells (Bischoff and Schnabel, 2006), it is therefore tempting to speculate that non-canonical Wnt signaling might be a common signaling pathway to relate AP axis information to LR patterning with and without motile cilia/Nodal signaling.
A Role for the Cell Division Furrow in Morphogenesis
Cell division is typically viewed as the vehicle for proliferation in development. Our results on the EMS division furrow suggest that cell division plays important roles in morphogenesis. First, we show that the EMS furrow triggers the formation of the ABpl ventral protrusion (Figure 4D). This suggests that there is a novel mechanism whereby the cytokinetic furrow can induce contact formation with a neighboring cell, although the signaling molecules and the nature of the protrusion remain to be identified. Second, with the EMS furrow serving as an instructive signal, the timing of EMS division functions as a novel developmental clock to orchestrate chiral morphogenesis. Cell cycle timing is highly reproducible during C. elegans (Bao et al., 2008) and is conserved in C. briggsae (Zhao et al., 2008). Our result suggests that morphogenesis may have provided the selective pressure on the accuracy of cell cycle length.
Experimental Procedures
C. elegans strains
Worms were cultured with standard procedures (Brenner, 1974) and were well fed for at least two generations and cultured at 20–25°C (or 16°C for ts mutants) before embryos were collected from young adults and imaged at 20°C or 25°C (restrictive condition for ts mutants) on a temperature-controlled stage. Genotypes of strains can be found in Supplemental Data. Several of the parental strains were obtained from the Caenorhabditis Genetics Center (CGC).
Embryo preparation and imaging
Embryos were dissected from gravid hermaphrodites, mounted in 2.5µl of a M9 buffer suspension containing 20 or 25µm poly-styrene microspheres (Polyscience, Warrington, PA), and sealed between two coverslips (Corning, Lowell, MA) with vaseline. To image uncompressed embryos, mounting was performed in a cavity of 8-well test slides (MP Biomedicals, Solon, OH) and the slide was sealed with a coverslip , or embryos were embedded in low-melting agarose (Invitrogen, Carlsbad, CA) and the agar block was covered with a drop of M9 buffer. Images were acquired on a Quorum Wave FX spinning disc confocal system (Quorum Technologies, Guelph, Canada) using a Zeiss Observer Z1 microscope with Zeiss PlanApo 40×/1.3 Oil or 63×/1.4 Oil objectives (Carl Zeiss Microimaging, Jena, Germany). Details on image acquisition and manipulation can be found in Supplemental Experimental Procedures. As embryos are normally imaged under mild compression, we assured that uncompressed embryos behave identically (Figure S1C).
Lineaging and data representation
Nuclear identification, tracking, and cell naming were performed as described previously (Bao et al., 2006; Murray et al., 2006). Data from lineaged embryos was visualized in AceTree as described (Boyle et al., 2006). Details on measurements can be found in Supplemental Experimental Procedures.
RNAi treatment
For RNAi treatment, we either used soaking according to published protocols (Ahringer, 2006) or feeding with clones from two commercially available libraries (Kamath et al., 2003; Rual et al., 2004) or prepared from cDNA (see Supplemental Materials and Methods for details). Larvae from L1 to L4 were used for feeding to obtain graded phenotypes and partial loss of function for the respective target gene.
Highlights
-
-
Novel strategy of left-right patterning: uncoupling midline and AP axis
-
-
Differential regulation of cortical forces between LR without fate differences
-
-
Non-canonical Wnt signaling activates cortical dynamics
-
-
Signaling from the contractile ring to neighboring cells
Supplementary Material
Acknowledgments
We are grateful to M. Tiongson for technical assistance and help in generating Lifeact::GFP expressing strains, and J. Moore and A. Santella for assistance with data analysis and preparation of figures. We thank A. Hall, B. Tsou, M. Jasin, M. Pellegrino and S. Pohl for advice and critical reading of the manuscript, and A. Hyman and A. Pozniakovsky for providing vector pTH304. Work in the laboratory of Z.B. is partly supported by an NIH grant (HG004643). C.P. is recipient of a long-term fellowship from the Human Frontier in Science Program Organization.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data includes 4 figures, 10 Movies, Supplemental Experimental Procedures and can be found with this article online at …
References
- Ahringer J. Reverse genetics. In: The C. elegans Research Community, editor. WormBook. 2006. doi/10.1895/wormbook.1891.1847.1891 ( www.wormbook.org) [Google Scholar]
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, 3rd, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Murray JI, Boyle T, Ooi SL, Sandel MJ, Waterston RH. Automated cell lineage tracing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2006;103:2707–2712. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Zhao Z, Boyle TJ, Murray JI, Waterston RH. Control of cell cycle timing during C. elegans embryogenesis. Dev Biol. 2008;318:65–72. doi: 10.1016/j.ydbio.2008.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3:113–125. doi: 10.1016/s1534-5807(02)00185-5. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lee M, Robertson B, Tsou MF, Rose LS, Wood WB. Embryonic handedness choice in C. elegans involves the Galpha protein GPA-16. Development. 2003;130:5731–5740. doi: 10.1242/dev.00839. [DOI] [PubMed] [Google Scholar]
- Bischoff M, Schnabel R. A posterior centre establishes and maintains polarity of the Caenorhabditis elegans embryo by a Wnt-dependent relay mechanism. PLoS Biol. 2006;4:e396. doi: 10.1371/journal.pbio.0040396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle TJ, Bao Z, Murray JI, Araya CL, Waterston RH. AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinformatics. 2006;7:275. doi: 10.1186/1471-2105-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NA, Wolpert L. The development of handedness in left/right asymmetry. Development. 1990;109:1–9. doi: 10.1242/dev.109.1.1. [DOI] [PubMed] [Google Scholar]
- Conti MA, Even-Ram S, Liu C, Yamada KM, Adelstein RS. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J Biol Chem. 2004;279:41263–41266. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- Cui C, Little CD, Rongish BJ. Rotation of organizer tissue contributes to left-right asymmetry. Anat. Rec. 2009;292:557–561. doi: 10.1002/ar.20872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui R, Fenteany G. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell Sci. 2005;118:51–63. doi: 10.1242/jcs.01577. [DOI] [PubMed] [Google Scholar]
- Gros J, Feistel K, Viebahn C, Blum M, Tabin CJ. Cell movements at Hensen's node establish left/right asymmetric gene expression in the chick. Science. 2009;324:941–944. doi: 10.1126/science.1172478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Shinohara K, Wang J, Ikeuchi S, Yoshiba S, Meno C, Nonaka S, Takada S, Hatta K, Wynshaw-Boris A, Hamada H. Planar polarization of node cells determines the rotational axis of node cilia. Nat. Cell Biol. 2010;12:170–176. doi: 10.1038/ncb2020. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Leung B, Priess JR. Left-right asymmetry in C. elegans intestine organogenesis involves a LIN-12/Notch signaling pathway. Development. 2000;127:3429–3440. doi: 10.1242/dev.127.16.3429. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125:33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Hutter H, Schnabel R. Establishment of left-right asymmetry in the Caenorhabditis elegans embryo: a multistep process involving a series of inductive events. Development. 1995;121:3417–3424. doi: 10.1242/dev.121.10.3417. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kuroda R, Endo B, Abe M, Shimizu M. Chiral blastomere arrangement dictates zygotic left-right asymmetry pathway in snails. Nature. 2009;462:790–794. doi: 10.1038/nature08597. [DOI] [PubMed] [Google Scholar]
- Lee JY, Marston DJ, Walston T, Hardin J, Halberstadt A, Goldstein B. Wnt/Frizzled signaling controls C. elegans gastrulation by activating actomyosin contractility. Curr Biol. 2006;16:1986–1997. doi: 10.1016/j.cub.2006.08.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mech Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92:229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Maddox AS, Lewellyn L, Desai A, Oegema K. Anillin and the septins promote asymmetric ingression of the cytokinetic furrow. Dev Cell. 2007;12:827–835. doi: 10.1016/j.devcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Waterston RH. The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat Protoc. 2006;1:1468–1476. doi: 10.1038/nprot.2006.222. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Priess JR. Notch signaling in the C. elegans embryo. In: The C. elegans Research Community, editor. WormBook. doi/10.1895/wormbook.1.4.1 ( www.wormbook.org) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, Mello CC. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–716. doi: 10.1016/s0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger A, Shelton CA, Maloof JN, Meneghini M, Bowerman B. Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev. 1999;13:2028–2038. doi: 10.1101/gad.13.15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R, Hutter H, Moerman D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev Biol. 1997;184:234–265. doi: 10.1006/dbio.1997.8509. [DOI] [PubMed] [Google Scholar]
- Seifert JR, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–138. doi: 10.1038/nrg2042. [DOI] [PubMed] [Google Scholar]
- Severson AF, Baillie DL, Bowerman B. A Formin Homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr Biol. 2002;12:2066–2075. doi: 10.1016/s0960-9822(02)01355-6. [DOI] [PubMed] [Google Scholar]
- Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- Speder P, Petzoldt A, Suzanne M, Noselli S. Strategies to establish left/right asymmetry in vertebrates and invertebrates. Curr Opin Genet Dev. 2007;17:351–358. doi: 10.1016/j.gde.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tabin C. Do we know anything about how left-right is first established in the vertebrate embryo? J Mol Histol. 2005;36:317–323. doi: 10.1007/s10735-005-9000-y. [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Hayashi A, Rose LS. LET-99 opposes Galpha/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development. 2003;130:5717–5730. doi: 10.1242/dev.00790. [DOI] [PubMed] [Google Scholar]
- Walston T, Tuskey C, Edgar L, Hawkins N, Ellis G, Bowerman B, Wood W, Hardin J. Multiple Wnt signaling pathways converge to orient the mitotic spindle in early C. elegans embryos. Dev Cell. 2004;7:831–841. doi: 10.1016/j.devcel.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Wood WB. Evidence from reversal of handedness in C. elegans embryos for early cell interactions determining cell fates. Nature. 1991;349:536–538. doi: 10.1038/349536a0. [DOI] [PubMed] [Google Scholar]
- Wood WB. Left-right asymmetry in animal development. Annu Rev Cell Dev Biol. 1997;13:53–82. doi: 10.1146/annurev.cellbio.13.1.53. [DOI] [PubMed] [Google Scholar]
- Wood WB, Bergmann D, Florance A. Maternal effect of low temperature on handedness determination in C. elegans embryos. Dev Genet. 1996;19:222–230. doi: 10.1002/(SICI)1520-6408(1996)19:3<222::AID-DVG5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Boyle TJ, Bao Z, Murray JI, Mericle B, Waterston RH. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev Biol. 2008;314:93–99. doi: 10.1016/j.ydbio.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.