SUMMARY
In inflammatory bowel disease, the relationship between a host and gut microbial community goes awry. We have characterized the fecal microbial communities in a mouse IBD model driven by T-bet deficiency in the innate immune system. 16S rRNA-based analysis of T-bet−/− × Rag2−/− and Rag2−/− mice revealed distinctive communities that correlate with host genotype. Culture-based surveys, invasion assays, antibiotic treatment, and TNF-α blockade disclosed that the presence of Klebsiella pneumoniae and Proteus mirabilis correlates with colitis in T-bet−/− × Rag2−/− animals, and that T-bet−/− × Rag2−/− derived strains can elicit colitis in Rag2−/− and wild-type adults. Cross-fostering experiments provided evidence for the role of these organisms in maternal transmission of disease. This model provides a foundation for defining how gut microbial communities work in concert with specific culturable colitogenic agents to cause IBD, and a foundation for conducting proof-of-concept tests of new preventative or therapeutic measures directed at components of the gut microbiota and/or host.
INTRODUCTION
The human intestine is populated with up to 1012 microbes per gram of luminal contents. Co-existence with this microbial community (microbiota) demands a well-regulated homeostasis between the host immune system and the microbiota (Duerkop et al., 2009; Hill and Artis, 2009). Inflammatory bowel disease (IBD) can occur when this homeostasis is disrupted (Sartor, 2009). Whether individual pathogenic species or entire microbial communities instigate inflammation still remains controversial (Frank and Pace, 2008; Hansen et al. 2010). Defining features of the microbiota and host that are associated with or initiate IBD is a long sought after goal (Peterson et al., 2008).
In the absence of an adaptive immune system, loss of the transcription factor T-bet in conventionally-raised T-bet−/− × Rag2−/− knockout mice results in a spontaneous and highly penetrant colitis that shares histologic features with ulcerative colitis in humans. T-bet−/− × Rag2−/− ulcerative colitis (TRUC) is associated with altered colonic barrier function, elevated TNF-α levels, and dysfunctional dendritic cells (Garrett et al., 2007; Garrett et al., 2009). It is transmissible to wild-type hosts when they are cross-fostered or co-housed with TRUC mice (Garrett et al., 2007). TRUC mice provide an opportunity to probe the host-microbe relationship in a model that displays both the immunodeficiency and hyper-immunity observed in humans with IBD.
Here we show that the presence of Proteus mirabilis and Klebsiella pneumoniae correlates with colitis in TRUC mice and that TRUC derived strains in conjunction with an endogenous microbial community can incite colitis in wild type mice. These studies revealed the utility of using both culture-independent and -dependent approaches to interrogate the contribution of community members to disease pathogenesis. This model also provides a foundation for defining how gut microbial communities work in concert with specific culturable colitogenic agents to cause IBD and creates an opportunity to evaluate preventative or therapeutic measures directed at components of the gut microbiota and/or host.
RESULTS
16S rRNA-based time series analysis of T-bet−/− × Rag2−/− (TRUC) vs. Rag2−/− fecal microbiota
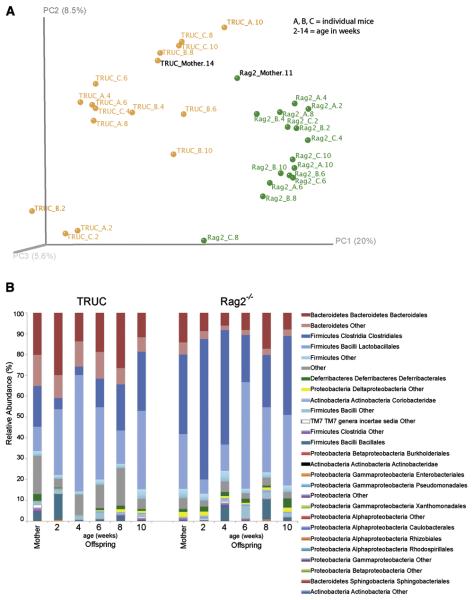
We began with a pilot experiment involving the offspring of a conventionally-raised, specified pathogen-free T-bet−/− × Rag2−/− mother and a Rag2−/− mother. Fecal samples were collected from mothers at a single time point, and their female pups (n=3/genotype) at multiple time points, beginning at two weeks of age and continuing at two week intervals until the animals were 10 weeks-old. A culture-independent survey of their fecal microbiota was carried out by multiplex pyrosequencing of amplicons generated from the V2 region of bacterial 16S rRNA genes (n=32 samples; 2,348±343 reads per sample). UniFrac is a metric that measures the degree of similarity of communities based on the degree to which they share branch length on a phylogenetic tree constructed from all 16S rRNA sequences generated from a survey. Principal coordinates analysis (PCoA) plots based on unweighted UniFrac measurements disclosed a correlation between host genotype and community phylogeny at all ages surveyed (Fig. 1A). A total of 69 species-level phylogenetic types, belonging to 4 major bacterial phyla, exhibited significant differences, at various ages, in the fecal communities of mice belonging to the two genotypes (Supplemental Table 1). Compared to Rag2−/− controls, TRUC mice had a significantly higher proportional representation of species-level operational taxonomic units (OTUs) belonging to the order Bacteroidales (phylum Bacteroidetes; p=0.00643, Mann-Whitney test with Bonferroni-correction) and significantly lower proportional representation of OTUs belonging to the orders Clostridiales (phylum Firmicutes; p=0.0201, Mann-Whitney test with Bonferroni-correction) and Deltaproteobacteria (phylum Proteobacteria; p=0.0299, Mann-Whitney test with Bonferroni-correction) (Fig. 1B).
Fig. 1. 16S rRNA-based time series analysis of T-bet−/− × Rag2−/− (TRUC) vs Rag2−/− fecal microbiota.
(A) Host genotype influences microbial community structure. Principal coordinates analysis of unweighted UniFrac distances from 2-10 week TRUC (n= 3) and Rag2−/− (n =3) mice and their mothers. Abbreviations: A, B, C, individual pups colored by genotype, followed over time (A.2, A.4, A.6, A.8, and A.10 refers to animal A sampled at 2,4,6, and 10 weeks of age). (B) The distribution of order-level phylotypes in TRUC and Rag2−/− fecal microbial communities. Percent relative abundance is plotted for each age group.
Klebsiella pneumoniae and Proteus mirabilis correlate with the presence of colitis in TRUC mice
We also performed quantitative cultures to obtain independent verification of differences in bacterial burden for defined species and to have culturable isolates available for the purposes of testing Koch's postulates regarding specific effects of individual strains on disease initiation and progression. A total of 57 bacterial species were recovered and identified from fecal pellets obtained from 3 TRUC and 3 Rag2−/− mice surveyed at 2, 4, 6, 8, and 10 weeks of age, and their mothers (Fig. 2A and Supplemental Table 2).
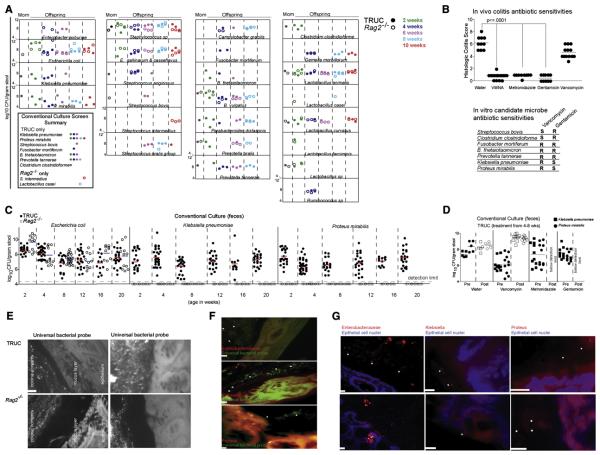
Fig 2. The presence of Klebsiella pneumoniae and Proteus mirabilis correlates with the presence of colitis in TRUC mice.
(A) Culture-based identification of bacterial species present in fecal samples from the same animals and time points as analyzed in Fig. 1. Species observed in more than one animal or in one animal at more than one time point are shown. Inset is a summary of bacterial species-level differences in the fecal microbiota of TRUC versus Rag2−/− mice observed in 2A. (B) Upper panel: Histologic colitis scores demonstrate the in vivo antibiotic sensitivities of TRUC colitis. Each dot represents an individual mouse treated for four weeks with the indicated antibiotics dissolved in drinking water. VMNA refers to treatment with a combination of vancomycin, metronidazole, neomycin, and ampicillin. Horizontal bars represent the mean. p-value ≤0.0001 defined by Mann-Whitney test. Lower panel: Summary of in vitro antibiotic sensitivities for several species selectively detected in TRUC fecal microbiota. (C) Culture-based survey of Gram-negative aerobes present in fecal samples from TRUC (shaded circles) and Rag2−/− (open circles) at 2-20 weeks of age. Fecal samples were collected and cultured twice at each time point from Rag2−/− mice. (D) In vivo sensitivity of Klebsiella pneumoniae (squares) and Proteus mirabilis (circles), as defined by culture-base surveys of TRUC fecal samples collected 1d before (shaded symbol) and 1d after (open symbol) treatment with antibiotic. Each dot represents data from a fecal sample obtained from one mouse. Horizontal bars represent the mean value. (E) Fluorescence In Situ Hybridization using an Oregon-Green® 488 conjugated “Universal bacterial” 16S rRNA-directed oligonucleotide probe (EUB 338) demonstrates the presence of bacteria in the mucus layer and directly adjacent to the epithelium in TRUC mice. Upper panels: TRUC, lower panels: Rag2−/−. A 10 micron scale bar for the panel is shown in the lower left of the first image. (F) Enterobacteriaceae (red), Klebsiella (red), and Proteus (red) were visualized adjacent to the epithelium in TRUC mice using fluor-conjugated 16S rRNA or 23S rRNA oligonucleotide probes [(pB-00914 (Enterobacteriaceae), pB-00352 (Klebsiella pneumoniae), pB-02110 (Proteus mirabilis)]. Sections were also hybridized with the EUB338 universal bacterial probe (green). Scale bars (10 micron) are shown for each image. (G) Enterobacteriaceae (red), Klebsiella (red), and Proteus (red) probe signals are seen adjacent to or along the epithelium in TRUC mice. Epithelial cell nuclei were stained with DAPI. White star symbols mark bacteria in (F) and (G). Scale bars (10 micron) are shown for each image.
Subsequent experiments administering oral antibiotics helped further refine potential classes of commensal organisms that contribute to the colitogenic phenotype. Orally administering gentamicin or metronidazole was highly effective in ameliorating TRUC colitis and resulted in clinically and statistically significant changes in colitis scores (mean colitis score 0.5±0.52, p<.0001 compared to water control). In contrast, mice treated with oral vancomycin still demonstrated clinically significant disease (mean colitis score 4.6±0.96, p<.0001 compared to gentamicin or metronidazole; p<.03 compared to water) (Fig. 2B). The ability of gentamicin or metronidazole to prevent the development of colitis in the TRUC mice suggested a role for Gram negative facultative organisms.
We next evaluated the in vitro antibiotic resistance profiles of the commensal strains selectively recovered from TRUC but not Rag2−/− fecal samples (Fig. 2A, Fig. 2B lower panel). Susceptibility testing demonstrated that Klebsiella pneumoniae and Proteus mirabilis, both facultative enterics, were sensitive to gentamicin but, not surprisingly, were resistant to vancomycin (Fig. 2B lower panel). These results corresponded to the in vivo antibiotic sensitivity of the colitis (Fig. 2B upper panel).
We then performed a more extensive, culture-based survey of a larger number of TRUC and Rag2−/− mice to determine if these bacteria were present in afflicted mice but absent from healthy mice. In our initial culture-based screen, Klebsiella pneumoniae counts were below our detection limit at a few time points sampled for the TRUC mice. We subsequently performed a more intensive analysis with more sample mass and found that Klebsiella pneumoniae and Proteus mirabilis were culturable in all TRUC mice tested (n = 126), at every time point examined (Fig. 2C). In contrast, both species were consistently below our limit of detection (4.4 log10 CFU/g fecal material) in every Rag2−/− mouse surveyed at each time point (Fig. 2C).
We treated 4-week old TRUC mice with antibiotics using the protocol shown previously to ameliorate colitis, and cultured feces obtained 1d before and 1d after antibiotic administration. After treatment with gentamicin or metronidazole, fecal levels of K. pneumoniae and P. mirabilis fell below our limit of detection (Fig. 2D). In contrast, treatment with vancomycin neither abolished colitis nor reduced levels of K. pneumoniae and P. mirabilis (Fig 2D).
These two findings suggested that K. pneumoniae and P. mirabilis may play a role in the disease pathogenesis of TRUC. To evaluate this hypothesis, we first determined the physical location of these species using 16S and 23S rDNA fluorescence in situ hybridization (FISH) oligonucleotide probes on whole colonic sections, with a focus on the degree of colonization in the lumen, mucus layer over the epithelium, and the mucosa. As has been reported in inflammatory bowel disease patients using a universal bacterial 16S rRNA FISH probe (Swidsinski et al., 2005), we observed that the colonic mucus of colitic TRUC mice harbored numerous bacteria and that there was a consistent loss of a ‘bacterial-free zone’ adjacent to the colonic epithelium (Fig. 2E). Healthy (non-colitic) Rag2−/− mice did not exhibit any of these phenotypes (Fig. 2E). We subsequently employed a set of probeBase consortium 23S and 16S rDNA probes to detect K. pneumoniae and P. mirabilis (Loy A et al., 2007). We were able to visualize a small number of organisms that reacted with these reagents adjacent to the epithelium (Fig. 2F and G). These data suggested that K. pneumonia and P. mirabilis may have invasive potential or that the proximity of their bacterial products to the apical epithelial surface may trigger inflammatory responses in the absence of frank invasion. Either could explain their role in TRUC colitis as access to the mucosa would increase the opportunity for eliciting a host pro-inflammatory response.
Klebsiella pneumoniae and Proteus mirabilis elicit colitis but require a maternally-transmitted endogenous microbial community for maximal intestinal inflammation
To further evaluate the relevance of K. pneumoniae and P. mirabilis, we took advantage of a transmissible model of TRUC (Garrett et al., 2007). Following postnatal exposure to a TRUC dam, wild-type (WT) and Rag2−/− mice develop histopathologic features of colitis (penetrance of phenotype: 94% at 8 weeks of age) (Garrett et al., 2007). We first asked if this maternally transmitted disease had a pattern of antibiotic sensitivity that was similar to spontaneous TRUC colitis. Therefore, we cross-fostered TRUC, Rag2−/−, and WT pups on TRUC mothers who received water, gentamicin, metronidazole, or vancomycin from pre-conception through weaning. Gentamicin and metronidazole markedly improved the colitis score for all mice in a statistically significant fashion while vancomycin did not-- similar to what we observed in spontaneous TRUC colitis [n = 2 foster mothers/genotype; 2-4 pups per litter surveyed] (Fig 3A).
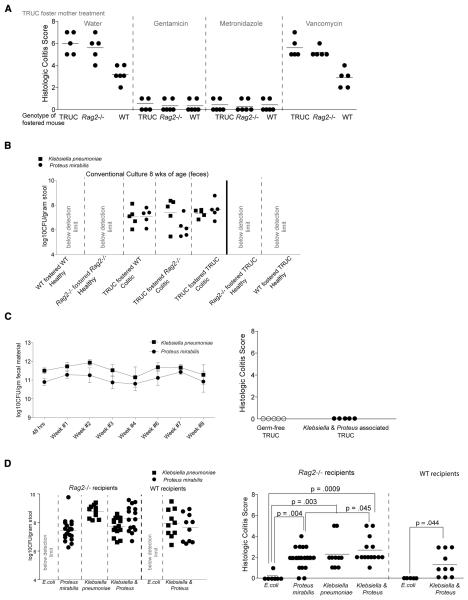
Fig 3. Klebsiella pneumoniae and Proteus mirabilis elicit colitis but require a maternally-transmitted endogenous microbial community for maximal intestinal inflammation.
(A) The antibiotic sensitivities of colitis transmitted via TRUC cross-fostering are the same as spontaneous TRUC colitis. Antibiotic-treated pregnant TRUC females were used as foster mothers and treated with antibiotics in their water until weaning. Histologic colitis scores are shown for the fostered mice at 8 weeks of age. (B) Klebsiella pneumoniae (squares) and Proteus mirabilis (circles) are detected in the fecal microbiota of TRUC cross-fostered Rag2−/− and WT mice at 8 weeks of age but not in 8 week old TRUC mice fostered by Rag2−/− or WT mice. TRUC-fostered TRUC, Rag2−/−-fostered Rag2−/−, and WT-fostered WT are shown as controls. Limits of detection; 104.4 cfu/g dry weight of feces. Each filled square or circle represents a fecal sample from a different animal. (C) Left panel: Fecal bacterial counts for co-colonized gnotobiotic TRUC mice. Mean values ±1 S.D. are shown for Klebsiella pneumoniae (squares) and Proteus mirabilis (circles) (n=5 mice). Right panel: Histologic colitis scores of germ-free TRUC and germ-free TRUC mice co-colonized with Klebsiella pneumoniae and Proteus mirabilis from the TRUC mother in Fig. 1. (D) Left panel: Klebsiellapneumoniae and Proteus mirabilis fecal cfu in Rag2−/− and WT mice treated every other day from 2-10 weeks of age with 107 cfu of E. coli, Proteus mirabilis, Klebsiella pneumoniae, or a combination of both added to their drinking water (all strains isolated from the TRUC mother in Fig. 1). Right panel: Histologic scores for colitis as assayed at sacrifice at 10 weeks of age. Each filled circle represents a separate animal in the treatment group. p-values shown were calculated using the Mann-Whitney test.
We cultured fecal samples obtained from WT and Rag2−/− mice that developed colitis as a result of TRUC cross-fostering (Fig. 3B). K. pneumoniae and P. mirabilis were detected in all fecal samples obtained from 8 week old TRUC-fostered Rag2−/− and WT pups, and at levels comparable to age-matched TRUC-fostered TRUC mice. In contrast, neither of these organisms was detected in any control Rag2−/−-fostered Rag2−/− or WT-fostered WT animals (n = 2 foster mothers/genotype; 2-3 pups per litter surveyed; Fig. 3B). Neither TRUC mice fostered on Rag2−/− nor WT mothers had evidence of colitis at 8 weeks of age and of K. pneumoniae or P. mirabilis. These Rag2−/− or WT fostered TRUC had no evidence of histologic colitis at 8 weeks of age and K. pneumoniae and P. mirabilis were not detectable at that time point (Fig. S2; Fig 3B).
The presence of K. pneumoniae and P. mirabilis in colitic TRUC mice and TRUC fostered Rag2−/− and WT mice and the lack of detectable levels of these bacteria in the fecal microbiota of healthy Rag2−/−, WT, and WT or Rag2−/− fostered TRUC provided additional evidence for an association between the presence of these bacteria and colitis.
One possibility is that the presence of K. pneumoniae and P. mirabilis is a consequence rather than a cause of inflammation. For example, intestinal inflammation caused by Citrobacter rodentium has been suggested to drive blooms of Enterobacteriaceae, although this result is controversial (Hoffmann et al., 2009; Lupp et al., 2007). To investigate the effects of inflammation on intestinal colonization by K. pneumoniae and P. mirabilis, we treated 8 week-old WT and Rag2−/− mice with dextran sulfate sodium, a mucosal disruptant and irritant, to induce colitis (n=8 mice/genotype). We did not detect culturable K. pneumoniae or P. mirabilis in the fecal microbiota of any of these mice during our period of surveillance (n=8 mice/genotype; samples collected before and 1d after the completion of a one week treatment course; Fig. S1) arguing against an inflammatory response per se causing expansion of K. pneumoniae and P. mirabilis in the gut microbiota of TRUC mice.
To directly test the colitogenic potential of K. pneumoniae and P. mirabilis, we re-derived conventionally-raised TRUC mice as germ-free and co-colonized the animals with these two Enterobacteriaceae at 8 weeks of age for 8 weeks (n = 5 mice). Both organisms established themselves in the guts of all recipients (mean value 1011.29±0.46 cfu/microbial species/g dry weight of feces; assayed 48h and weekly after the initial gavage) (Fig. S2). Colonic inflammation did not develop in these co-colonized gnotobiotic TRUC mice, suggesting that interactions among K. pneumoniae and P.mirabilis and other members of a gut microbial community are required to ignite the immuno-inflammatory cascade that leads to colitis. To evaluate this possibility, we colonized 2 week-old specified pathogen-free WT and Rag2−/− mice with Klebsiella pneumoniae, P. mirabilis, or a combination of K. pneumoniae and P. mirabilis (these strains were recovered from feces obtained from the female TRUC mother in Fig. 2A and were administered by direct oral instillation of 107 cfu, and by addition of 107 cfu to the drinking water every other day for 8 weeks; n= 5-18 mice/treatment group). Control groups of mice received a TRUC-derived E. coli strain. Both K. pneumoniae and P. mirabilis established themselves in the gut microbiota of both Rag2−/− and WT (as defined by cfu assays of feces obtained 2d after the completion of an 8-week course of treatment; Fig. 3C). Feces from WT and Rag2−/− hosts contain E. coli, but we did not have the tools to distinguish these indigenous strains from the exogenously administered TRUC-associated E. coli strain. While no colonic inflammation was observed after inoculation with E. coli (Fig. 3D), treatment with P. mirabilis, K. pneumoniae, or a combination of the two organisms, induced inflammation in both WT and Rag2−/− mice with the severity of the colitis being significantly greater in Rag2−/− mice exposed to both species, compared to P. mirabilis alone (Fig. 3D). Taken together, these results suggest that two Enterobacteriaceae in concert with members of the microbiota are able to elicit colitis, even in mice that are not genetically predisposed to developing immunopathologic responses.
The penetrance and severity of colitis observed in the co-colonization experiments with K. pneumoniae and P. mirabilis were decreased compared to what we had previously observed in the spontaneous TRUC model (e.g. Fig. 2C) and in neonatal cross-fostering experiments (TRUC-fostered Rag2−/− mean colitis score 5.6±1.14 and TRUC-fostered WT 3.17±.75 (Fig. 3A)). Instead, it resembled what we had observed in experiments where adult TRUC mice were co-housed with adult Rag2−/− or WT mice (Garrett et al., 2007), speaking to a possible role of maternal/foster bacterial and non-bacterial factors in structuring microbial communities in the neonate. Consistent with this, we found that TRUC milk has a pro-inflammatory cytokine profile (Fig. S5) and that the microbiota of 2 week old TRUC mice clusters in a distinct group as judged by PCoA plots of UniFrac measurements of 16S rRNA-defined communities (Fig. 1A).
Klebsiella pneumoniae and Proteus mirabilis colonization pattern change in response to immunotherapy and both strains induce TNF-α production in T-bet−/− Rag2−/− MyD88−/− bone marrow derived DCs
We next asked whether K. pneumoniae and P. mirabilis colonization patterns might change in response to two immunotherapeutic interventions previously shown to cure TRUC colitis; i.e., TNF-α neutralization and T-regulatory cell (T-reg) transfer (Garrett et al., 2007). We used quantitative culture-based methods to assay K. pneumoniae and P. mirabilis levels in feces prior to treatment of 4 week-old TRUC mice with anti-TNF-α, during weekly treatment for 4 weeks, and for six weeks after the last dose (Fig. 4A) (n=10 mice treated with anti-TNF-α and n=10 treated with an isotype control; histologic colitis scores are shown in Fig. S3). The differences in fecal K. pneumoniae levels between the TNF-α neutralization and isotype control groups were significantly different after animals had been treated for 7 weeks (i.e. were 11 weeks-old; p = 0.0172; Mann-Whitney test) and for P. mirabilis after a shorter period of treatment (p=0.008, p=0.0012, p=0.0004, and p=0.0403 at 7, 8, 9 and 10 weeks of age). Two way ANOVA revealed that anti-TNF-α neutralization accounted for 10.7% of the total variance observed in fecal P. mirabilis levels (after adjusting for matching: F = 22.83. DFn=1 DFd=18, p = 0.0002). Control experiments showed that TNF-α did not affect the growth kinetics of either K. pneumoniae or P. mirabilis under in vitro mono-culture conditions (Fig. S4).
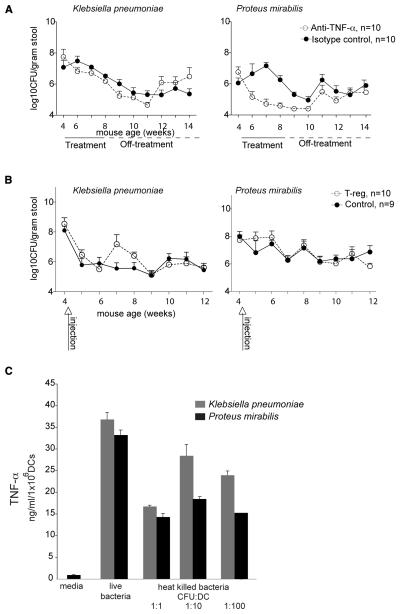
Figure 4. Klebsiella pneumoniae and Proteus mirabilis colonization patterns change in response to immunotherapies and both strains induce TNF-α production in T-bet−/− Rag2−/− MyD88−/− bone marrow derived DCs.
(A) Successful immunotherapy by TNF-α blockade alters levels of culturable Enterobacteriaceae in the feces. TRUC mice were treated with curative anti-TNF-α (15mg/kg every week) (open circles) or isotype control (shaded circles) for four weeks, after which time therapy was stopped. Enterobacteriaceae levels were defined by culture of fecal samples obtained 1 day before, during, and after treatment (up to 14 weeks of age). Circles represent the mean value of anti-TNF-α mice (n=10) and isotype controls (n=10). Error bars represent the standard deviation. (B) Successful immunotherapy by T-reg infusion does not produce statistically significant differences in the levels of culturable Enterobacteriaceae species compared to vehicle-treated controls. TRUC mice were injected once with 75,000 CD4+CD62LhiCD25+ T-regulatory cells (n=10) or PBS (n=9). (C) TNF-α production from T-bet−/− Rag2−/− MyD88−/− bone marrow derived DCs co-cultured with heat-killed and live K. pneumoniae and P. mirabilis strains. Bars represent the mean value of triplicate determinations/sample. Error bars are 1 S.D. Data are representative of three independent experiments.
We performed a similar analysis in TRUC mice that had received 75,000 purified wild type T-reg cells at 4 weeks of age (histologic colitis scores at 12 weeks of age are shown in Fig. S3). Surprisingly, while T-reg infusion ameloriated this colitis (Garrett et al., 2007), it did not affect fecal levels of either of these two bacterial species (Fig. 4B). These results further demonstrate that K. pneumoniae and P. mirabilis levels are not simply associated with inflammation per se, as both these modalities reduced host inflammation but did not uniformly alter Enterobacteriaceal representation. Our results illustrate that certain host-directed treatments may exert their effects not only by altering host inflammatory pathways but also by directly impacting the microbiota itself.
Finally, to begin to identify cell-based mechanisms by which TRUC-derived Enterobacteriaceae elicit a host immune response, TNF-α production was measured in T-bet−/− Rag2−/− MyD88−/− bone marrow-derived dendritic cells (DC) co-cultured with the K. pneumoniae and P. mirabilis TRUC strains; as DCs and TNF-α production are key features of the immunopathogenesis in TRUC mice and the TRUC inflammatory response is independent of MyD88 (Garrett et al., 2007 and 2009). Both live and heat-killed bacteria stimulated TNF-α production from DCs that were deficient in T-bet, Rag2 and MyD88 (Fig. 4C). These latter findings set the stage for future bacterial cell fractionation experiments where the microbial molecular determinants of host responses can be characterized using in vitro systems composed of immune cells that have been subjected to defined genetic manipulations.
DISCUSSION
Defining microbial features that are associated with or initiate IBD is complicated by host genetics, inflammatory state, and diet (Peterson et al., 2008). Designing prospective studies in human IBD to identify microbial communities that instigate inflammation has not been feasible, even in genetically susceptible populations. Thus we performed a time series screen in a mouse model of IBD that shares several pathophysiologic features of human IBD, including immunodeficiency, compromised host barrier function, and hyper-immunity to characterize a colitogenic microbiota. We established that host genotype influenced the global structure of the associated microbial community detected in feces and observed a number of significant order- and species- level differences. Combined with culture-dependent methods, we were able to identify bacterial species whose role we could test in the development of disease. Our experiments demonstrate that K. pneumoniae and P. mirabilis, together with other members of the endogenous microbiota can elicit colitis even in wild-type animals. It will be important to determine if significant associations are noted between these Enterobacteriaceae species and ulcerative colitis or Crohn's disease in ongoing (e.g., Qin et al., 2010) and future metagenomic studies of gut microbial ecology in various populations of patients with IBD.
K. pneumoniae and P. mirabilis can colonize the intestines of mice and humans (Lau et al., 2008). Notably, we only recovered these microbes from TRUC mice in our colony but not in Rag2−/− or WT animals. While there was individual variation in bacterial counts, the colonization pattern of these species across the TRUC population over time was not significantly different and did not vary as colitis worsened with age. Inciting inflammation with the colitogenic agent, dextran sulfate sodium, in wild type mice in the colony did not result in an emergence of these bacteria. In contrast, antibiotic treatment had a dramatic effect on the degree of host colonization with K. pneumoniae and P. mirabilis, which is to be expected. Pyrosequencing technology has begun to shed light on the sweeping effects of antibiotics on distal gut microbes and the differential resiliency of these communities (Hill DA et al., 2010, Dethlefsen et al., 2008). The increased counts observed in response to vancomycin suggest that in the untreated host, members of the Gram positive flora affect the degree of colonization by members of the Enterobacteriaceae.
A key feature of the colonic inflammation in TRUC mice is elevated TNF-α. While both neutralizing antibodies and T-reg infusion reduce mucosal TNF-α levels; these interventions had disparate effects on K. pneumoniae and P. mirabilis fecal counts. Cytokines may interact with bacteria and TNF-α has been shown to affect Salmonella typhimurium replication in vivo (Romanova et al., 2002). While TNF-α did not appear to affect the proliferation of TRUC-derived K. pneumoniae and P. mirabilis in vitro; in vivo there were significant effects in response to TNF-neutralizing antibodies. Neutralizing antibodies and infusion of T-regs both lower TNF-α levels in TRUC mice but through different mechanisms. T-regs also produce both IL-10 and TGF-β (Izcue et al., 2009). T-regs and neutralizing antibodies may have direct but distinct effects on microbial populations or indirect effects through their differential effects on colonic dendritic cells.
Opportunistic infection with K. pneumoniae and P. mirabilis is well recognized in the respiratory and urinary tracts. However, Klebsiellaoxytoca but not Klebsiella pneumoniae has been tied to intestinal pathology, specifically, antibiotic-associated diarrhea (Hogenauer et al., 2006). Klebsiella and Proteus species are observed more frequently in the stool of patients with ulcerative colitis than healthy controls (Dorofeyev et al., 2009; Kanareykina et al., 1987). There are also numerous reports of elevated titers of Enterobacteriaceal antibodies in patients with IBD (Cooper et al., 1988; Ibbotson et al., 1987; Tiwana et al., 1998).
As to the mechanism by which TRUC derived K. pneumoniae and P. mirabilis drive intestinal inflammation in TRUC, WT, and Rag2−/− mice, many factors are likely involved. Genome sequencing of these isolates and comparisons to other sequenced isolates obtained from other body and environmental habitats could yield testable hypotheses about genetic determinants that may underlie their ability to drive development of an IBD phenotype. As noted above, both heat-killed and live K. pneumoniae and P. mirabilis are able to induce TNF-α in T-bet−/− Rag2−/− MyD88−/− DCs, allowing for future mechanistic studies directed at identifying the responsible bacterial molecules and their host receptors. However, it is important to also emphasize that an endogenous microbial community is required for K. pneumoniae and P. mirabilis to exert their colitogenic effects. Both K. pneumoniae and P. mirabilis share an ability to occupy luminal and mucosal habitats in the gut. Visualization of small numbers of these bacterium adjacent to the epithelium suggest they may have invasive capacity or that their bacterial products near the apical surface of the epithelium may trigger pro-inflammatory responses. Proximity to the apical surface may also increase the likelihood that they are sampled by dendritic cells and from our previous work dendritic cells appear to be a key effector cell in TRUC immunopathogenesis (Garrett et al., 2007, 2009).
The host's genetic background also clearly influences the development of inflammatory responses to the microbiota. Isogenic WT mice consistently had lower histologic colitis scores than cross-fostered or Klebsiella pneumoniae and Proteus mirabilis treated Rag2−/− mice. While T-regulatory cell infusion into TRUC mice did not affect K.pneumoniae or P. mirabilis counts in a statistically significant manner, other adaptive factors such as commensal-specific secretory IgA might provide host defense mechanisms to better control the overall localization of K. pneumoniae and P. mirabilis in the gut lumen. IgA has been shown to both contribute to sequestration of bacteria in the lumen and to alter bacterial gene expression (MacPherson AJ et al., 2000, Cerrutti and Rescigno 2008, and Peterson DA et al., 2007).
Gut microbes help to structure the mucosal immune system and the mucosal immune system shapes microbial community structure (Smith K et al., 2007; Duerkop BA et al., 2009; Hooper LV and Macpherson AJ 2010). Microbial community members may be needed for the development of particular immune subsets or appropriate localization of immune cell subsets within the mucosa to generate pro-inflammatory responses to K. pneumoniae and P. mirabilis. For example, adherent cecal segmented filamentous bacteria have recently been shown to play a central role in the development of IL-17 producing CD4+ T helper cells in mice (Ivanov et al., 2009; Gaboriau-Routhiau et al., 2009). We have previously demonstrated that CD11c+ dendritic cells are necessary for TRUC colitis (Garrett et al., 2007, 2009) and a recent study has shown that lamina propria CD11c+ CX3CR1+ dendritic cells are markedly reduced in germ-free mice (Niess and Adler 2010). In addition, interactions between K. pneumoniae and P. mirabilis and microbial community members may result in the acquisition of traits by these two Enterobacteriaceae (e.g. invasion) or by other community members that elicit intestinal inflammation. Convergence of host genetic susceptibility and microbial community features could also affect the behavior of these Enterobacteriaceae and the immune response to them, as we have observed in the TRUC model.
Elevated TNF-α and beneficial responses to TNF-α neutralizing antibodies are common to both human IBD and several experimental colitis models. We hypothesize that host factors, like elevated TNF–α, may have virulence-promoting effects on these microbes. This notion is not without precedent as the Pseudomonas aeruginosa protein OprF binds the pro-inflammatory cytokine IFNγ resulting in expression of PA-I lectin, a quorum-sensing dependent virulence determinant (Wu et al., 2005).
In summary, future studies need to be directed at defining the genomic features of TRUC-associated K. pneumoniae and P. mirabilis strains, identifying co-occurring culturable members of the microbiota that contribute to disease pathogenesis in conventionally-raised and gnotobiotic mouse models, characterizing host factors that drive these microbes to become colitogenic, and determining the microbial associated molecular patterns and pattern recognition receptors involved in spontaneous and transmitted TRUC colitis. Together, these efforts may provide new mechanistic insights about how gut microbial communities, working in concert with specific colitogenic agents, contribute to initiation and perpetuation of IBD in susceptible human hosts and provide the foundation for proof-of-concept tests of new preventative or therapeutic measures. An additional benefit may be to help elucidate the association between IBD and increased risk for tumorigenesis since the majority of TRUC mice spontaneously develop colonic dysplasia and rectal adenocarcinoma (Garrett et al., 2009).
EXPERIMENTAL PROCEDURES
Husbandry of conventionally-raised mice
Rag2−/−, T-bet−/− × Rag2−/−, and MyD88−/− × T-bet−/− × Rag2−/− mice and their genotyping have been described previously (Lugo-Villarino et al., 2005 and Garrett et al., 2009). Mice were housed in micro-isolator cages in a barrier facility located in the Harvard School of Public Health, under a 12h light cycle. Animals were fed Pico 20 Lab Diet 5058 (Purina) ad libitum. Animal studies and experiments were approved and carried out according to Harvard University's Standing Committee on Animals and NIH guidelines for animal use and care. Mice in the colony were specified pathogen-free, and negative for Helicobacter hepaticus, H. bilis, and H. muridarum but have 16S rRNA and qPCR evidence of Helicobacter ganmani in their cecal contents and feces.
16S rRNA-based analyses of fecal microbial communities
Community DNA preparation
Fecal samples were flash frozen immediately after collection and stored at −80°C before processing. DNA was extracted by bead-beating followed by phenol-chloroform extraction as described previously (Turnbaugh et al., 2009).
Sequencing and analysis of 16S rRNA gene amplicons
The V2 region (primers 8F-338R) of bacterial 16S rRNA genes was targeted for amplification and multiplex pyrosequencing with error-correcting barcodes (Hamady et al., 2008). A total of 75,145 high-quality reads were generated from 32 samples (2,348±343 reads per sample). V2 16S rRNA gene sequencing data were pre-processed to remove sequences with low quality scores, sequences with ambiguous characters, and sequences outside the length bounds (200-300 nucleotides). All subsequent data processing and analyses were done using QIIME software (http://qiime.sourceforge.net/). In summary, 16S rRNA reads were binned according to their sample-specific, error-correcting barcode incorporated into the reverse primer. Similar sequences were binned into operational taxonomic units (OTUs) using cd-hit with minimum pairwise identity of 97% (Li et al., 2001). A total of 3,229 species-level OTUs were identified in the 32 fecal communities. A phylogenetic tree was built from one representative sequence from each OTU by using FastTree's approximately-maximum-likelihood implementation, and the tree used for unweighted UniFrac analysis (Price et al., 2009). A matrix of UniFrac distance measurements for all pairwise comparisons of communities was constructed and used to generate Principal Coordinates Analysis (PCoA) plots. Taxonomy was assigned using the RDP database (Cole et al., 2009). The Mann-Whitney test was used to calculate which of the 3,229 OTUs were significantly different in their proportional representation in the fecal communities of mice belonging to the two genotypes. Raw p-values were adjusted for multiple comparisons using Bonferroni and False Discovery Rate (FDR) methods.
Culture-based studies of fecal microbial community structure
Stool Collection
A minimum of three fecal pellets were collected directly upon expulsion from each individual mouse, in a laminar flow hood. Each mouse (3 per genotype; TRUC and Rag2−/−, all female) was sampled every two weeks, at the same time of day from two weeks of age through ten weeks of age. Mothers were sampled once when their pups were two weeks old.
Culture
Fecal pellets were collected into tubes containing PBS supplemented with 0.05% cysteine HCl. Serial 10-fold dilutions were made and plated on non-selective media (tryptic soy agar with 5% sheep blood, and Brucella blood agar (Remel)), selective media (MacConkey, Bile Esculin, and Bacteroides Bile Esculin agar (Remel)), and differential media (Rogosa (Difco), Brucella agar with laked blood, kanamycin, and vancomycin (Remel)) and Columbia medium containing colistin, naldixic acid, and aztreonam (Remel)) for recovery of aerobic and anaerobic bacteria. Anaerobes were incubated at 37°C in a Coy Anaerobic chamber for a minimum of 5d. Aerobes were incubated for 24-48 h at 37°C.
Colonies were described, enumerated, and sub-cultured. All bacterial concentrations are expressed as colony forming units (cfu) per gram fecal dry weight. Gram-stain analysis was performed and identification methods included: long chain fatty acid analysis using the Sherlock GC-FAME platform from MIDI (Newark, DE), API kits from bioMerieux (Marcy l'Etoile, France), and/or Vitek2 from bioMerieux. Gas chromatography-based short chain fatty acid analysis was also employed to confirm the identification of several Gram-positive anaerobes. A.B.O reviewed all final identifications. The identities of Klebsiella pneumoniae and Proteus mirabilis were cross-validated using several of these methods. In vitro antibiotic sensitivities were performed as per Clinical Laboratory and Standards Institute guidelines, on the Vitek 2 system using the AST-GN 13 card (bioMerieux).
Fecal collection and culture of Gram-negative aerobes
Individual mice were placed in autoclaved plastic cages. Fecal pellets were transferred immediately upon expulsion into capped microfuge tubes. 4-6 pellets were collected per mouse per time point. All Rag2−/−mice in Fig. 2F were sampled twice over a 3d period for each weekly time point. Pellets were resuspended in sterile PBS and 10-fold serial dilutions were generated, plated on MacConkey's medium, and incubated in ambient air at 37°C overnight. Biochemical assays with the API-20E panel (bioMerieux) confirmed that colony morphology correlated with Klebsiella pneumoniae, Proteus mirabilis, or E. coli: therefore, these organisms were subsequently identified based on their colony morphotypes.
The lower limit of detection for these studies was 104.4 cfu/gram fecal dry weight. A minimum of 30 colonies or a maximum of 300 colonies was counted at a given dilution and the serial dilution had to appropriately reflect the colony counts for the quantitative counts to pass our quality control standards.
Histology
Colons were harvested upon sacrifice and colonic contents were removed prior to fixation in 4% paraformaldehyde. Following paraffin embedding, sections (0.5 μm thick) were cut and stained with hematoxylin and eosin. Histopathology was evaluated in a blinded fashion (with respect to genotype and experimental protocol) by J.N.G. using four parameters: mononuclear cell infiltration, polymorphonuclear cell infiltration, epithelial hyperplasia, and epithelial injury that were scored as absent (0), mild (1), moderate (2), or severe (3) as described previously (Neurath et al., 2002).
Antibiotic treatment of TRUC colitis
Mice were treated with the following antibiotics dissolved in their autoclaved drinking water as indicated: ampicillin (1g/L; Roche), vancomycin (500mg/L; Sigma), neomycin sulfate (1g/L; Sigma), metronidazole (1g/L; Sigma, solubilized with 15ml of 0.1N acetic acid/L), and gentamicin (2g/L; Cell Gro); and fluid intake monitored.
Fluorescence In Situ Hybridization (FISH)
Colons were harvested from 16 Rag2−/− and 15 TRUC (3-8 week old) mice and fixed in Carnoy's solution overnight, embedded in paraffin, and 5μm thick sections were prepared (Swidsinksi et al., 2005). The sequences of the following FISH probes were obtained from probeBase (http://www.microbial-ecology.net/probebase/) (Loy A et al., 2007): the ‘universal’ bacterial probe-EUB338 (pB-00159), the Enterobacteriaceae targeted probe (pB-00914), the Klebsiella pneumoniae-directed probe (pB-00352), and the P. mirabilis probe (pB-02110). While several of these probes have been validated for FISH by investigators (Kemp VA et al., 2000, Friedrich U et al., 2003), searches of the probeBase probecheck, Greengenes, and RDP2 databases indicated that the K. pneumoniae and P. mirabilis probes are not species-specific. Therefore, for the FISH analysis described in this report, we performed control experiments using a using a 10 fold molar excess of unlabeled probe and checked for loss of specific fluorescence signal. We also co-incubated sections with the EUB338 probe to determine whether the signal produced by the K. pneumoniae and P. mirabilis oligonucleotides co-localized with the signal from the universal bacterial probe.
Cross-Fostering
On the day of birth, the mother was removed from the birthing cage and placed in a clean cage. A litter of pups with the designated genotype was then transferred into the cage. Pups were weaned on postnatal day 21 (Garrett et al., 2007).
Dextran sulfate sodium treatment
Dextran sulfate sodium (M.W. 40-50,000; USB Cat# 14489) was dissolved in drinking water at a final concentration of 4% (w/v) and provided for 7d.
Gnotobiotic mouse experiments
All protocols related to the generation and husbandry of germ-free mice were approved by the Washington University Animal Studies Committee. Conventionally-raised, specified pathogen free T-bet−/− × Rag2−/− mice were re-derived as germ-free in the gnotobiotic facility at Washington University. At 6 weeks of age, germ-free mice were transported in a germ-free state using a specialized shipping apparatus (Taconic Laboratories), to the Harvard Digestive Disease Center (HDDC) gnotobiotic mouse facility. After transfer to flexible film gnotobiotic isolators, mice were monitored for one week and their germ-free status confirmed by culture and qPCR of fecal bacterial cDNA (16S rRNA primer set (FOR: 5′ TCCTACGGGAGGCAGCAGT and REV: 5′ GGACTACCAGGGTATCTAATCCTGTT) (Nadkarni et al., 2002). One cohort of five mice was maintained germ-free. Another cohort of five mice (3 female and 2 male) was co-colonized by introducing 4.8 ×108 cfu of Klebsiella pneumoniae and 9.2 ×108 cfu of Proteus mirabilis into their oral cavity and by simultaneously spreading an equivalent amount of organisms on their fur and anus. All animals were maintained on the Rodent NIH-31 Modified Auto diet (Ziegler Brothers, Inc). Fecal samples were collected to define levels of colonization of these organisms 48h after inoculation, and weekly thereafter for 8 weeks.
Invasion experiments
2×107 cfu of Klebsiella pneumoniae, Proteus mirabilis, E. coli, or both Klebsiella pneumoniae and Proteus mirabilis (all isolated from the TRUC mother in Fig. 1) were gently instilled into the oral cavity of each mouse using a sterile pipette tip, and 1×107 cfu was placed into a new container of their drinking water every other day.
anti-TNF-α treatment
anti-TNF-α (clone TN3-19.12), a hamster anti-mouse TNF-α neutralizing IgG1 antibody and control Ab (hamster anti-GST IgG1) were purchased from Leinco Technologies. Mice were injected with these reagents (15mg/kg) on a weekly basis for four weeks (Garrett et al., 2007).
Adoptive Transfer of T-regulatory cells
Peripheral lymph nodes were harvested and cell suspensions were generated as previously described (Garrett et al., 2007). Fluorescence activated cell sorted CD4+ CD62Lhi CD25+ cells were collected and resuspended in PBS. 75,000 cells were injected per mouse. An equivalent amount of PBS was injected in the control group. 10 mice were injected with T-regulatory cells and nine mice with PBS. All mice were 4 weeks of age at the time of injection. This experiment was stopped when the mice were 12 weeks old (as opposed to 14 weeks of age in the TNF-α blockade experiment), because two mice in the control group became quite moribund from their colitis, and euthanasia was indicated as per our animal protocol.
Co-culturing bone marrow-derived dendritic cells and bacterial strains
Generation of mouse bone marrow-derived dendritic cells was performed as previously described (Garrett et al., 2007). DCs were purified using MACS bead positive selection with anti-mouse CD11c coupled magnetic beads. K. pneumoniae or P. mirabilis were co-cultured with the DCs at a ratio of 1 CFU per dendritic cell for 4h at 37°C in a cell culture incubator with an atmosphere of 5% CO2. Gentamicin (50 μg/ml) was then added to the culture medium for 1 h, cells were collected and washed extensively, and then incubated in the presence of medium containing gentamicin (20 μg /ml) for an additional 16 h. Bacteria were also heat-killed (incubation at 100°C for 3 min followed by plating to confirm killing) and added to cultures of DCs at ratios of 1:1, 10:1. 100:1. Cells were co-cultured for 20 h. Supernatants were collected from the live and heat killed co-cultures, centrifuged to pellet cells, and TNF-α levels in the resulting supernatants were determined using the mouse OptEIA ELISA kit (BD Biosciences) according to the manufacturer's instructions. TNF-α levels are expressed as ng/ml/1×106 DCs
TNF-α bacterial co-culture
TNF-α (final concentration 100μg/mL culture medium), or PBS alone, was added to cultures of the designated microbes and bacterial concentrations (cfu/ml) were followed during a 6h incubation (37°C with agitation in ambient air) at 2h intervals. Bacteria were cultured in Luria Broth and plated on MacConkey's medium.
Cytokine assays in milk
Lactating mice were injected with 2U oxytocin (Sigma-Aldrich) i.p. and milk was collected using a suction-powered milking apparatus (Wilson and Butcher, 2004). Milk was then centrifuged (14,000 × g for 5 min) at room temperature, fat was discarded, and the remaining material was stored at −20°C until use (Wilson and Butcher, 2004).
Cytokines were measured in defatted fractions using SearchLight high dynamic range imaging and analysis service unless otherwise indicated. For TNF-α, the mouse OptEIA ELISA kit (BD Biosciences) was used according to the manufacturer's instructions. Low levels of TNF-α (10pg/ml) were detected in the milk of TRUC but not in Rag2−/− mice (Fig. S5); these low levels in TRUC animals are likely attributable TNF-α sequestration by TNF-R, as milk is known to contain high levels of TNF-R1 and TNF-R2 (Buescher and Malinowska, 1996).
Statistical Analysis
The Prism graphing and analysis program was used for calculation of statistical measures including mean values, standard deviations, p-values (Mann-Whitney test), and two-way ANOVA.
Supplementary Material
Acknowledgements
We thank Jonathan Braun (UCLA), David Relman (Stanford University), Alexander Swidsinski (Charite Humboldt University), Gunnar Hansson (University of Gothenburg) and members of the Gordon and Glimcher labs for helpful discussions. We thank Jacobo Ramirez and Diana Pascual for their care of our specified pathogen free colony and Sabrina Wagoner, Vladimir Yesliseyev and Clara Belzer for their technical assistance and care of germ-free TRUC mice. This work was supported by from the NIH (CA112663 to LHG), Danone Research (LHG), and the Crohn's and Colitis Foundation of America (JIG), plus career development awards from the Burroughs Wellcome Fund and the NIH (K08AI078942) to WSG. The HDDC germ-free core is supported by P30-DK03485. 454 pyrosequencing reads have been deposited in the NCBI Short Read Archive.
LHG declares that she is a member of the Board of Directors of the Bristol Myers Squibb Corporation (BMSC) and holds equity in BMSC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Buescher ES, Malinowska I. Soluble receptors and cytokine antagonists in human milk. Pediatr Res. 1996;40:839–844. doi: 10.1203/00006450-199612000-00011. [DOI] [PubMed] [Google Scholar]
- Cerrutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R, Fraser SM, Sturrock RD, Gemmell CG. Raised titres of anti-Klebsiella IgA in ankylosing spondylitis, rheumatoid arthritis, and inflammatory bowel disease. Br Med J (Clin Res Ed) 1988;296:1432–1434. doi: 10.1136/bmj.296.6634.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorofeyev AE, Vasilenko IV, Rassokhina OA. Joint extraintestinal manifestations in ulcerative colitis. Dig Dis. 2009;27:502–510. doi: 10.1159/000233289. [DOI] [PubMed] [Google Scholar]
- Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24:4–10. doi: 10.1097/MOG.0b013e3282f2b0e8. [DOI] [PubMed] [Google Scholar]
- Friedrich U, Van Langenhove A, Altendorf K, Lipski A. Microbial community and physicochemical analysis of an industrial waste gas biofilter and design of 16S rRNA-targeting oligonucleotide probes. Environ Microbiol. 2003;5:183–201. doi: 10.1046/j.1462-2920.2003.00397.x. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Punit S, Gallini CA, Michaud M, Zhang D, Sigrist KS, Lord GM, Glickman JN, Glimcher LH. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16:208–219. doi: 10.1016/j.ccr.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen R, Thomson JM, El-Omar EM, Hold GL. The role of infection in the aetiology of inflammatory bowel disease. J Gastroenterol. 2010 doi: 10.1007/s00535-009-0191-y. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Artis D. Maintaining diplomatic relations between mammals and beneficial microbial communities. Sci Signal. 2009;2:pe77. doi: 10.1126/scisignal.298pe77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–58. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C, Hill DA, Minkah N, Kirn T, Troy A, Artis D, Bushman F. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun. 2009;77:4668–4678. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenauer C, Langner C, Beubler E, Lippe IT, Schicho R, Gorkiewicz G, Krause R, Gerstgrasser N, Krejs GJ, Hinterleitner TA. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N Engl J Med. 2006;355:2418–2426. doi: 10.1056/NEJMoa054765. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. 2010;10:159–69. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- Ibbotson JP, Pease PE, Allan RN. Serological studies in Crohn's disease. Eur J Clin Microbiol. 1987;6:286–290. doi: 10.1007/BF02017614. [DOI] [PubMed] [Google Scholar]
- Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- Kanareykina SK, Misautova AA, Zlatkina AR, Levina EN. Proteus dysbioses in patients with ulcerative colitis. Nahrung. 1987;31:557–561. doi: 10.1002/food.19870310570. [DOI] [PubMed] [Google Scholar]
- Kempf VA, Trebesius K, Autenrieth IB. Fluorescence in situ hybridization allows for rapid identification of microorganisms in blood cultures. J Clin Microbiol. 2000;38:830–8. doi: 10.1128/jcm.38.2.830-838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HY, Huffnagle GB, Moore TA. Host and microbiota factors that control Klebsiella pneumoniae mucosal colonization in mice. Microbes Infect. 2008;10:1283–1290. doi: 10.1016/j.micinf.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- Lugo-Villarino G, Ito S, Klinman DM, Glimcher LH. The adjuvant activity of CpG DNA requires T-bet expression in dendritic cells. Proc Natl Acad Sci U S A. 2005;102:13248–13253. doi: 10.1073/pnas.0506638102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Maxiner F, Wagner M, Horn M. probeBase- an online resource rRNA-tageted oligonucleotide probes: new features 2007. Nucleic Acids Res. 2007;35:D800–804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- MacPherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–6. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Niess JH, Adler G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. Journal of Immunology. 2010;184:2026–37. doi: 10.4049/jimmunol.0901936. [DOI] [PubMed] [Google Scholar]
- Neurath MF, Weigmann B, Finotto S, Glickman J, Nieuwenhuis E, Iijima H, Mizoguchi A, Mizoguchi E, Mudter J, Galle PR, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J Exp Med. 2002;195:1129–1143. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Peterson DA, Frank DN, Pace NR, Gordon JI. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe. 2008;3:417–427. doi: 10.1016/j.chom.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova YM, Scheglovitova ON, Boshnakov RH, Alekseeva NV, Stepanova TV, Tomova AS, Gintsburg AL. TNF-alpha and gamma-irradiation induced activation of the Salmonella typhimurium reproduction in the organs of infected animals. Russ J Immunol. 2002;7:129–134. [PubMed] [Google Scholar]
- Sartor RB. Microbial-host interactions in inflammatory bowel diseases and experimental colitis. Nestle Nutr Workshop Ser Pediatr Program. 2009;64:121–132. doi: 10.1159/000235787. discussion 132-127, 251-127. [DOI] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. Journa of Clinical Microbiology. 2005;43:3380–9. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwana H, Walmsley RS, Wilson C, Yiannakou JY, Ciclitira PJ, Wakefield AJ, Ebringer A. Characterization of the humoral immune response to Klebsiella species in inflammatory bowel disease and ankylosing spondylitis. Br J Rheumatol. 1998;37:525–531. doi: 10.1093/rheumatology/37.5.525. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium. Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser MM. Intestinal Epithelial Cell Surface Membrane Glycoprotein Synthesis: I. An Indicator of Cellular Differentiation. Journal of Biochemistry. 1973;248:2536–41. [PubMed] [Google Scholar]
- Wilson E, Butcher EC. CCL28 controls immunoglobulin (Ig)A plasma cell accumulation in the lactating mammary gland and IgA antibody transfer to the neonate. J Exp Med. 2004;200:805–809. doi: 10.1084/jem.20041069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, Patel N, Musch MW, Chang EB, Fu YX, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.