Abstract
In Drosophila, Iroquois (Irx) genes have various functions including the specification of the identity of wing veins. Vertebrate Iroquois (Irx) genes have been reported to be expressed in the developing digits of mouse limbs. Here we carry out a phylogenetic analysis of vertebrate Irx genes and compare expression in developing limbs of mouse, chick and human embryos and in zebrafish pectoral fin buds. We confirm that the six Irx gene families in vertebrates are well defined and that Clusters A and B are duplicates; in contrast, Irx1 and 3, Irx2 and 5, and Irx4 and 6 are paralogs. All Irx genes in mouse and chick are expressed in developing limbs. Detailed comparison of the expression patterns in mouse and chick shows that expression patterns of genes in the same cluster are generally similar but paralogous genes have different expression patterns. Mouse and chick Irx1 are expressed in digit condensations, whereas mouse and chick Irx6 are expressed interdigitally. The timing of Irx1 expression in individual digits in mouse and chick is different. Irx1 is also expressed in digit condensations in developing human limbs, thus showing conservation of expression of this gene in higher vertebrates. In zebrafish, Irx genes of all but six of the families are expressed in early stage pectoral fin buds but not at later stages, suggesting that these genes are not involved in patterning distal structures in zebrafish fins.
Keywords: chick limb, human embryo, Iroquois, mouse limb, zebrafish pectoral fin bud
Introduction
Iroquois (Irx) genes encode homeodomain-containing proteins belonging to the TALE (three amino acid loop extension) family. In Drosophila, where Irx genes were first discovered, there is a cluster of three closely related genes, Araucan, Caupolican and Mirror. Among the several roles now uncovered for these Drosophila genes are specification of the dorsal-most region of the wing imaginal disc to form notum (Diez del Corral et al., 1999; Cavodeassi et al., 2001) and later specification of the longitudinal wing veins L1, L3 and L5 (Gomez-Skarmeta et al., 1996). Vertebrate Irx genes have been identified and shown to be expressed in developing digits in mouse limbs (Houweling et al., 2001). A number of other genes encoding transcription factors involved in vein specification in Drosophila wings are also expressed in vertebrate limbs.
In most vertebrates, an ancestral Irx gene cluster of three genes appears to have been duplicated (Kerner et al., 2009). Thus there are six Irx genes in mouse and human (Irx1–6) in two clusters, the IrxA cluster containing Irx1, 2 and 4, and the IrxB cluster Irx3 5, and 6 (Peters et al., 2000). Xenopus also has six Irx genes (De la Calle-Mustienes et al., 2005). Ogura et al. (2001) described five chick Irx genes, Irx1, 2 and 4, in cluster A and Irx3 and 5 in cluster B, but recently a fragment of a sixth chicken Iroquois gene has been identified (Kerner et al., 2009). Eleven Irx genes have been found in the zebrafish genome, where they are organized into four clusters apart from one isolated gene, zIrx7. The four zebrafish clusters appear to have originated by duplication of ancestors of the two mammalian clusters and have therefore been named IrxAa (Irx1a, 2a, 4a), IrxAb (Irx1b, 4b), IrxBa (Irx3a, 5a, 6a) and IrxBb (Irx3b, 5b) (Dildrop & Ruther, 2004; Feijoo et al., 2004; Lecaudey et al., 2005).
In developing mouse limbs, Zulch et al. (2001) have shown that mIrx1 is expressed in the condensations of digits 2, 3 and 4 (the middle three digits of the paw) first, then mIrx2 is later expressed strongly in digits 1 and 2 (the most anterior and most posterior digits, respectively) and digits 2, 3 and 4 express both mIrx1 and Irx2. The expression of Irx genes in developing chick digits has not been described. In the chick wing and leg, three and four digits respectively develop, but their identity in relation to the pentadactyl limb plan is still debated (Vargas & Fallon, 2005; Xu et al., 2009). Therefore examination of Irx expression in the chick might not only reveal whether expression of these genes is conserved in digit development in different vertebrates but also cast light on digit identity. We found that Irx1 becomes expressed in all the digit condensations in both mouse and chick limbs, although the timing of expression differs in individual digits. We also examined Irx1 expression in human limbs to see whether the pattern of expression of this gene is further conserved among higher vertebrates. We found in zebrafish that Irx genes are expressed in early pectoral fin buds but, unlike mouse, chick and human limbs are not expressed at later stages when distal structures are developing.
Materials and methods
Mouse embryos
Pregnant CD1 mice were obtained from either the Wellcome Trust Resource Centre (University of Dundee) or the Resource Centre (University of Bath). Embryos of different developmental stages (E11.5–E14.5) were dissected from amniotic sacs and all membranes were removed before fixing in 4% paraformaldehyde overnight at 4 °C. Embryos were then dehydrated, stored in 100% methanol at −20 °C and processed for in situ hybridization.
Chick embryos
Fertilized White Leghorn chicken eggs were obtained from Henry Stewart (Lincolnshire, UK). Eggs were incubated on their sides at 38 °C in a humidified incubator for various lengths of time until embryos had reached the required developmental stage (Hamburger & Hamilton, 1992). Embryos were dissected from eggs and fixed in 4% paraformaldehyde (PFA) overnight at 4 °C, then gradually dehydrated through a methanol series. Embryos were stored at −20 °C for periods of up to several months and then processed for in situ hybridization.
Human embryonic tissue
The Human Developmental Biology Resource (HDBR) is a collection of human embryonic and fetal material ranging from 4 to 12 weeks of development; tissue from this resource was used to study expression of our gene of interest (Irx1) in a project registered with the In-House Gene Expression Service at the Institute of Child Health (ICH), London. Human embryonic limb tissue from the HDBR was processed for in situ hybridization at the ICH. Limbs from Carnegie Stage 18 (CS18) and CS19 embryos were used for section in situ hybridization to detect hIrx1 (IMAGE clone 1706829). Digital images of Irx1 gene expression patterns produced from the study were provided by the HDBR.
Zebrafish embryos
Danio rerio (zebrafish) embryos were randomly bred naturally, and fertilized eggs were allowed to develop at 28 °C. Some embryos were treated with 1-phenyl-2-thiourea (PTU) to inhibit pigmentation and allow in situ patterns to be seen more clearly. Zebrafish embryos were staged according to Kimmel et al. (1995), and embryos at 30 h post fertilization (hpf), 36 or 48 hpf were then fixed in 4% PFA. Embryos were fixed overnight at 4 °C, dehydrated, stored in 100% methanol at −20 °C and then processed for in situ hybridization.
Whole mount in situ hybridization
Plasmid DNA for the synthesis of RNA probes for in situ hybridization were obtained as follows: cIrx1, cIrx2, cIrx3 (kind gift from Dr A. Munsterberg), cIrx4 (chEST360a5), cIrx5 (chEST413l15), mIrx1-6 (IMAGE clones 2615958, 5347048, 1255751, 40089986, 3469768, 4512181, respectively), zIrx1a-7 (kind gift from Dr S. Schnieder-Maunoury), hIrx1 (IMAGE clone 1706829). All plasmids used for in situ hybridization were sequenced using the DNA sequencing service at the Wellcome Trust Biocentre (University of Dundee). Whole mount in situ hybridization of chick, mouse and zebrafish embryos was performed as previously described (Wilkinson & Nieto, 1993; Lecaudey et al., 2004).
Section in situ hybridization
Limbs were isolated from HH27 and HH28 chicks and E12.5 and E13.5 mice which had been stored in 100% MeOH. The limbs were embedded in paraffin wax. Sections were cut at a thickness of 14 μm and mounted onto aminoalkylsilane-coated slides. Sections were de-waxed by baking the slides at 60 °C for 1 h and then allowing them to cool to room temperature. In situ hybridization was then carried out on sections as previously described (Moorman et al., 2001).
Photography
A dissection microscope with a Jenoptic C14 digital camera (Laser Optic System) was used to take photographs of whole mount specimens using openlab software. In situ hybridization specimens were photographed in phosphate-buffered saline on 1% agarose plates using Drosophila pins to orientate embryos into the correct positions. Sections were photographed using a compound Leica DMR microscope with a Nikon D1X digital camera.
Phylogenetic analysis
Accession numbers of sequences belonging to the Iroquois gene families were extracted from Hovergen (Duret et al., 1994). Four families were employed: HBG006181, HBG006180, HBG073961 and HBG093209. These were supplemented with sequences identified as putative orthologs within the Hologene framework. Complete sequences were employed where possible. For a list of genes and accession numbers see Table S1. Translations of all sequences were extracted. Alignment was performed on the translated sequences using m-coffee (Wallace et al., 2006; Moretti et al., 2007).
Results
Phylogeny of vertebrate Iroquois genes
To study the phylogeny of vertebrate Irx genes including those from chick, mouse, human and zebrafish, accession numbers of sequences belonging to the Irx gene families were extracted from Hovergen (a database of homologous vertebrate genes). These were supplemented with sequences identified as putative orthologues. Complete sequences were used where possible, and alignment was performed using m-coffee software.
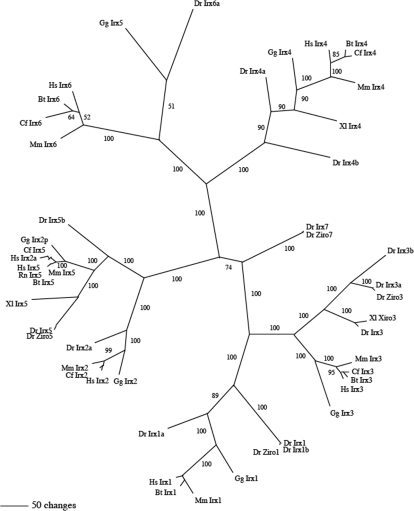
Phylogeny was constructed by two methods. First, phylogeny was constructed using paup* beta 4 (Swofford, 2003). A distance metric was employed with the quartet puzzling method. Hundred replicates were performed to generate support values. This tree is shown in Fig. S1. The second method, a Bayesian approach (Huelsenbeck & Ronquist, 2001), employed MrBayes (v 3.1.2) to perform two parallel runs of one million searches. A burn in of 5000 of the 10 000 resolved trees was employed to determine the 50% majority consensus rule unrooted tree. This and appropriate support values are shown in Fig. 1.
Fig. 1.
An unrooted Bayesian phylogram of Iroquois genes. Numbers indicate support values. Note that each of the six Irx genes form well defined groupings, e.g. Irx1 gene family is well defined with all genes in expected phylogenetic locations. Chick Irx5 is misidentified, as it belongs to Irx6 groupings. Species are as follows: Hs, Homo sapiens; Mm, Mus musculus; Bt, Bos taurus; Cf, Canis familiaris; Gg, Gallus gallus; Dr, Danio rerio; Xl, Xenopus laevis; Rn, Rattus norvegicus.
The main point of the phylogeny reconstruction was to ensure that genes were assigned correctly to their ortholog groups. Overall, the six Irx gene families define themselves very well, with both reconstruction methods agreeing. For example, the Irx1 gene family is well defined with all genes in expected phylogenetic locations. A few genes appear to have been mis-named: chicken Irx5 (XM_001234058) should be Irx6, human Irx2a (U90304) is a variety of Irx5, as is the sequence identified as a possible chicken Irx2 [Irx2p, p for possible, (XM_001234100]; Figs 1, S1). We will refer to the previously named cIrx5 as cIrx6 in the following description. A complete sequence with a properly annotated protein for chicken Irx5 was not found in the Hovergen database. The tree nicely shows the paralogous Iroquois genes in each cluster; Irx1and Irx3, Irx2 and Irx5 and Irx4 and Irx6 in clusters A and B, respectively (see also Kerner et al., 2009). The branch lengths from the root to each duplication are about the same, suggesting that the ancestral set of three genes all duplicated at one time.
Mouse limb Iroquois expression
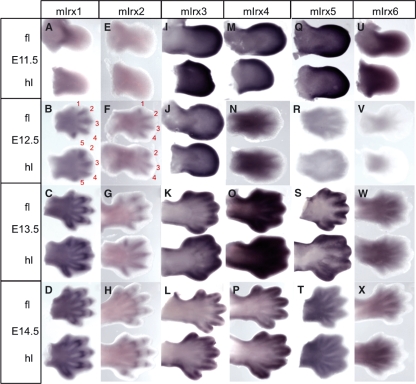
Expression patterns of mIrx1-6 were studied in the limbs of mouse embryos from E11.5 to E14.5. mIrx1 and mIrx2 have similar patterns of expression in limbs throughout development (although mIrx2 expression is much weaker than mIrx1), consistent with previous reports (Houweling et al., 2001; Zulch et al., 2001). At E11.5, mIrx1 and mIrx2 are expressed in the proximal regions of the forelimb and hindlimb (Fig. 2A,E). At E12.5, expression is stronger in the developing digit condensations. Expression of both genes is seen first and is strongest in digits 2–4, with weaker expression in digits 1 and 5 (Fig. 2B,F). At later stages of digit development (E13.5), expression of mIrx1 and mIrx2 is stronger in the joint-forming regions of digits 2, 3 and 4 (Fig. 2C,G). By E14.5, expression is seen in joint-forming regions in all digits in both forelimb and hindlimb (Fig. 2D,H).
Fig. 2.
mIrx1-6 expression in E11.5–E14.5 mouse limbs. mIrx1 and mIrx2 have the same pattern of expression in limbs throughout development, although mIrx2 expression is much weaker. (A,E) mIrx1 and mIrx2 are proximally expressed in forelimb and hindlimb at E11.5. (B,F) Expression of mIrx1 and mIrx2 is restricted to condensing digits at E12.5, with stronger expression in digits 2–4. (C,G,D,H) mIrx1 and mIrx2 are expressed in joint-forming regions at E13.5 and E14.5. (I,J) mIrx3 is expressed at the distal edge of both forelimb and hindlimb at E11.5 and E12.5. (K,L) Strong expression of mIrx3 at distal edges of digits at E13.5 and E14.5 and also interdigitally at E13.5. (M) mIrx4 distally expressed at E11.5. (N) At E12.5, mIrx4 expression is reduced; expression does not reach the tip and appears to be weakly expressed in proximal interdigital regions. (O) mIrx4 strong in interdigital regions at E13.5, and then at the distal edges of the digits at E14.5 (P). (Q) mIrx5 stronger in distal limb regions at E11.5. (R) At E12.5, mIrx5 expression very weak, but faint expression can be seen in forelimb interdigits. (S) mIrx5 restricted to interdigital regions at E13.5, with stronger expression around digit tips in the forelimb. (T) At E14.5, mIrx5 expressed between and around the edges of the digits. (U) mIrx6 expressed in the middle of forelimb and proximally in hindlimb at E11.5. (V) Weak mIrx6 expression seen at E12.5 in region just proximal to digital plate. (W,X) mIrx6 expression in proximal interdigital regions at E13.5 and E14.5.
Like mIrx1 and mIrx2, mIrx6 is also expressed proximally during early stages of limb development (Fig. 2U), whereas mIrx3, 4 and 5 appear to be expressed distally at E11.5 (Fig. 2I–J, M–N and Q–R, respectively). mIrx3 and mIrx4 show similar expression patterns at later stages of digit formation (E13.5 and E14.5). mIrx3 is expressed distally in interdigital spaces and then around the tips of the digits (Fig. 2K,L), and mIrx4 shows a similar pattern (Fig. 2O,P). mIrx5 and mIrx6 are both expressed in the proximal interdigital spaces of the forelimb and hindlimb at late stages of limb development (Fig. 2T; W–X, respectively).
Chick limb Iroquois expression
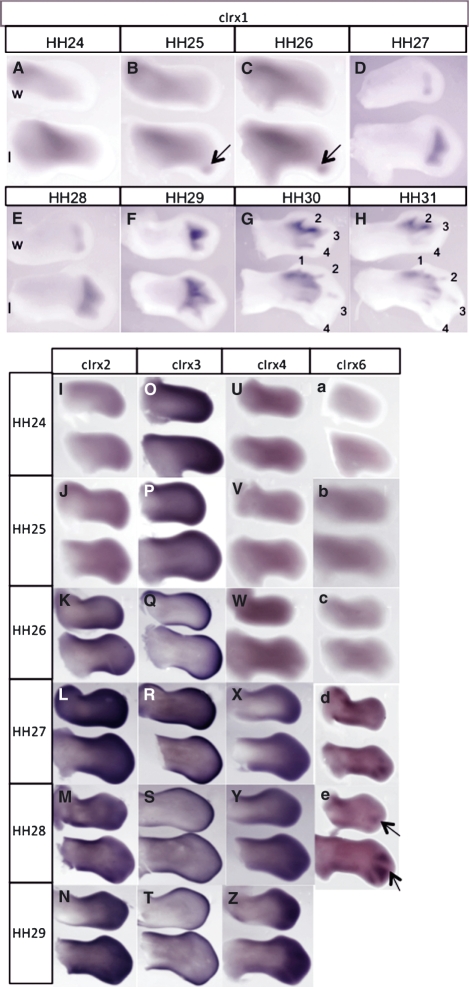
In situ hybridization was carried out to determine the expression pattern of the five chicken Irx genes described by Ogura et al. (2001), which according to our phylogenetic analysis are cIrx1, cIrx2, cIrx3, cIrx4 and cIrx6. Limbs at stages of development HH24–29+ were studied.
The gene with the strongest, most specific expression in the digit-forming region was found to be cIrx1. cIrx1 is expressed at low levels in the proximal region of both wing and leg at HH24 (Fig. 3A). By HH25/26, weak proximal expression remains, but this is now accompanied by a strong, posterior distal domain of expression in the leg (Fig. 3B,C arrowed). By HH27, cIrx1 is also detected in the posterior region of the hand plate in the wing (Fig. 3D). As the limbs develop, cIrx1 expression spreads across the digital plate and comes to be expressed in two distinct spots in the wing and three in the leg; the spots appearing to correspond to developing digit condensations (Fig. 3E). These spots of expression then extend across the limb anteriorly, with expression later becoming stronger and more restricted to the developing digits (Fig. 3F). At later stages of development, cIrx1 expression is reduced posteriorly and becomes restricted to anterior digits, particularly digits 2 and 3 in both the wing and leg (Fig. 3G,H). It should be noted that cIrx1 expression never extends right up to the apical ectodermal ridge, the thickened epithelium at the tip of the wing bud.
Fig. 3.
cIrx1-6 expression in chick limbs. (A–C) cIrx1 expressed proximally in HH24–26 limbs, with distal expression initiated posteriorly in hindlimbs at HH25. (D) cIrx1 is initiated in distal wing at HH27 posteriorly, whilst Irx1 in leg has extended anteriorly at this stage to become expressed in two spots. (E,F) cIrx1 expression extends anteriorly in wings and legs, becoming more strongly expressed in developing digits at HH29. (G,H) At late stages of digit development (HH30/31), expression of cIrx1 is restricted to anterior digits. (I,J) cIrx2 is expressed distally in both wing and leg at HH24/25, with high expression developing at anterior and posterior edges of the limbs between HH26 and 28 (K–M). (N) cIrx2 is strongly expressed in distal digit-forming regions at HH29. (O) cIrx3 is highly expressed in distal limb at early stages before becoming restricted to the outside edges of the limbs at HH25 (P). (Q–T) From HH26 to 29, cIrx3 expression is confined to the distal rim of wing and leg. (U–W) cIrx4 expression is not detected in limbs from HH24 to 26. (X,Y) Expression of cIrx4 is detected in the distal limb at HH27 and HH28. (Z) At HH29, strong expression of cIrx4 is seen distally in both wing and leg. (a–c) Like cIrx4, cIrx6 expression is not detected in early (HH24–26) limb buds. (d) At HH27, anterior and posterior regions of expression in wing and in posterior interdigital regions in leg. (e) cIrx6 strongly expressed in leg interdigital regions, with weaker expression in wing interdigital regions at HH28.
cIrx2 and cIrx3 are both expressed distally at HH24-25 in both wing and leg buds (Fig. 3I,J; Fig. 3O,P, respectively) but there is no detectable expression of cIrx4 and cIrx6 (Fig. 3U–W; Fig. 3a–c, respectively). cIrx2, cIrx3, cIrx4 and cIrx6 are then all expressed distally from HH26/27 onwards. cIrx2 is highly expressed throughout the digital plate (Fig. 3K–N), whilst cIrx3 expression is strongest around the distal rim of the limbs (Fig. 3Q–T). cIrx4 is also strongly expressed distally during later digit-forming stages (Fig. 3X–Z). cIrx6 can be seen in anterior and posterior proximal regions in the wing, and in posterior interdigital areas in the leg at HH27 (Fig. 3d). By HH28, cIrx6 expression is clearly visible in the interdigital spaces, with expression stronger in the interdigital spaces of the leg (Fig. 3e arrows).
Chick, mouse and human Irx1 section in situ
As our expression studies showed Irx1 to be strongly expressed in the digit condensations of both chick and mouse embryos, we examined expression of this gene in more detail by carrying out section in situ hybridization. We also compared Irx1 expression in sections of human embryonic limbs to investigate whether the pattern of expression of this gene is conserved amongst higher vertebrates. Comparable chick and mouse developmental stages were chosen (Martin, 1990). The appropriate human developmental stages were chosen with reference to the University of New South Wales human embryology website (http://embryology.med.unsw.edu.au).
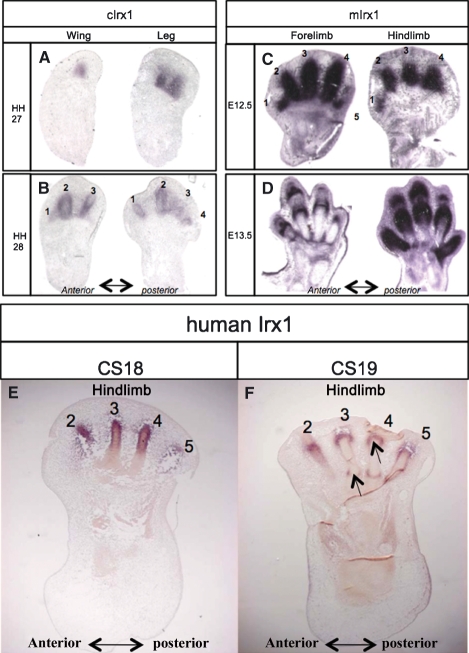
In the chick at HH27, cIrx1 is expressed in the posterior digit condensation in the wing and restricted to posterior digit-condensations in the leg (Fig. 4A). This expression in condensing cells at early digit-forming stages is consistent with patterns shown in in situ whole mounts for cIrx1, and confirms that the spots of cIrx1 expression correspond to the developing digits. At HH28, in situ sections show that cIrx1 is expressed more strongly around the edges of the digit condensations and in presumptive joint regions but more weakly in the centre of the condensations, details not apparent from the whole mount in situ (Fig. 4B).
Fig. 4.
Section in situ hybridization of Irx1 in chick, mouse and human limbs. (A) At HH27, cIrx1 is expressed in the posterior digit condensation(s) in wing and leg. (B) At HH28, cIrx1 is not expressed throughout the cartilage digit rudiments but is expressed at the edges in both wing and leg and in developing joints. (C) mIrx1 strongly expressed in all digit condensations of forelimb and hindlimb at E12.5. (D) At E13.5, expression in hindlimb throughout the skeleton of digits 1 and 5 and in joint-forming regions of digits 2, 3 and 4; in the more developmentally advanced forelimb, expression in joint-forming regions. Absence of digit 5 in hindlimb at E12.5 and digit 5 in forelimb at E13.5 is due to plane of section; additional serial sections showed Irx1 expression in these digits. (E) hIrx1 expressed in digits 2–5 of the hindlimb at CS18. (F) At CS19, expression in joint-forming regions of digits (arrows). Absence of digit 1 in E and F due to plane of section; additional serial sections showed expression in digit 1.
In E12.5 mouse limbs, very strong mIrx1 expression was seen in all developing digits of the forelimb and hindlimb (Fig. 4C and data from serial sections). At E13.5, expression is confined to the joint areas in all the digits in the section of the forelimb (Fig. 4D), whereas in the hindlimb, strong expression is seen throughout digits 1 and 5, and in the joint-forming regions of digits 2, 3 and 4, consistent with the patterns of expression observed in in situ whole mounts.
In CS18 human embryos, hIrx1 is expressed in all the condensations of the toes in the hindlimb (Fig. 4E and data from serial sections). At CS19, hIrx1 expression was clearly seen in the joint-forming regions of the digits (Fig. 4F; toe 1 is not present due to the plane of section). These Irx1 expression patterns are almost identical to those seen in mouse limbs of equivalent stages (compare with Fig. 4C,D), showing that expression of this gene in the tissues of developing digits is highly conserved.
Zebrafish pectoral fin Iroquois expression
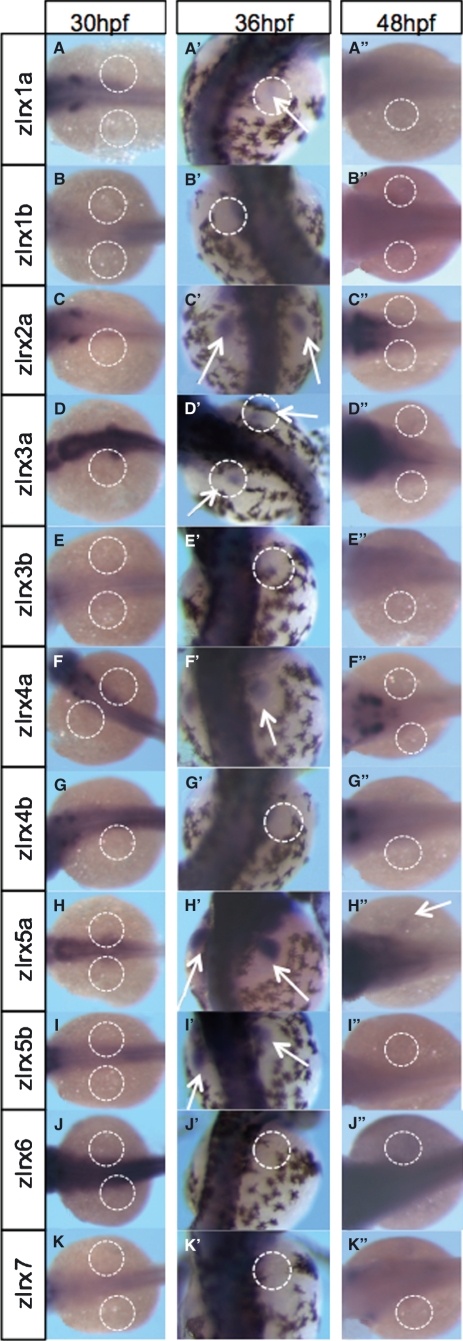
In situ hybridization was carried out to examine the expression of the 11 known zebrafish Iroquois genes in the pectoral fin buds of zebrafish embryos at different stages of development (30, 36 and 48 hpf). At 30 hpf, no Iroquois expression was detected in the pectoral fin buds (Fig. 5A–K). All probes were known to be working due to the strong expression of Irx genes seen in developing head and brain regions of embryos as previously described (Lecaudey et al., 2005). At 36 hpf, six of 11 genes were shown to be expressed in the pectoral fin buds. zIrx1a is faintly expressed throughout the pectoral fin, as shown in Fig. 5A’. zIrx2a shows clear expression throughout the pectoral fins (Fig. 5C’), and zIrx3a and zIrx4a are also expressed throughout the pectoral fins (Fig. 5D’, F’ respectively). zIrx5a shows the strongest expression in the pectoral fins (Fig. 5H’). Fig. 5I’’ shows that zIrx5b is also expressed in the pectoral fins. At 48 hpf, none of these genes was expressed in the pectoral fins, with the exception of zIrx5a, which is expressed very faintly (Fig. 5H’’).
Fig. 5.
Zebrafish Iroquois pectoral fin bud expression at 30, 36 and 48 hpf. Position of pectoral fin buds indicated by white-dashed circles; arrows indicate expression in pectoral fins. At 36 hpf, expression of six of 11 zIrx genes detected in pectoral fins. (A’) zIrx1a faintly expressed in pectoral fins at 36 hpf. (C’,D’,F’, respectively) zIrx2a, zIrx3a and zIrx4a expressed throughout developing pectoral fins (white arrows). (H’) zIrx5a displays strongest pectoral fin expression (arrows in H’). (I’) zIrx5b also expressed in pectoral fins, albeit with a weaker expression than zIrx5a. (H’’) By 48 hpf, only zIrx5a shows pectoral fin expression (weak expression, arrow in H’’). The 10 remaining zIrx genes are not expressed in pectoral fins at 48 hpf (A’’–G’’,I’’–K’’).
Discussion
Phylogenetic analysis of vertebrate Irx genes, including those from chick, mouse, human and zebrafish showed that the six Irx gene families define themselves very well with all genes in expected phylogenetic locations. Chick Irx5 has been previously misidentified, as it belongs to the Irx6 grouping; we used the name cIrx6 to refer to this gene. Our analysis also clearly showed that Irx1 and 3, Irx2 and 5, and Irx4 and 6 are the nearest paralogs, confirming the results of Kerner et al. (2009).
We compared Irx expression patterns in the forming digits of mouse and chick limbs (Table 1). This comparison shows that genes in the same cluster are generally expressed in more similar patterns than are the paralogs (Table 1). In the mouse, for example, there are particularly striking similarities in expression between the Cluster A genes mIrx1 and mIrx2, which are first expressed in digit condensations and then later very strongly in developing joints. But in contrast, for example, the expression of the paralogous B cluster gene to mIr2, Irx5, is at the edges of the digits and located interdigitally. In the chick, the similarity of the expression patterns of genes in the same clusters is not as striking. For example, the two cluster A genes, cIrx2 and cIrx4, appear to be expressed in a broad distal region of the limb buds, whereas the cluster B genes, cIrx3 and cIrx6, have very different expression patterns, with cIrx6 having marked interdigital expression. Furthermore there are no similarities in the expression patterns of paralogous chick genes. It therefore appears that duplication of the Irx cluster in vertebrates has led to a divergence in expression in the distal part of the limb which may then have allowed functional specialization.
Table 1.
Summary of expression of Irx genes in the developing digits of mouse and chick embryos.
| Cluster A |
Cluster B |
|||||
|---|---|---|---|---|---|---|
| Irx1 | Irx2 | Irx4 | Irx3 | Irx5 | Irx6 | |
| Mouse E12.5 | Digit condensations | Digit condensations | Interdigital | Distal | Weak interdigital | Weak proximal |
| Mouse E13.5 | Joints | Joints | Stronger interdigital | Distal edges of digits Interdigital | Edges of digits Interdigital | Interdigital |
| Mouse E14.5 | Joints | Joints | Distal edges of digits | Distal edges of digits | Edges of digits Interdigital | Interdigital |
| Chick HH27 | Digit condensations | Broad distal | Broad distal | Distal rim | N/A | Interdigital (leg) |
| Chick HH28 | Digit condensations Joints | Broad distal | Broad distal | Distal rim | N/A | Interdigital (wing and leg) |
Comparison of expression patterns of corresponding genes in mouse and chick revealed some similarities. Our results generally confirmed reported patterns of Irx gene expression in mouse limbs (Houweling et al., 2001; Mummenhoff et al., 2001; Zulch et al., 2001), except that we observed Irx4 expression in the digit-forming region of the limb which had not previously been detected (Houweling et al., 2001). Cluster A genes, mIrx1 and mIrx2, were expressed in digit condensations and Cluster B genes, mIrx5 and mIrx6, interdigitally. Similarly, in the chick we found that cIrx1, 2, 3, 4 and 6 are all expressed in the digit-forming region of wing and leg buds. Also similar to the expression patterns in the mouse limb, cIrx1 is strongly expressed in digit condensations, whereas cIrx6 is expressed interdigitally.
A previous report on Irx1 and Irx2 gene expression in mouse limbs highlighted a difference in the timing of expression in different digits, with mIrx1 being expressed more strongly initially in the middle digits 2, 3 and 4 and mIrx2 more strongly initially in digits 1 (anterior) and 5 (posterior) (Zulch et al., 2001). However, in our analysis, mIrx2 also appeared to be expressed first in the middle digits 2, 3 and 4. In the chick, the timing of cIrx1 expression in the condensations of the digits in both the wing and leg is very different to that in the mouse and appears in sequence from posterior to anterior. cIrx2 is broadly expressed in the distal region of the limb. In mouse, it has been suggested based on Alcian Green staining that the middle three digits form first (Martin, 1990; Zhu et al., 2008), whereas in the chick the condensations appear in a posterior to anterior sequence. Therefore our observations suggest that the timing of expression of Irx genes in both mouse and chick limbs is likely related to the order in which the digits form rather than to the identity of a particular digit.
We found that the pattern of Irx1 expression is conserved in the tissues of developing digits of chick, mouse and human. In all cases, Irx1 is expressed in digit condensations at equivalent stages, then, later, expression is reduced in the centre of the skeletal elements, remaining around the edges and also in the joints. Expression of Irx1 in the joints is similar to genes such as Gdf5 and Wnt14 (Storm & Kingsley, 1999; Guo et al., 2004), although these genes are not initially expressed throughout the digit condensations. The similarity between mouse Irx1 and human Irx1 is striking. There is evidence that there are differences and similarities in the expression of other important developmental genes (such as Wnt7a, Calpain3, Lhx3/4, Wnt8b) in humans compared with model organisms such as the mouse (Lako et al., 1998; Fougerousse et al., 2000; Sobrier et al., 2004). It remains to be investigated whether the other human Irx genes are expressed in the same patterns as in the mouse.
The patterns of Irx gene expression in zebrafish pectoral fin buds differ from those in tetrapod limb buds. In zebrafish, representatives of all the Irxa genes (except Irx6a) together with Irx5b are expressed in early pectoral fin buds (36 hpf), but Irx gene expression is generally undetectable in later fin buds (48 hpf). This absence of Irx gene expression in later pectoral fin buds contrasts with the strong expression of most of the Irx genes in the distal regions of chick and mouse limb buds at comparable stages; chick stage 26 and mouse E11.5. These observations are intriguing in the context of previous work which compared Hox gene expression in zebrafish pectoral fin buds and tetrapod limb buds. Three stages of Hox gene expression have been recognized in tetrapod limbs with stage III associated with digit development (Nelson et al., 1996). Studies in zebrafish have shown that although similar patterns of phases II and III Hox gene expression can be recognized, the third phase (from 36 hpf onwards) may be absent (Sordino et al., 1995) or be somewhat different (Ahn & Ho, 2008). The fact that Irx genes are not detectably expressed in fin buds after 36 hpf is strikingly different to what is seen in chick and mouse limbs, in which Irx genes are expressed during phase III Hox gene expression. It remains to be seen whether these patterns of Irx genes are truly characteristic of fish fins or just a peculiarity of zebrafish.
Acknowledgments
The authors would like to thank Robert Kelsh for providing some of the zebrafish embryos, Dr S. Schneider-Maunoury for providing template DNA for zebrafish Irx genes and Matthew Towers for help with the images. Human embryonic and fetal material was provided by the MRC/Wellcome-funded Human Developmental Biology Resource. L.M. was sponsored by an ASGBI studentship, performed most of the experimental work and helped write the manuscript. C.T. is a Royal Society Professor and helped write the manuscript L.D.H. is a Royal Society Wolfson Research Merit Award holder and performed the bioinformatic analysis. D.G. and Y.F. carried out the work on the human material.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. An unrooted distance-based phylogram of Irx genes. Numbers indicate quartet puzzling support values.
Fig. S2. mIrx1-6 expression in E11.5–E14.5 mouse limbs. These panels show unmanipulated images for comparison with images in Fig. 2.
Fig. S3. cIrx1-6 expression in chick limbs. These panels show unmanipulated images for comparison with images in Fig. 3.
Table S1. List of genes and accession numbers used in construction of unrooted phylogram of vertebrate Irx genes.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Ahn D, Ho RK. Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages. Dev Biol. 2008;322:220–233. doi: 10.1016/j.ydbio.2008.06.032. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- De la Calle-Mustienes E, et al. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez del Corral R, et al. The Iroquois homeodomain proteins are required to specify body wall identity in Drosophila. Genes Dev. 1999;13:1754–1761. doi: 10.1101/gad.13.13.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dildrop R, Ruther U. Organization of Iroquois genes in fish. Dev Genes Evol. 2004;214:267–276. doi: 10.1007/s00427-004-0402-8. [DOI] [PubMed] [Google Scholar]
- Duret L, et al. HOVERGEN – a database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijoo CG, et al. The Irx gene family in zebrafish: genomic structure, evolution and initial characterization of irx5b. Dev Genes Evol. 2004;214:277–284. doi: 10.1007/s00427-004-0401-9. [DOI] [PubMed] [Google Scholar]
- Fougerousse F, et al. Human-mouse differences in the embryonic expression patterns of developmental control genes and disease genes. Hum Mol Genet. 2000;9:165–173. doi: 10.1093/hmg/9.2.165. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, et al. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- Guo X, et al. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Houweling AC, et al. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev. 2001;107:169–174. doi: 10.1016/s0925-4773(01)00451-8. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kerner P, et al. Evolutionary history of the iroquois/Irx genes in metazoans. BMC Evol Biol. 2009;9:74. doi: 10.1186/1471-2148-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, et al. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lako M, et al. A novel mammalian wnt gene, WNT8B, shows brain-restricted expression in early development, with sharply delimited expression boundaries in the developing forebrain. Hum Mol Genet. 1998;7:813–822. doi: 10.1093/hmg/7.5.813. [DOI] [PubMed] [Google Scholar]
- Lecaudey V, et al. The zebrafish Iroquois gene iro7 positions the r4/r5 boundary and controls neurogenesis in the rostral hindbrain. Development. 2004;131:3121–3131. doi: 10.1242/dev.01190. [DOI] [PubMed] [Google Scholar]
- Lecaudey V, et al. Expression of the zebrafish Iroquois genes during early nervous system formation and patterning. J Comp Neurol. 2005;492:289–302. doi: 10.1002/cne.20765. [DOI] [PubMed] [Google Scholar]
- Martin P. Tissue patterning in the developing mouse limb. Int J Dev Biol. 1990;34:323–336. [PubMed] [Google Scholar]
- Moorman AF, et al. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J Histochem Cytochem. 2001;49:1–8. doi: 10.1177/002215540104900101. [DOI] [PubMed] [Google Scholar]
- Moretti S, et al. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 2007;35:W645–W648. doi: 10.1093/nar/gkm333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummenhoff J, et al. Expression of Irx6 during mouse morphogenesis. Mech Dev. 2001;103:193–195. doi: 10.1016/s0925-4773(01)00353-7. [DOI] [PubMed] [Google Scholar]
- Nelson CE, et al. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- Ogura K, et al. Cloning and chromosome mapping of human and chicken Iroquois (IRX) genes. Cytogenet Cell Genet. 2001;92:320–325. doi: 10.1159/000056921. [DOI] [PubMed] [Google Scholar]
- Peters T, et al. Organization of mouse Iroquois homeobox genes in two clusters suggests a conserved regulation and function in vertebrate development. Genome Res. 2000;10:1453–1462. doi: 10.1101/gr.144100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrier ML, et al. Pathophysiology of syndromic combined pituitary hormone deficiency due to a LHX3 defect in light of LHX3 and LHX4 expression during early human development. Gene Expr Patterns. 2004;5:279–284. doi: 10.1016/j.modgep.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Sordino P, et al. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- Storm EE, Kingsley DM. GDF5 coordinates bone and joint formation during digit development. Dev Biol. 1999;209:11–27. doi: 10.1006/dbio.1999.9241. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2003. Version 4. [Google Scholar]
- Vargas AO, Fallon JF. The digits of the wing of birds are 1, 2, and 3. A review. J Exp Zool B Mol Dev Evol. 2005;304:206–219. doi: 10.1002/jez.b.21051. [DOI] [PubMed] [Google Scholar]
- Wallace IM, et al. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–1699. doi: 10.1093/nar/gkl091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Xu X, et al. A Jurassic ceratosaur from China helps clarify avian digital homologies. Nature. 2009;459:940–944. doi: 10.1038/nature08124. [DOI] [PubMed] [Google Scholar]
- Zhu J, et al. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14:624–632. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulch A, et al. Expression pattern of Irx1 and Irx2 during mouse digit development. Mech Dev. 2001;106:159–162. doi: 10.1016/s0925-4773(01)00411-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.