Abstract
We used CD, UV resonance Raman spectroscopy, and molecular dynamics simulation to examine the impact of salts on the conformational equilibria and the Ramachandran Ψ angle (un)folding Gibbs free energy landscape coordinate of a mainly polyalanine α-helical peptide, AP of sequence AAAAA-(AAARA)3A. NaClO4 stabilizes α-helical-like conformations more than does NaCl, which stabilizes more than Na2SO4 at identical ionic strengths. This α-helix stabilization ordering is the reverse of the Hofmeister series of anions in their ability to disorder water hydrogen bonding. Much of the NaClO4 α-helix stabilization results from ClO4− association with the AP terminal –NH3+ groups and Arg side chains. ClO4− stabilizes 310-helix conformations but destabilizes turn conformations. The decreased Cl− and SO42− AP α-helix stabilization probably results from a decreased association with the Arg and terminal –NH3+ groups. Cl− is expected to have a smaller binding affinity and thus stabilizes α-helical conformations intermediately between NaClO4 and Na2SO4. Electrostatic screening stabilizes π-bulge conformations.
The mechanism(s) whereby peptides and proteins fold into their native states are poorly understand (1–6). The well-known Levinthal paradox (7) clearly demonstrates that proteins do not fold through a random search of their conformational space since this would take longer than the age of our universe. Recent energy landscape models (1, 3, 8, 9) propose that funnel-shaped folding energy landscapes occur, where the native state is accessed via a strategically sloped energy landscape that funnels unfolded conformations toward the native folded state (3, 10, 11).
In the work here we use CD, UV resonance Raman spectroscopy (UVRR), and molecular dynamics simulations to examine the Gibbs free energy landscape along the Ψ Ramachandran angle folding coordinate of a mainly polyalanine peptide, AP of sequence AAAAA(AAARA)3A, in pure water and in the presence of NaClO4, NaCl, and Na2SO4. AP-like peptides have been the subject of intensive experimental (12–30) and theoretical (31–42) studies which have probed the mechanism(s) of α-helix folding and unfolding. The AP peptide is ~50%α-helical-like at 0 °C and melts to PPII-like conformations at higher temperatures (43–47). We previously found that AP (un)folding is not a simple two-state process because it involves other secondary structure conformations such as π-bulge and 310-helix and turn structures (22, 31, 32, 41, 42, 48, 49). We examined the dependence of the AP conformational equilibrium and melting on the presence of different salts and find that ion binding and electrostatic screening significantly modulate the Gibbs free energy landscape and stabilize α-helix conformations. We also find that the ordering of salt stabilization of the α-helical content can be explained by Collins et al. (law of matching water affinities) (50–52).
EXPERIMENTAL PROCEDURES
The 21-residue peptide AP of sequence AAAAA(AAARA)3A was purchased from AnaSpec Inc. (>95% purity). Anhydrous NaCl, NaClO4, and Na2SO4 were purchased from J. T. Baker (>99% purity). All AP samples were prepared at 1.0 mg/mL concentrations at pH 7.
The CD spectra were measured by using a Jasco-715 spectro-polarimeter, using a 200 μm path length cuvette. We coadded ten individual spectra.
The UVRR spectrometer was described in detail by Bykov et al. (53). Briefly, 204 nm UV light was obtained by generating the fifth anti-Stokes Raman harmonic of the third harmonic of a Nd:YAG laser (Coherent, Infinity). We used a spectral accumulation time of 5 min for each measurement and coadded four accumulations.
Molecular Dynamics Simulation
We performed replica exchange molecular dynamics (REMD) studies of AP immersed in pure water and in a 0.2 M NaClO4 aqueous solution. The temperatures studied range from 270 to 505 K. The simulation details are given in the Supporting Information. To investigate the mechanisms which govern helix stabilization, we used Chimera (54) to calculate the average ion occupancy surrounding the AP peptide.
RESULTS
CD Results
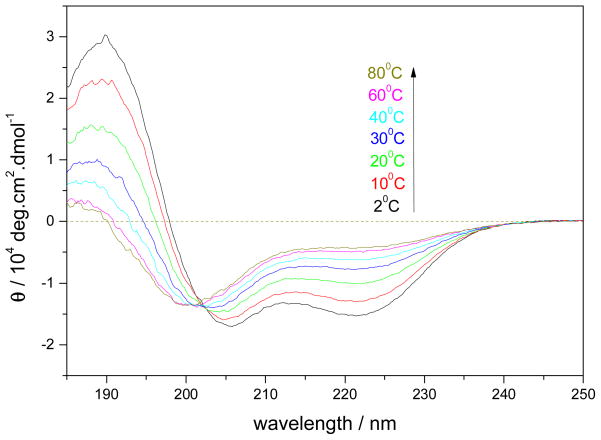
Figure 1 shows the temperature dependence of the CD spectra of AP in pure water. The lower temperature CD spectra show two troughs at 222 and 206 nm that are characteristic of α-helix conformations (55). As the temperature increases, the ellipticity at 222 nm, Θ222, becomes less negative, indicating α-helix melting. The isosbestic point at 202 nm indicates that this melting appears spectroscopically as a “two-state” process. Previous work by our group demonstrated that the AP α-helix conformation melts to a dominantly PII-like conformation (56).
Figure 1.
Temperature dependence of the CD spectra of AP in pure water at pH 7.
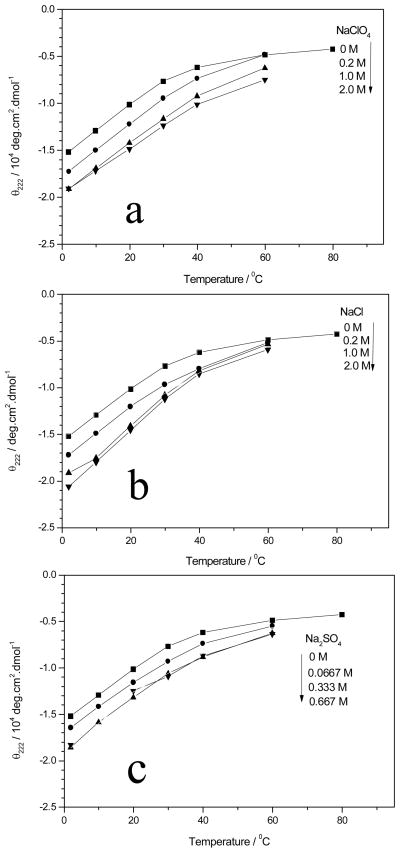
Addition of NaClO4 (Figure 2a) increases the AP α-helical content at all temperatures, as evident from the more negative values of Θ222; however, the CD changes are relatively small between 1 and 2MNaClO4 concentrations. As discussed below, Tm also increases as the NaClO4 concentration increases.
Figure 2.
Θ222 melting curve of AP (a) in NaClO4 at different ionic strengths, (■) pure water, (●) 0.2 M NaClO4, (■) 1.0 M NaClO4, and (▼) 2.0 M NaClO4, (b) in NaCl at different ionic strengths, (■) pure water, (●) 0.2 M NaCl, (■) 1.0 M NaCl, and (▼) 2.0 M NaCl, and (c) in Na2SO4 at various ionic strengths, (■) pure water, (●) 0.0667 M Na2SO4, (■) 0.333 M Na2SO4, and (▼) 0.667 M Na2SO4.
Panels b and c of Figure 2 show similar AP melting curves for NaCl and Na2SO4. Again, the α-helix fractions increase as evident from the more negative Θ222 values as the salt concentrations increase. Again, little increase occurs between 1 and 2M salt concentrations.
To quantitatively model α-helix melting, we calculated the α-helical conformational fraction, fα, using a two-state model (eq 1) by utilizing the reported Θ222 values for the pure α-helix ([θ]α=−26000 deg · cm2 · dmol−1) and the “pure melted” conformations ([θ]r = −3500 deg · cm2 · dmol−1) (57).
| (1) |
As shown below, the structure is actually more complex than a two-state transition, but significant useful thermodynamics information associated with the resulting quasi-two-state transition can be extracted from the CD data.
We also calculated the melting thermodynamic parameters by fitting the calculated equilibrium α-helix fraction, fα to
| (2) |
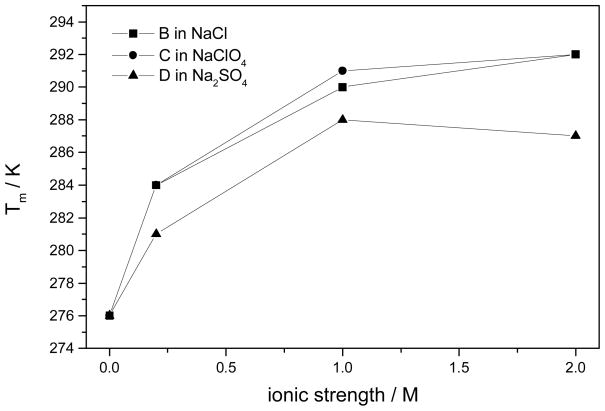
The fits to eq 2 versus T−1 are highly linear with R2 values of >0.995. Table 1 lists the calculated values for ΔH, ΔS, and the resulting estimated Tm values. Figure 3 shows that Tm increases for all salts as their concentrations increase (except at the highest Na2SO4 concentration). NaClO4 and NaCl stabilize α-helices more than does Na2SO4. Tm increases as the Na2SO4 concentration increases to 0.333M(ionic strength of 1.0 M) but then begins to decrease at a concentration of ~0.667 M (ionic strength of 2.0 M).
Table 1.
Thermodynamic Parameters Calculated from CD Data solution
| solution | ΔH/kJ · mol−1 | ΔS/J · mol−1 · K−1 | Tm/Ka | R2 |
|---|---|---|---|---|
| pure water | −33.4 ± 1.4 | −121 ± 5 | 276 | 0.995 |
| 0.2 M NaCl | −36.4 ± 1.1 | −128 ± 4 | 284 | 0.998 |
| 1.0 M NaCl | −41.8 ± 1.4 | −144 ± 5 | 290 | 0.998 |
| 2.0 M NaCl | −42.1 ± 1.0 | −144 ± 3 | 292 | 0.999 |
| 0.2 M NaClO4 | −39.7 ± 1.5 | −140 ± 5 | 284 | 0.997 |
| 1.0 M NaClO4 | −35.5 ± 0.5 | −122 ± 2 | 291 | 0.999 |
| 2.0 M NaClO4 | −30.4 ± 0.5 | −104 ± 2 | 292 | 0.999 |
| 0.0667 M Na2SO4 | −33.5 ± 0.5 | −119 ± 2 | 281 | 0.999 |
| 0.333 M Na2SO4 | −33.7 ± 0.6 | −117 ± 2 | 288 | 0.999 |
| 0.667 M Na2SO4 | −32.4 ± 1.5 | −113 ± 5 | 287 | 0.997 |
We did not calculate the Tm standard error because it is likely that its error is not dominated by random processes but instead is dominated by bias due to the ignored temperature dependencies of ΔH and ΔS.
Figure 3.
Calculated Tm of AP α-helix melting in different salts.
ΔH becomes more negative as the NaClO4 concentration increases to 0.2 M but then becomes less negative at higher NaClO4 concentrations. A similar trend occurs for ΔS. Thus, the α-helix becomes more (less) favored enthalpically (entropically) as the salt concentration increases. ΔG, the difference between ΔH and TΔS, for α-helix formation becomes more negative as the NaClO4 concentration increases.
In contrast, ΔH and ΔS both become more negative as the NaCl concentration increases to 1.0 M, but the values saturate upon concentration increases to 2.0 M.
Na2SO4 also stabilizes the α-helix giving rise to an increasingly negative ΔG. However, ΔH and ΔS only show a modest dependence on the Na2SO4 concentration. They both change together to make ΔG increasingly negative as evident from the Figure 2c melting data for the lower concentrations.
UV Resonance Raman Measurements
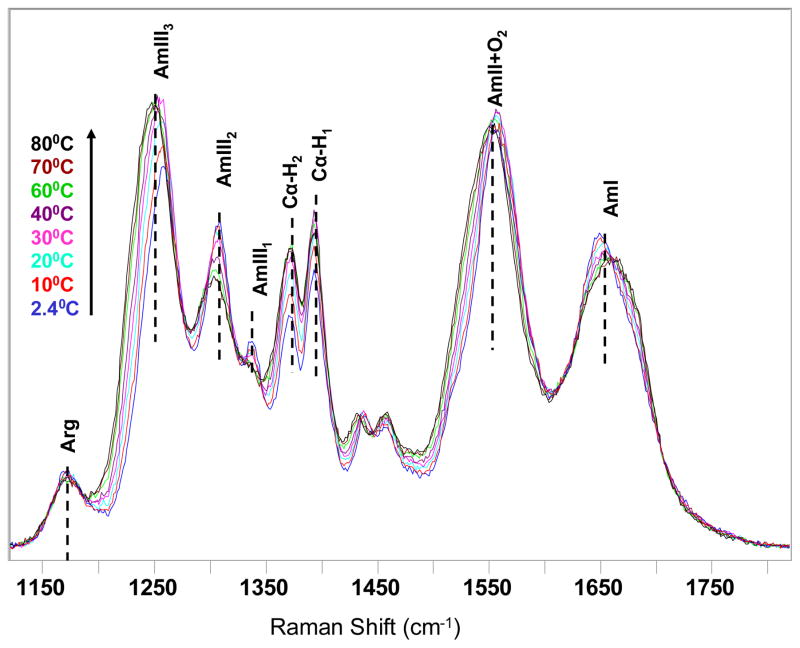
Figure 4 shows the temperature dependence of the AP 204 nm UVRR spectra. The AmI band (~1660 cm−1) results from a mainly C=O stretching vibration. The AmII band (~1550 cm−1) derives from out-of-phase motion of C–N stretching and N–H bending. The Cα–H doublet (~1372 and ~1393 cm−1) derives from a Cα–H bending vibration which is resonance enhanced because of coupling of Cα–H bending to N–H bending. The intensities of the Cα–H bending bands increase as the concentration of nonhelical conformations increases (58). The AmIII bands arise from vibrations which involve in-phase contributions of C–N stretching and N–H bending. The AmIII region contains three subbands: the AmIII1 band (~1336 cm−1), the AmIII2 band (~1306 cm−1), and the AmIII3 band (~1250 cm−1).
Figure 4.
Temperature dependence of 204 nm excited UVRR spectra of AP in pure water.
The AmIII3 band is the most conformationally sensitive because it involves Ψ-angle-dependent coupling between N–H bending and the Cα–H bending motions (59). For example, the AmIII3 band of the α-helix appears at 1258 cm−1 and contains little Cα–H bending. However, it shifts to 1247 cm−1 in the PPII conformation (60) and contains significant Cα–H bending. The α-helixAmIII3 band cross section is roughly half that of the PPII-like conformation because of the α-helix conformation electronic transition hypochromism (61). As the temperature increases, the intensities of the Cα–H bands increase, indicating α-helix melting. Also, the AmIII3 band frequency shifts from ~1258 cm−1 at 2 °C to ~1247 cm−1 at 80 °C.
The UVRR of AP in 0.2 M NaCl, 0.2 M NaClO4, and 0.0667 M Na2SO4 solutions all indicate α-helix melting as the temperature increases (spectra not shown). The spectra of samples without ClO4− or SO42− were normalized to the AmI band integrated intensity, which shows little variation upon peptide conformational changes (62).
To calculate the α-helical fractions, we calculated the temperature-dependent basis spectra of the PPII-like conformation by using the method of Lednev et al. (57). We then digitally smoothed and then subtracted the appropriate amount of the PPII-like conformation basis spectra from the measured and smoothed UVRR of AP.
The relative amount of the PPII conformation subtracted is the maximum amount of the PPII basis spectrum which minimized the Cα–H region intensity, with the constraint that no negative features occur in the difference spectrum. The basis spectral intensities subtracted are directly proportional to the concentration of the PPII conformation at each temperature (57). The resulting difference spectra should result only from non-PPII conformations and appear to be mainly α-helix-like.
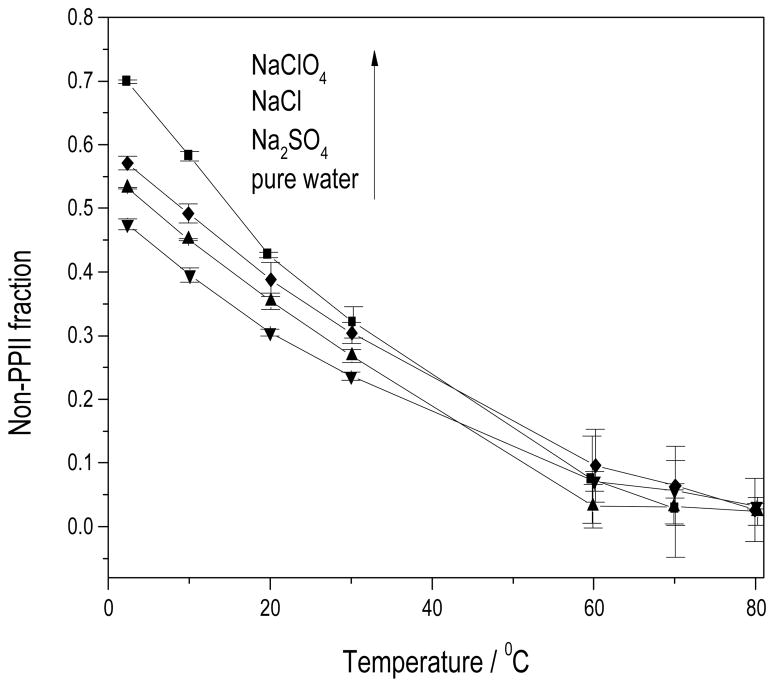
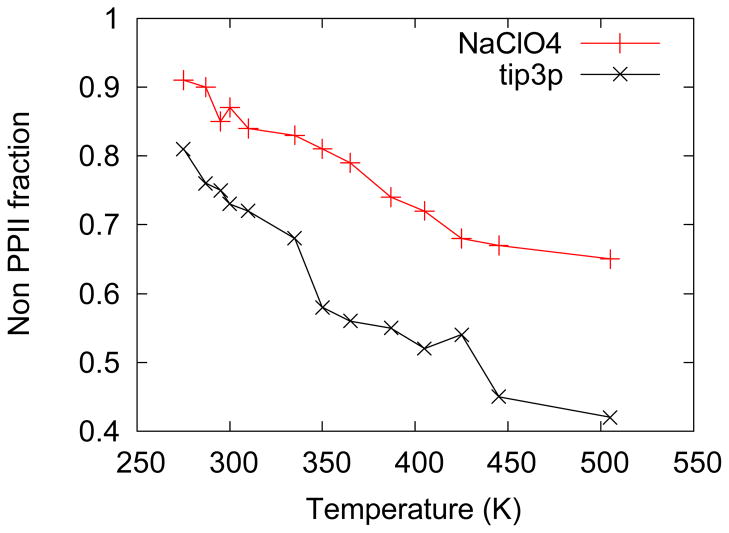
Figure 5, which shows the temperature dependence of the non-PPII fraction, indicates that NaClO4 is the most “helix”-stabilizing salt, followed by NaCl, Na2SO4, and then pure water. These results agree with the CD results above and with the salt ordering previously observed by others (63).
Figure 5.
Non-PPII (primarily α-+310-+π-helix) fractions of AP in different salt solutions: (■) in 0.2MNaClO4; (◆) in 0.2MNaCl; (■) in 0.0667MNa2SO4; (▼) in pure water.
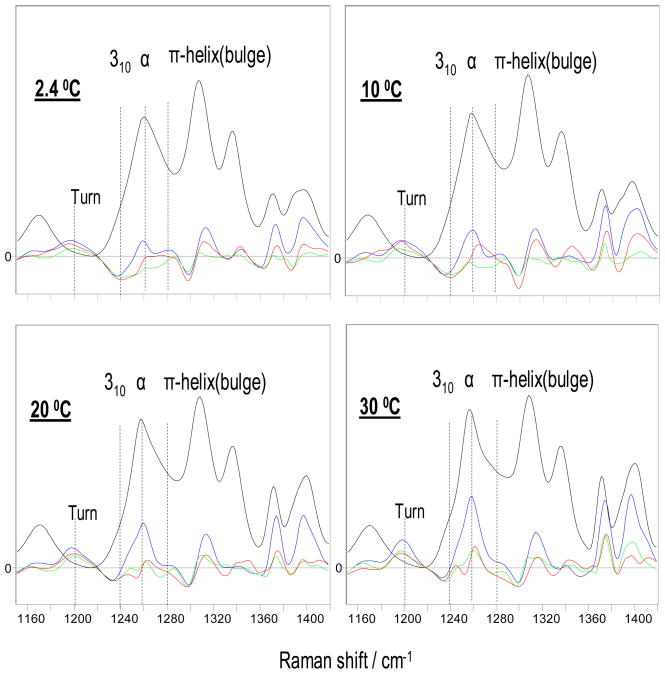
Figure 6 shows the temperature dependence of the calculated non-PPII, α-helix-like spectra of AP in NaClO4, NaCl, and Na2SO4 and the difference spectra between the different salt solution spectra. At all temperatures we observe a triplet of bands which are the hallmark of α-helix-like UVRR spectra. The 30 °C spectrum shows a change in the AmIII3 band shape as earlier noted by Mikhonin and Asher (48). The 30 °C AmIII3 band slightly narrows, while the maximum becomes more sharply peaked. This band shape change, which appeared as a more simple band narrowing in our previous poorer S/N spectra, was ascribed to a decrease in the concentrations of 310-helix and π-bulge conformations relative to the α-helix concentration as the temperature increases.
Figure 6.
Temperature dependence of calculated α-helix-like spectra in 0.2 M NaClO4 and difference spectra between different salt solutions: red, 0.2 M NaCl minus 0.2 M NaClO4; green, 0.0667MNa2SO4 minus 0.2 M NaClO4; blue, pure water minus 0.2 M NaClO4. All displayed calculated α-helix-like spectra were normalized to the intensity of the AmIII1 band of the 2.4 °C α-helix-like spectrum in NaClO4. The difference spectra between salts were calculated from these normalized spectra.
The most prominent difference between the different salt spectra is that an ~1200 cm−1 band that occurs in pure water, in NaCl, and in Na2SO4 disappears in NaClO4 as indicated by the ~1200 cm−1 troughs in the difference spectra. A new band occurs at ~1240 cm−1 in NaClO4 solution, as evident by the trough in the difference spectra between the NaClO4 solution and the other salts and pure water (most clearly at temperatures below 20 °C). Previous work (48, 62) indicated that an AmIII3 band at ~1200 cm−1 derives from turn structures, while the AmIII3 band at ~1240 cm−1 derives from 310-helix conformations. Therefore, we conclude that NaClO4 selectively stabilizes 310-helix conformations which are replaced in pure water, in NaCl, and in Na2SO4 by turn conformations.
We calculated the Gibbs free energy landscapes of AP along the Ψ-folding coordinate from the UVRR (Figures 7 and 8) by using the methodology of Mikhonin et al. (48, 53, 60, 64). We calculate the Ψ angle probability distribution from the AmIII3 band shape and utilize the Boltzmann relationship to calculate the Gibbs free energy landscape.
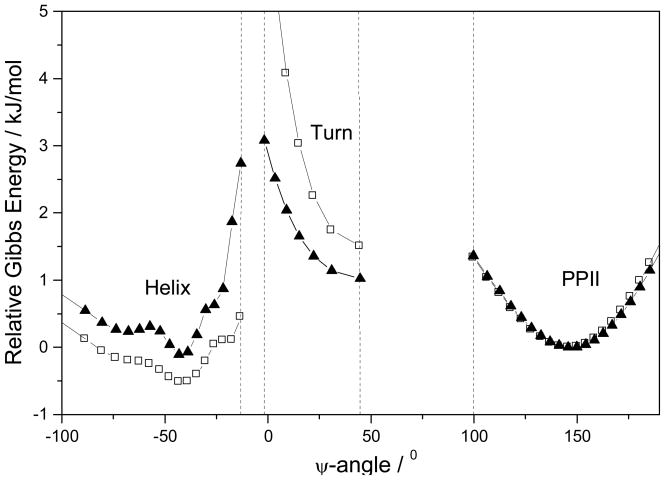
Figure 7.
Calculated 2.4 °C Gibbs free energy landscape of AP in pure water (▲) and in 0.2 M NaClO4 (□). We use the PPII-like conformation as the reference state.
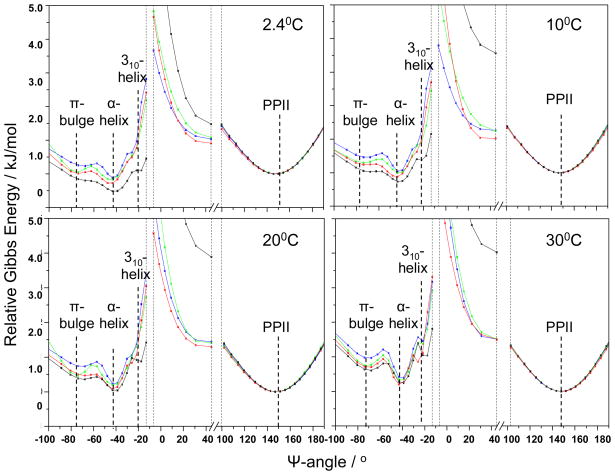
Figure 8.
Calculated Gibbs free energy landscape of AP along the Ramachandran Ψ angle coordinate: in pure water (blue); in 0.0667 M Na2SO4 (green); in 0.2 M NaCl (red); in 0.2 M NaClO4 (black). The PPII-like conformation is the reference state.
The calculated AP Gibbs free energy landscapes in pure water and in 0.2 M NaClO4 at 2.4 °C (Figure 7) show a broad α-helix-like conformational region which includes Ψ angles corresponding to 310-helices and π-bulges and the broad PPII region. The presence of 0.2 M NaClO4 selectively decreases the Gibbs free energies of the 310-helix conformations but increases the free energy of the turn conformations. The α-helix-like region, the turn region, and the PPII region are separated by high activation barriers due to steric clashes. These results are consistent with the calculated Φ and Ψ dependence of the Gibbs free energies of peptide conformations (65). High activation energies are expected between α-helix and turn conformations and PPII conformations. Our ability to monitor turn conformations is important for insight into the mechanisms of α-helix melting since these turn conformations are likely to serve as intermediates along the reaction coordinate that links α-helix-like to the melted PPII-like conformations (66, 67).
The energy landscape (Figure 8) is bumpy within the α-helix-like basin. Within this basin the pure α-helix conformation (Ψ ~ −45°) is always lowest in energy, followed by the π-bulge conformation (Ψ ~ −70°). The 310-helix conformation (Ψ ~ −20°) lies at a slightly higher energy. Both the π-bulge and 310-helix conformations appear to show activation barriers between their minima and that of the α-helix conformation. The relative energy of the π-bulge conformation compared to the α-helix conformation is highest in pure water. As the temperature increases, the α-helix basin Gibbs free energy increases, indicating that the α-helix is destabilized relative to the PPII conformation. We have drawn the energy landscape as a projection onto the Ψ angle coordinate. Traversing the energy landscape from one conformation to another could involve complex dynamics and involve significant dynamics involving Φ angle excursions and excursions in other coordinates.
Figure 8 also shows the dependence of the conformational energies as a function of the salts dissolved in the AP solution at 0.2 M ionic strengths. For all temperatures, the lowest α-helix Gibbs free energies occur in the presence ofNaClO4, followed by NaCl, Na2SO4, and pure water. The 310-helix conformation is selectively stabilized by NaClO4.
Molecular Dynamics Simulation
The NaClO4 α-helix stabilization was qualitatively reproduced by REMD simulations. Figure 9 shows the theoretically calculated AP non-PPII fraction as a function of temperature in 0.2 M NaClO4 solution and in pure water (all conformations with dihedral angles Ψ < +50° are counted as non-PPII conformations). The AP α-helical-like conformation (non-PPII conformations) concentrations in NaClO4 are greater than in pure water at all temperatures.
Figure 9.
Molecular dynamics calculated AP non-PPII fraction in 0.2 M NaClO4 solution and in pure water by REMD simulations. The molecular dynamics predicted AP α-helical conformation melting temperatures are higher than the experimental values because current force fields overstabilize the α-helical conformation (68). Also, REMD simulations often predict much higher melting temperatures than standard MD simulations because the dynamical information is distorted by the REMD simulation temperature exchange process (69).
To investigate the mechanisms governing the helical stabilization, we studied the equilibrium ClO4− distribution around AP. The average ClO4− concentrations around AP were calculated by using Chimera (54), where we calculated the average occupancy of ClO4− within a 3D grid over the trajectory frames. Figure 10 shows the concentration distribution where a green volume indicates a region of increased ClO4− occupancy.
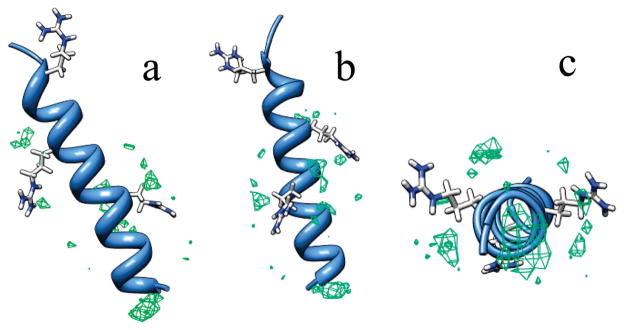
Figure 10.
ClO4− occupancy around AP. The green contour shapes represent a higher than normal probability of finding ClO4− in a volume element near AP. (b) results from rotating (a) 180° about the helix axis; (c) looking down the helix axis from the N-terminus to the C-terminus.
The largest increased ClO4− occupancy occurs around the N-terminus. Panels a and b of Figure 10 also show increased ClO4− occupancies around Arg 9 and Arg 14, with no increased ClO4− occupancy around Arg 19, presumably due to the neutralization of Arg 19 charge by the carboxylate. Figure 10c shows that the ClO4− occupancy increases in the region between the Arg side chain and the peptide backbone.
Panels a and b of Figure 11, which show the radial distribution functions for ClO4−, for the average β-carbon of Ala, the terminal –NH3+, and the three different Arg ζ-carbons, indicate that ClO4− is located on the average 4.5A ° from the Ala β carbon and ~4 A° from the terminal –NH3+ nitrogen, while it is on the average ~5 A° from the Arg ζ carbon. The distribution of ClO4− around the terminal –NH3+ is highest, indicating that the largest association of ClO4− with AP occurs around the terminal –NH3+. Arg 9 has the highest affinity for ClO4−, followed by Arg 14 and then Arg 19.
Figure 11.
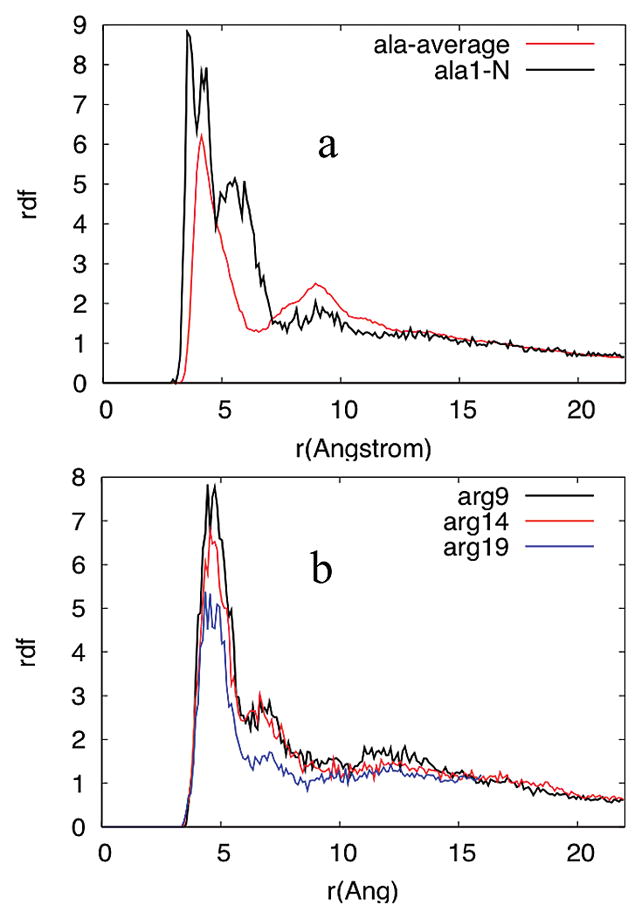
Average radial distribution functions of ClO4− with (a) the average Ala residue and the terminal –NH3+ and (b) the different Args. r is the distance between the Cl inClO4− and the β carbon of Ala or the nitrogen of the terminal –NH3+ or the Arg ζ carbon.
ClO4− binding to the terminal –NH3+ and to Arg is expected from Collion’s matching water affinity model (50–52) where ion pair formation occurs preferentially between oppositely charged ions of similar charge densities. In this model small ions of high charge density tend to preferentially form ion pairs. Large ions of low charge density also preferentially form ion pairs. Ion pairing between oppositely charged ions of high and low charge density is less favorable. Thus, for AP we expect that the weakly hydrated (with low charge density) N-terminal and Arg side chains (50–52) will most strongly bind to the weakly hydrated, low charge density ClO4− which will promote neutralization of the electrostatic interactions within AP which will significantly stabilize the α-helix conformation.
DISCUSSION
Both the CD and UVRR results show that salts stabilize α-helical-like conformations of AP, with efficiencies: NaClO4 > NaCl ~Na2SO4. Numerous previous studies (68, 69) have proposed that these salts affect the protein/peptide stability through three main effects.
The Hofmeister effect (70) phenomenon proposes that salts differentially “salt-in” or “salt-out” proteins/peptides by differentially interacting with water molecules, leaving less water available for protein/peptide hydration. The Hofmeister series orders ions in their decreasing ability to perturb water structure. For example, SO42− preceeds Cl− in the Hofmeister series and thus will more efficiently “salt-out” proteins/peptides. SO42− will preferentially dehydrate the AP backbone and should stabilize α-helix-like conformations (14, 71–73). ClO4− follows Cl− and will “salt-in” proteins/peptides by interacting weakly with water, which leaves more water available for protein/peptide hydration. This predicts that ClO4− should better stabilize melted, water – hydrogen-bonded PPII-like conformations. Cl− should have an intermediate dehydration impact compared to SO42 and ClO4−.
Ionic screening decreases electrostatic interactions between protein and peptide charges (74). Higher ionic strengths increasingly screen electrostatic interactions between charges, as well as between charges and the helix dipole which can impact α-helix stability. For AP at pH7, interactions between the α-helix dipole and the N-terminal positive charge, as well as the anionic carboxylate C-terminal charge, destabilize the α-helical conformation (75–77). In addition, electrostatic repulsions between the three Arg side chains should destabilize the α-helix. Electrostatic screening by high ionic strength solutions will decrease these unfavorable interactions and will thus stabilize α-helical-like conformations.
Specific ion binding between solution ions and peptide and protein side chains can impact the α-helix stability according to the Collins et al. (50–52) model of matching water affinities where ion pair formation is predicted on the basis of preferential formation between oppositely charged ions of similar charge densities. Small ions of high charge density (kosmotropes) tend to preferentially form ion pairs. In contrast, large ions of low charge density (chaotropes) form ion pairs between themselves. Hydrated ion pairing between kosmotropes and chaotropes is less favorable.
Thus, for AP we expect that the weakly hydrated N-terminal and Arg side chains (50–52) most strongly bind to the weakly hydrated chaotrope ClO4−. The resulting charge neutralization will decrease electrostatic interactions within AP which should significantly stabilize the α-helix conformation. Our molecular dynamics simulations directly observe association of ClO4− with the terminalNH3+ group and the Arg. SO42− is expected to show the least ion pairing with NH3+ and Arg, while Cl− should show intermediate ion pairing. These ion pairing propensities predict that NaClO4 should be the most helix stabilizing, followed by NaCl and then Na2SO4.
Table 2 summarizes the expected impact of ions on AP α-helix stability from different effects. Our observations show that at identical ionic strengths α-helical-like conformations are most stabilized by NaClO4, followed by NaCl, then Na2SO4, and then pure water. For NaClO4 to exhibit the most stabilization, the positive impact from the specific ion binding effect must override the negative impact of the Hofmeister effect. The fact that NaCl stabilizes α-helical-like conformations less than does NaClO4 but more than Na2SO4 suggests that the Hofmeister effect (water structure modification) has little impact.
Table 2.
Impact of Ions on AP α-Helix Stabilitya
| solution | ionic screening | Hofmeister effect | specific ion binding |
|---|---|---|---|
| pure water | 0 | 0 | 0 |
| NaCl | + | 0 | + |
| NaClO4 | + | − | + |
| Na2SO4 | + | + | 0 |
(+) helix stabilizing; (−) helix destabilizing; (0) no impact.
The fact that π-helix (bulge) is disfavored in pure water but is stabilized in the presence of these three salts demonstrates that importance of electrostatic screening, where formation of the π-bulge must overcome repulsion between Arg side chains spaced at i, i + 5 positions. In contrast, the Arg side chains are spaced further apart for the α-helix and the 310-helix. We are continuing to study why NaClO4 stabilizes the 310-helix but destabilizes the turn structure.
CONCLUSION
We used CD and UV resonance Raman spectroscopy and molecular dyamcis to study the solution conformation of a mainly polyalanine peptide containing Arg groups for solubility. We calculated the Gibbs free energy landscape along the Ramachandran Ψ angle folding coordinate. We observe that at identical ionic strengths α-helical-like conformations are stabilized most byNaClO4 due to preferential ion binding of ClO4− to the terminal NH3+ and Arg side chains. ClO4− stabilizes 310-helices but destabilizes turn conformations. Cl− has a smaller binding affinity and thus stabilizes α-helical conformations intermediate between NaClO4 and Na2SO4. Electrostatic screening stabilizes π-bulge conformations. We find that we can understand ion association to the peptide through the Collins “laws of matching water affinities model”.
Supplementary Material
Acknowledgments
We thank Aleksandr V. Mikhonin, Zeeshan Ahmed, Bhavya Sharma, Lu Ma, and Zhenmin Hong for useful discussions.
Footnotes
This work was supported by NIH Grant GM8RO1EB002053.
SUPPORTING INFORMATION AVAILABLE
Molecular dynamics simulation details. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivarsson Y, Travaglini-Allocatelli C, Brunori M, Gianni S. Mechanisms of protein folding. Eur Biophys J Biophys Lett. 2008;37(6):721–728. doi: 10.1007/s00249-007-0256-x. [DOI] [PubMed] [Google Scholar]
- 3.Dobson CM, Sali A, Karplus M. Protein folding: A perspective from theory and experiment. Angew Chem, Int Ed. 1998;37(7):868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin RL, Rose GD. Is protein folding hierarchic? II. Folding intermediates and transition states. Trends Biochem Sci. 1999;24(2):77–83. doi: 10.1016/s0968-0004(98)01345-0. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin RL, Rose GD. Is protein folding hierarchic? I. Local structure and peptide folding. Trends Biochem Sci. 1999;24(1):26–33. doi: 10.1016/s0968-0004(98)01346-2. [DOI] [PubMed] [Google Scholar]
- 6.Jahn TR, Radford SE. The Yin and Yang of protein folding. FEBS J. 2005;272(23):5962–5970. doi: 10.1111/j.1742-4658.2005.05021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CL Are there pathways for protein folding? J Chem Phys. 1968;65:44–45. [Google Scholar]
- 8.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4(1):10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 9.Dill KA. Polymer principles and protein folding. Protein Sci. 1999;8(6):1166–1180. doi: 10.1110/ps.8.6.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mello CC, Barrick D. An experimentally determined protein folding energy landscape. Proc Natl Acad Sci USA. 2004;101(39):14102–14107. doi: 10.1073/pnas.0403386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy Y, Jortner J, Becker OM. Solvent effects on the energy landscapes and folding kinetics of polyalanine. Proc Natl Acad Sci USA. 2001;98(5):2188–2193. doi: 10.1073/pnas.041611998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ihalainen J, Bredenbeck J, Pfister R, Helbing J, Chi L, van Stokkum I, Woolley G, Hamm P. Folding and unfolding of a photoswitchable peptide from picoseconds to microseconds. Proc Natl Acad Sci USA. 2007;104(13):5383–5388. doi: 10.1073/pnas.0607748104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balakrishnan G, Hu Y, Bender G, Getahun Z, DeGrado W, Spiro T. Enthalpic and entropic stages in alpha-helical peptide unfolding, from laser T-Jump/UV Raman spectroscopy. J Am Chem Soc. 2007;129(42):12801–12808. doi: 10.1021/ja073366l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Starzyk A, Barber-Armstrong W, Sridharan M, Decatur S. Spectroscopic evidence for backbone desolvation of helical peptides by 2,2,2-trifluoroethanol: An isotope-edited FTIR study. Biochemistry. 2005;44(1):369–376. doi: 10.1021/bi0481444. [DOI] [PubMed] [Google Scholar]
- 15.Wang T, Zhu Y, Getahun Z, Du D, Huang C, DeGrado W, Gai F. Length dependent helix-coil transition kinetics of nine alanine-based peptides. J Phys Chem B. 2004;108(39):15301–15310. doi: 10.1021/jp037272j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C, Getahun Z, Zhu Y, Klemke J, DeGrado W, Gai F. Helix formation via conformation diffusion search. Proc Natl Acad Sci USA. 2002;99(5):2788–2793. doi: 10.1073/pnas.052700099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Klemke J, Getahun Z, DeGrado W, Gai F. Temperature-dependent helix-coil transition of an alanine based peptide. J Am Chem Soc. 2001;123(38):9235–9238. doi: 10.1021/ja0158814. [DOI] [PubMed] [Google Scholar]
- 18.Thompson P, Munoz V, Jas G, Henry E, Eaton W, Hofrichter J. The helix-coil kinetics of a heteropeptide. J Phys Chem B. 2000;104(2):378–389. [Google Scholar]
- 19.Yoder G, Pancoska P, Keiderling T. Characterization of alanine-rich peptides, Ac-(AAKAA)(n)-GY-NH2 (n=1–4), using vibrational circular dichroism and Fourier transform infrared. Conformational determination and thermal unfolding. Biochemistry. 1997;36(49):15123–15133. doi: 10.1021/bi971460g. [DOI] [PubMed] [Google Scholar]
- 20.Thompson P, Eaton W, Hofrichter J. Laser temperature jump study of the helix reversible arrow coil kinetics of an alanine peptide interpreted with a “kinetic zipper” model. Biochemistry. 1997;36(30):9200–9210. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 21.Thompson P, Eaton A, Hofrichter J. Laser temperature jump studies of the helix-coil transition of an alanine-based peptide. Biophys J. 1997;722:WP377–WP377. [Google Scholar]
- 22.Millhauser G, Stenland C, Hanson P, Bolin K, vandeVen F. Estimating the relative populations of 3(10)-helix and alpha-helix in Ala-rich peptides: A hydrogen exchange and high field NMR study. J Mol Biol. 1997;267(4):963–974. doi: 10.1006/jmbi.1997.0923. [DOI] [PubMed] [Google Scholar]
- 23.Williams S, Causgrove T, Gilmanshin R, Fang K, Callender R, Woodruff W, Dyer R. Fast events in protein folding: Helix melting and formation in a small peptide. Biochemistry. 1996;35(3):691–697. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 24.Shalongo W, Dugad L, Stellwagen E. Distribution of helicity within the model peptide acetyl(AAQAA)3amide. J Am Chem Soc. 1994;116(18):8288–8293. [Google Scholar]
- 25.Fiori W, Miick S, Millhauser G. Increasing sequence length favors alpha-helix over 3(10)-helix in alanine-based peptides-evidence for a length-dependent structural transition. Biochemistry. 1993;32(45):11957–11962. doi: 10.1021/bi00096a003. [DOI] [PubMed] [Google Scholar]
- 26.Scholtz JM, York EJ, Stewart JM, Baldwin RL. A neutral, water-soluble, alpha-helical peptide: The effect of ionic strength on helix-coil equilibrium. J Am Chem Soc. 1991;113:5102–5104. [Google Scholar]
- 27.Marqusee S, Robbins VH, Baldwin RL. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramajo AP, Petty SA, Volk M. Fast folding dynamics of alpha-helical peptides—Effect of solvent additives and pH. Chem Phys. 2006;323(1):11–20. [Google Scholar]
- 29.Silva RAGD, Kubelka J, Bour P, Decatur SM, Keiderling TA. Site-specific conformational determination in thermal unfolding studies of helical peptides using vibrational circular dichroism with isotopic substitution. Proc Natl Acad Sci USA. 2000;97(15):8318–8323. doi: 10.1073/pnas.140161997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long HW, Tycko R. Biopolymer conformational distributions from solid-state NMR: alpha-helix and 3(10)-helix contents of a helical peptide. J Am Chem Soc. 1998;120(28):7039–7048. [Google Scholar]
- 31.Wang W, Lin T, Sun Y. Examination of the folding of a short alanine-based helical peptide with salt bridges using molecular dynamics simulation. J Phys Chem B. 2007;111(13):3508–3514. doi: 10.1021/jp067637a. [DOI] [PubMed] [Google Scholar]
- 32.Sorin E, Rhee Y, Shirts M, Pande V. The solvation interface is a determining factor in peptide conformational preferences. J Mol Biol. 2006;356(1):248–256. doi: 10.1016/j.jmb.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 33.Morozov A, Lin S. Modeling of folding and unfolding mechanisms in alanine-based alpha-helical polypeptides. J Phys Chem B. 2006;110(41):20555–20561. doi: 10.1021/jp061781e. [DOI] [PubMed] [Google Scholar]
- 34.Sorin E, Pande V. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J. 2005;88(4):2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jas G, Kuczera K. Equilibrium structure and folding of a helix-forming peptide: Circular dichroism measurements and replica-exchange molecular dynamics simulations. Biophys J. 2004;87(6):3786–3798. doi: 10.1529/biophysj.104.045419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gnanakaran S, Hochstrasser R, Garcia A. Nature of structural inhomogeneities on folding a helix and their influence on spectral measurements. Proc Natl Acad Sci USA. 2004;101(25):9229–9234. doi: 10.1073/pnas.0402933101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gnanakaran S, Hochstrasser R, Garcia A. Nature of structural inhomogeneities during folding of a helix: Manifestation in IR. Abstr Paper Am Chem Soc. 2003;226:U335–U335. [Google Scholar]
- 38.Ghosh T, Garde S, Garcia A. Role of backbone hydration and salt-bridge formation in stability of alpha-helix in solution. Biophys J. 2003;85(5):3187–3193. doi: 10.1016/S0006-3495(03)74736-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armen R, Alonso D, Daggett V. The role of alpha-, 3(10)-, and pi-helix in helix → coil transitions. Protein Sci. 2003;12(6):1145–1157. doi: 10.1110/ps.0240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vila J, Ripoll D, Scheraga H. Influence of lysine content and pH on the stability of alanine-based copolypeptides. Biopolymers. 2001;58(3):235–246. doi: 10.1002/1097-0282(200103)58:3<235::AID-BIP1001>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 41.Shirley W, Brooks C. Curious structure in “canonical” alanine based peptides. Proteins: Struct, Funct, Genet. 1997;28(1):59–71. doi: 10.1002/(sici)1097-0134(199705)28:1<59::aid-prot6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Lei HX, Chowdhury S, Duan Y. Fs-21 peptides can form both single helix and helix-turn-helix. J Phys Chem B. 2004;108(22):7479–7489. [Google Scholar]
- 43.Asher S, Mikhonin A, Bykov S. UV Raman demonstrates that alpha-helical polyalanine peptides melt to polyproline II conformations. J Am Chem Soc. 2004;126(27):8433–8440. doi: 10.1021/ja049518j. [DOI] [PubMed] [Google Scholar]
- 44.Kentsis A, Mezei M, Gindin T, Osman R. Unfolded state of polyalanine is a segmented polyproline II helix. Proteins: Struct, Funct, Bioinf. 2004;55(3):493–501. doi: 10.1002/prot.20051. [DOI] [PubMed] [Google Scholar]
- 45.Mezei M, Fleming P, Srinivasan R, Rose G. Polyproline II helix is the preferred conformation for unfolded polyalanine in water. Proteins: Struct, Funct, Genet. 2004;55(3):502–507. doi: 10.1002/prot.20050. [DOI] [PubMed] [Google Scholar]
- 46.Pappu R, Rose G. A simple model for polyproline II structure in unfolded states of alanine-based peptides. Protein Sci. 2002;11(10):2437–2455. doi: 10.1110/ps.0217402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi Z, Olson C, Rose G, Baldwin R, Kallenbach N. Polyproline II structure in a sequence of seven alanine residues. Proc Natl Acad Sci USA. 2002;99(14):9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikhonin AV, Asher SA. Direct UV Raman monitoring of 3(10)-helix and pi-bulge premelting during alpha-helix unfolding. J Am Chem Soc. 2006;128(42):13789–13795. doi: 10.1021/ja062269+. [DOI] [PubMed] [Google Scholar]
- 49.Mikhonin A, Asher S, Bykov S, Murza A. UV Raman spatially resolved melting dynamics of isotopically labeled polyalanyl peptide: Slow alpha-helix melting follows 3(10)-helices and pi-bulges premelting. J Phys Chem B. 2007;111(12):3280–3292. doi: 10.1021/jp0654009. [DOI] [PubMed] [Google Scholar]
- 50.Collins KD. Charge density-dependent strength of hydration and biological structure. Biophys J. 1997;72(1):65–76. doi: 10.1016/S0006-3495(97)78647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins KD. Ions from the Hofmeister series and osmolytes: Effects on proteins in solution and in the crystallization process. Methods. 2004;34(3):300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 52.Collins KD, Neilson GW, Enderby JE. Ions in water: Characterizing the forces that control chemical processes and biological structure. Biophys Chem. 2007;128(2–3):95–104. doi: 10.1016/j.bpc.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 53.Bykov S, Lednev I, Ianoul A, Mikhonin A, Munro C, Asher SA. Steady-state and transient ultraviolet resonance Raman spectrometer for the 193–270 nm spectral region. Appl Spectrosc. 2005;59(12):1541–1552. doi: 10.1366/000370205775142511. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Manning MC, Woody RW. Theoretical CD studies of polypeptide helices: Examination of important electronic and geometric factors. Biopolymers. 1991;31:569–586. doi: 10.1002/bip.360310511. [DOI] [PubMed] [Google Scholar]
- 56.Asher SA, Mikhonin AV, Bykov S. UV Raman demonstrates that alpha-helical polyalanine peptides melt to polyproline II conformations. J Am Chem Soc. 2004;126(27):8433–8440. doi: 10.1021/ja049518j. [DOI] [PubMed] [Google Scholar]
- 57.Lednev I, Karnoup A, Sparrow M, Asher S. alpha-helix peptide folding and unfolding activation barriers: A nanosecond UV resonance Raman study. J Am Chem Soc. 1999;121(35):8074–8086. [Google Scholar]
- 58.Wang Y, Purrello R, jordan T, Spiro TG. UVRR spectroscopy of the peptide bond. 1 Amide S, a nonhelical structure marker, is a CαH bending mode. J Am Chem Soc. 1991;113:6359–6368. [Google Scholar]
- 59.Asher SA, Ianoul A, Mix G, Boyden MN, Karnoup A, Diem M, Schweitzer-Stenner R. Dihedral psi angle dependence of the amide III vibration: A uniquely sensitive UV resonance Raman secondary structural probe. J Am Chem Soc. 2001;123(47):11775–11781. doi: 10.1021/ja0039738. [DOI] [PubMed] [Google Scholar]
- 60.Mikhonin AV, Bykov SV, Myshakina NS, Asher SA. Peptide secondary structure folding reaction coordinate: Correlation between UV Raman amide III frequency, Psi Ramachandran angle, and hydrogen bonding. J Phys Chem B. 2006;110(4):1928–1943. doi: 10.1021/jp054593h. [DOI] [PubMed] [Google Scholar]
- 61.Sharma B, Bykov SV, Asher SA. UV resonance Raman investigation of electronic transitions in alpha-helical and polyproline II-like conformations. J Phys Chem B. 2008;112(37):11762–11769. doi: 10.1021/jp801110q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma L, Ahmed Z, Mikhonin AV, Asher SA. UV resonance raman measurements of poly-L-lysine’s conformational energy landscapes: Dependence on perchlorate concentration and temperature. J Phys Chem B. 2007;111(26):7675–7680. doi: 10.1021/jp0703758. [DOI] [PubMed] [Google Scholar]
- 63.Damodaran S. Influence of protein conformation on its adaptability under chaotropic conditions. Int J Biol Macromol. 1988:11. doi: 10.1016/0141-8130(89)90030-5. [DOI] [PubMed] [Google Scholar]
- 64.Ma L, Ahmed Z, Mikhonin A, Asher S. UV resonance raman measurements of poly-L-lysine’s conformational energy landscapes: Dependence on perchlorate concentration and temperature. J Phys Chem B. 2007;111(26):7675–7680. doi: 10.1021/jp0703758. [DOI] [PubMed] [Google Scholar]
- 65.Ramachandran GN, Sasisekharan V. Conformation of Polypeptides and Proteins. Vol. 23. Academic Press; New York: 1968. pp. 283–437. [DOI] [PubMed] [Google Scholar]
- 66.Sundarlingam M, Sekharudu YC. Water-inserted alpha-helical segments implicate reverse turns as folding intermediates. Science. 1989;244:1333–1337. doi: 10.1126/science.2734612. [DOI] [PubMed] [Google Scholar]
- 67.Millhauser GL. Views of helical peptides—A proposal for the position of 3(10)-helix along the thermodynamic folding pathway. Biochemistry. 1995;34(32):10318–10318. doi: 10.1021/bi00012a001. [DOI] [PubMed] [Google Scholar]
- 68.Beck DAC, White GWN, Daggett V. Exploring the energy landscape of protein folding using replica-exchange and conventional molecular dynamics simulations. J Struct Biol. 2007;157(3):514–523. doi: 10.1016/j.jsb.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins: Struct, Funct, Bioinf. 2006;65(3):712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goto Y, Takahashi N, Fink AL. Mechanism of acid-induced folding of proteins. Biochemistry. 1990;29:3480–3488. doi: 10.1021/bi00466a009. [DOI] [PubMed] [Google Scholar]
- 71.Thomas AS, Elcock AH. Molecular dynamics simulations of hydrophobic associations in aqueous salt solutions indicate a connection between water hydrogen bonding and the Hofmeister effect. J Am Chem Soc. 2007;129(48):14887–14898. doi: 10.1021/ja073097z. [DOI] [PubMed] [Google Scholar]
- 72.Collins KD, Washabaugh MW. The Hofemister effect and the behavior of water at interfaces. Q Rev Biophys. 1985;18:323–422. doi: 10.1017/s0033583500005369. [DOI] [PubMed] [Google Scholar]
- 73.Vila JA, Ripoll DR, Scheraga HA. Physical reasons for the unusual alpha-helix stabilization afforded by charged or neutral polar residues in alanine-rich peptides. Proc Natl Acad Sci USA. 2000;97:13075–13079. doi: 10.1073/pnas.240455797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garcia AE, Sanbonmatsu KY. alpha-helical stabilization by side chain shielding of backbone hydrogen bonds. Proc Natl Acad Sci USA. 2002;99:2782–2787. doi: 10.1073/pnas.042496899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mukherjee S, Chowdhury P, Gai F. Tuning the cooperativity of the helix-coil transition by aqueous reverse michelles. J Phys Chem B. 2006;110:11615–11619. doi: 10.1021/jp062362k. [DOI] [PubMed] [Google Scholar]
- 76.Debye P, Huckel E. Phys Z. 1923;24:185–206. [Google Scholar]
- 77.Hol WGJ. The role of the alpha-helix dipole in protein fuction and structure. Prog Biophys Mol Biol. 1985;45:149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 78.Hol WGJ, Duijnen PTv, Berendsen HJC. The alpha-helix dipole and the properties of proteins. Nature. 1978;273:443–446. doi: 10.1038/273443a0. [DOI] [PubMed] [Google Scholar]
- 79.Shoemaker KR, Kim PS, York EJ, Stewart JM, Baldwin RL. Tests of the helix dipole model for stabilization of alpha-helices. Nature. 1987;326:573–567. doi: 10.1038/326563a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.