Abstract
Objectives/Hypothesis
Although many proinflammatory cytokines have been identified in nasal polyp tissue, the initial trigger that causes this inflammation characterized by edema, lymphocytosis, and eosinophilia, is still unknown. The purpose of the present study is to identify the presence of genetic polymorphisms in proinflammatory, anti-inflammatory, and chemokine genes that might contribute to genetic susceptibility to chronic hyperplastic sinusitis with nasal polyposis (CHSwNP).
Study Design
Case control study.
Methods
Buccal swabs were taken from the left and right oral mucosal surfaces from 179 patients with CHSwNP and 153 nonpolyposis controls with the Purgene DNA purification protocol (Gentra). Genotyping assays for cytokine gene loci were performed on 14 cytokine genes using the iPlex Gold and the Mass Array Compact system (Sequenom, San Diego, CA). Tests of Hardy-Weinberg equilibrium proportions were performed separately in the cases and controls. Tests for evidence of association between alleles at each single-nucleotide polymorphism (SNP) and case-control status were performed using unconditional logistic regression.
Results
The frequency of the A allele in a SNP located in tumor necrosis factor (TNF)-α (rs1800629) is significantly different in patients with nasal polyposis versus controls without nasal polyposis, 18.6% and 11.5%, respectively with an individuals’ odds of susceptibility to nasal polyps increasing almost twofold (odds ratio, 1.86; confidence interval, 1.4–3.09) given at least one copy of the A allele at this SNP. All other cytokine gene polymorphisms of both inflammatory, anti-inflammatory, and chemokine genes were not statistically different between the two groups.
Conclusions
TNF-α-308, a SNP in the promoter region of this cytokine gene is associated with increased odds of developing nasal polyposis. TNF-α is a potent immuno-mediator and proinflammatory cytokine that has been implicated in the pathogenesis of a large number of human diseases. The location of this gene on the short arm of chromosome 6, with the major histocompatibility complex genes and complement, has raised the probability that polymorphism within this locus may contribute to a genetic association of this region of the genome with a wide variety of infectious and autoimmune diseases.
Keywords: Nasal polyps, genes, single-nucleotide polymorphism, tumor necrosis factor-α-308, chronic inflammation
INTRODUCTION
A growing body of evidence is mounting to suggest that much of the host response to infectious disease is a direct reflection of heritable traits, accounting for interpersonal differences, and allowing for genetic detection.1 The role of the innate and acquired immune response in chronic inflammatory diseases is regulated and sustained by the activation of an acquired immune response to antigen-presenting cells, such as macrophages and dendritic cells with the subsequent release of inflammatory mediators.2 The inflammatory response is sustained by the overproduction of inflammatory mediators of T-lymphocytes and macrophage-derived cytokines.
When these mediators are produced in excess, or when their counteracting receptor antagonists or antagonistic-binding proteins are deficient, the protective role of the immune system devolves into a harmful one capable of tissue injury in the host.3
It is now clear that chronic hyperplastic sinusitis with nasal polyposis (CHSwNP) is a chronic inflammatory disease, in which proinflammatory cytokines play a major role.4 In an effort to understand the mechanisms of this disorder and to develop new therapeutic strategies, specific biochemical mediators and cytokines involved in CHSwNP, as well as their genetic control have been targeted.5
The genetic predisposition of patients who develop CHSwNP following exposure to bacteria, fungus, allergy, virus, or traumatic insult has only begun to be explored experimentally. If a relationship between the genetic constitution of a patient as a response to a major inflammatory stimulus could be established, better injury-scoring systems could be devised that more accurately predict the clinical trajectory of a patient. Whether or not the triggering events are microbial, fungal, or allergic products, the key initiators of the inflammatory responses are cytokines.
Areas of variability, called polymorphisms within a gene, are stable. Strictly defined, a polymorphism is the existence of two or more sequence variances occurring at significant frequencies within a population. The most common example is a single-nucleotide polymorphism (SNP). SNPs are single nucleotides that are substituted for other nucleotides within a sequence of the allele.
Certain polymorphisms, in either the promoter or core region of a gene, may be associated with such chronic inflammatory diseases as periodontitis, inflammatory bowel disease, cancer, septic shock, otitis media, and other inflammatory diseases.6–10 At the present time, several investigations have reported an association between nasal polyposis and SNPs of proinflammatory cytokine genes.11–15 These reported investigations have looked at susceptibility to nasal polyposis measuring one or two single polymorphisms.
We sought to investigate the role of seven proinflammatory, four anti-inflammatory, one Toll receptor, and two chemokine polymorphisms in patients with massive nasal polyposis previously shown to be associated with chronic inflammation.
MATERIALS AND METHODS
Study Population
Buccal swabs were obtained from 332 participants (179 patients with CHSwNP and 153 control patients). The control patients did not have a history of rhinosinusitis, chronic periodontal disease, inflammatory bowel disease, cancer, sepsis, or otitis media or any other chronic inflammatory disorder. Sinus cases were selected on the basis of a diagnosis of CHSwNP. The study was approved by the institutional review board of the State University of New York at Buffalo, and all patients consented at the time the buccal sample was taken. Demographic summary statistics for the total population, the cases only, and the controls only were calculated. The swabs were inserted into normal saline and frozen until SNP genotyping.
Selection of Patients
The subject patients all had chronic hyperplastic sinusitis with nasal polyposis. On clinical examination, all patients had visible nasal polyps with the unaided eye. Ninety-eight percent of these patients had bilateral disease, and 2% had unilateral disease. Using the scoring system of Gaskins16 for the extent of polyposis, all patients were in the classification of two or three, indicating that the nasal polyps were well below the middle turbinate and some almost extending toward the external nares. In regard to radiological score, the patients had a mean score of greater than 12 (Lund-MacKay),17 indicating bilateral opacification of the anterior and posterior ethmoids, and bilateral opacification of maxillary sinuses and obliteration of the infundibulum bilaterally.
The diagnosis of allergy was made by intradermal dilutional testing.
The presence of acetylsalicylic acid (ASA) intolerance was made primarily by history, in which the patients were told by their allergists or pulmonologists that they were not to take any aspirin product for fear of the development of severe asthma. Therefore, the majority of patients gave a history of aspirin intolerance and had a combination of asthma and chronic hyperplastic sinusitis with nasal polyposis.
SNP Selection and Genotyping
SNP genotyping of 14 markers that have been documented in the literature to play a role in chronic inflammation was performed using iPLEX Gold and the MassARRAY Compact system (Sequenom, San Diego, CA).18 Polymerase chain reaction (PCR) primers are summarized in Table I.
TABLE I.
Genotyping Assays for Cytokine Gene Loci.
| Gene | Locus | SNP ID | PCR Forward Primer (5′-3′) | PCR Reverse Primer (5′-3′) | Mass Extend Primer |
|---|---|---|---|---|---|
| TNFα | −308 | rs1800629 | ACGTTGGATGGATTT | ACGTTGGATGGGTCC | AGGTTTTGA |
| GTGTGTAGGACCCTG | CCAAAAGAAATGGAG | GGGGCATG | |||
| IL-1α | −889 | rs3783521 | ACGTTGGATGAGGAG | ACGTTGGATGCAACT | GCTACATTTCT |
| GAGAGGGTTAATCAG | GGCTACATTTCTGC | GCTTACCTTG | |||
| IL-1α | +4845 | rs1756 | ACGTTGGATGATCTG | ACGTTGGATGCACAT | TTGCTCAGGAA |
| CACTTGTGATCATGG | TGCTCAGGAAGCTAA | GCTAAAAGGTG | |||
| IL-1β | −511 | rs3087258 | ACGTTGGATGTCTGT | ACGTTGGATGCAGAG | TCCTGCAATT |
| ATTGAGGGTGTGGGT | GCTCCTGCAATTGAC | GACAGAGAG | |||
| IL-1 β | +3935 | rs1143634 | ACGTTGGATGAGTGA | ACGTTGGATGGTGCT | CACATTTCAGAA |
| TCGTAGAGGTGCATC | CCACATTTCAGAACC | CCTATCTTCTT | |||
| IL-6 | −174 | rs13447445 | ACGTTGGATGACCCT | ACGTTGGATGGGGTTG | TGAGACTCTAAT |
| CACCCTCCAACAAAG | AGACTCTCTAATATTG | ATTGAGACTCAT | |||
| IFNγ | +874 | rs2430561 | ACGTTGGATGGATAG | ACGTTGGATGTGATT | TTTTATTCTTACAA |
| TTCCAAACATGTGCG | TTATTCTTACAACAC | CACAAAATCAAATC | |||
| TGF β1 | −509 | rs11466315 | ACGTTGGATGTTCAA | ACGTTGGATGGGGG | GCTGTCTG |
| AACCCCCTGCCGAC | CCCTCGCTGTCTGG | GCTGCTC | |||
| IL-10 | −592 | rs1800895 | ACGTTGGATGACTTC | ACGTTGGATGCCCCG | TCATTAGGAT |
| CCCCAAGCACAGTTG | ATTTCATTAGGATTC | TCTCAGGCA | |||
| IL-10 | −819 | rs1800894 | ACGTTGGATGCTGGA | ACGTTGGATGTCACC | AAGGGTAC |
| GATGGTGTACAGTAG | TGTACAAGGGTACAC | ACCAGTGC | |||
| IL-10 | −1082 | rs1800896 | ACGTTGGATGTGGAT | ACGTTGGATGGACAA | AATACTAAGGC |
| AGGAGGTCCCTTACT | CACTACTAAGGCTTC | TTCTTTGGGA | |||
| CD14 | −159 | rs2569190 | ACGTTGGATGGCAAT | ACGTTGGATGAGACA | TGCAGAATCCT |
| GAAGGATGTTTCAGG | CAGAACCCTAGATGC | TCCTGTTACGG | |||
| CCL5 (RANTES) | −403 | rs2107538 | ACGTTGGATGAACAT | ACGTTGGATGCACTG | TCCTGCTTATTC |
| CCTTCCATGGATGAG | AGTCTTCAAAGTTCC | ATTACAGATCTTA | |||
| CCL2 (MCP1) | −2518 | rs3917882 | ACGTTGGATGTCTTG | ACGTTGGATGAAACAC | GGTATGAATCAG |
| ACAGAGCAGAAGTGG | AGGGAAGGTGAAGG | AAAAGAAAGTCTT |
SNP = single-nucleotide polymorphism; PCR = polymerase chain reaction; TNF = tumor necrosis factor; rs = reference SNP; IL = interleukin; IFN = interferon; TGF = transforming growth factor; RANTES = regulated on activation, normal T cell expressed and secreted; CCL = cysteinyl chemokine ligand; MCP = monocyte chemotactic protein.
Genotyping assays were designed using RealSNP and MassARRAY Designer (Sequenom).
Forward and reverse printers were used to PCR amplify the gene region containing the specific SNP followed by a single-base extension across the SNP using the mass extend primer.
The SNP500 Cancer Resource Panel obtained from the Coriell Cell Repositories (Camden, NJ) was used as the quality control DNA source (n = 95) (http://ccr.coriell.org/Sections/Search/Panel_Detail.aspx?Ref=SNP500V?&Pgld=202).
DNA was extracted from buccal cells of 179 polyposis patients and 153 controls with the Puregene DNA purification protocol (Gentra Systems, Inc., Minneapolis, MN). All SNP specific and mass extended oligonucleotides were designed using RealSNP and MassARRAY Assay Designer (Sequenom).18 Five nanograms of DNA were added to each 5-µL PCR reaction mixture in 384-W plates. PCR primers were assigned into appropriate multiplex groups by RealSNP and used at a 200 ng final concentration. Genotype confidence scores of conservative (A), moderate (B), and aggressive (C) were used for analysis and evaluated for quality. Genotype confidence scores below C were not included in the analysis.
Statistical Analyses
Using Fisher exact test, each SNP was tested to determine if the population under investigation showed deviation from genotype frequencies expected under Hardy-Weinberg equilibrium (HWE) proportions.19 Tests were performed separately in the cases and controls. Tests for evidence of association between alleles at each SNP and case-control status were performed using unconditional logistic regression.20,21 Specifically, we tested whether carriers of each genotyped SNP have different risk than noncarriers (0 copies) by testing for independence in 2 × 3 tables using standard odds ratio analysis using a test for trend. Covariates age and sex (all patients were self-identified Caucasians and thus race specific analyses were not performed) were only included in the final model if they were found to increase the odds of disease, increase/decrease the effect of the allele, or other covariates by at least 10%.
RESULTS
Study Participants
Demographic characteristics of the population are presented in Table II. The 179 cases (110 males and 69 females) ranged in age from 18 years to 87 years, with a mean (standard deviation [SD]) age of 55.4 (15.6) years. The 153 controls (49 males and 104 females) ranged in age from 12 years to 86 years, with a mean (SD) age of 46.5 (14.04) years. The median age of the cases and controls was 54 and 50, respectively.
TABLE II.
Demographics of Polyposis Study.
| Variable | Overall (N = 332) |
Polyposis (n = 179) |
Control (n = 153) |
|---|---|---|---|
| Sex | |||
| Female | 171 (51.4%) | 69 (36.8%) | 102 (68.0%) |
| Male | 161 (48.6%) | 110 (63.2%) | 51 (32.0%) |
| Allergies | |||
| No | 178 (53.5%) | 53 (29.6%) | 125 (81.7%) |
| Yes | 154 (46.5%) | 126 (70.4%) | 28 (18.3%) |
| Asthma | |||
| No | 259 (78.5%) | 109 (61.6%) | 150 (98.0%) |
| Yes | 73 (21.5%) | 70 (38.4%) | 3 (2.0%) |
| ASA intolerance | |||
| No | 299 (90.0%) | 148 (83.6%) | 151 (98.7%) |
| Yes | 33 (10.0%) | 31 (16.4%) | 2 (1.3%) |
ASA = acetylsalicylic acid.
Genetic Analyses
The genotyping call rate for each SNP ranged from 83% to 96%, with an average call rate of 93%. Eleven of the 14 SNPs genotyped were in HWE proportions in the control population. Two SNPs, rs3087258 (IL-1β-511) and rs1800894 (IL-10-819), showed significant (P < .01) deviation from HWE proportions; one SNP, rs1344735 (IL-6) was monomorphic (e.g., one only copy of the allele was present). These three SNPs were eliminated from the analysis.
The frequency of the A allele in a SNP located in tumor necrosis factor (TNF)-α (rs1800629) is significantly different in patients with nasal polyposis versus controls without nasal polyposis, 18.6% and 11.5%, respectively, with an individuals’ odds of susceptibility to nasal polyps increasing almost two-fold (odds ratio, 1.86; confidence interval, 1.14–3.09) given at least one copy of the A allele at this SNP (Table III). We did not find statistically significant results for any other SNPs evaluated. The multiple SNPs tested on chromosome 1 or chromosome 2 were not in pairwise linkage disequilibrium with one another.
TABLE III.
Statistical Analysis of SNP.
| Gene Symbol (Location) | SNP | Chr | Position in Base Pairs From Chr Start | Alleles | HWE P Value* | OR (CI) | OR P Value |
|---|---|---|---|---|---|---|---|
| IL-10 (−592) | rs1800895 | 1 | 205013095 | A/G | .87 | † | † |
| IL-10 (−819) | rs1800894 | 1 | 205013289 | A/G | .0009 | † | † |
| IL-10 (−1082) | rs1800896 | 1 | 205013520 | A/G | .64 | † | † |
| IL-1α (+4845) | rs17561 | 2 | 113253694 | G/T | .51 | † | † |
| IL-1α (−819) | rs3783521 | 2 | 113260048 | C/T | .38 | † | † |
| IL-β (+3935) | rs1143634 | 2 | 113306861 | C/T | .73 | † | † |
| IL-β (−511) | rs3087258 | 2 | 113311342 | C/T | .0001 | † | † |
| CD14 (−159) | rs2569190 | 5 | 139993100 | A/G | .16 | † | † |
| TNF-α (−308) | rs1800629 | 6 | 31651010 | A‡/G | .93 | 1.86 (1.14,3.09)§ | .01 |
| IL-6 (−174) | rs13447445 | 7 | 22733281 | C/G | ‖ | † | † |
| IFNγ (−874) | rs2430561 | 12 | 66838789 | A/T | .71 | † | † |
| CCL2 (−2518) | rs3917882 | 17 | 29603912 | A/G | .93 | † | † |
| CCL5 (−403) | rs2107538 | 17 | 31231893 | C/T | .81 | † | † |
| TGF-β1 (509) | rs11466315 | 19 | 46551616 | C/G | .86 | † | † |
SNP = single-nucleotide polymorphism; Chr = chromosome; HWE = Hardy Weinberg equilibrium; OR = odds ratio; CI = confidence interval; rs = reference SNP; IL = interleukin; TNF = tumor necrosis factor; IFN = interferon; TGF = transforming growth factor; CCL = cysteinyl chemokine ligand.
Fisher exact test P values for tests of Hardy Weinberg equilibrium proportions in controls.
Confidence interval includes 1.00/P values not significant.
Risk allele.
Dominant genetic model.
SNP is monomorphic.
An analysis of the A allele at position −308 of the TNF-α gene in the polyp patients was not significantly different in regard to gender, allergy, or ASA intolerance.
DISCUSSION
Our results suggest that the frequency of the A allele, whether homozygous or heterozygous in the −308 position of the promoter region of the cytokine gene TNF-α, is significantly higher in patients with nasal polyposis than in control patients. The A allele in this position of the TNF-α gene is associated with increased transcription of the protein TNF-α.22,23 These findings would then support the concept that this polymorphism has direct effect on TNF-α gene regulation and possibly lead to more severe disease in patients with CHSwNP.
Table II demonstrates a significant difference in the number of male patients having nasal polyposis compared with female patients. This difference is about 2:1 in favor of male patients having nasal polyposis in this study.
There has been support for this male to female predominance for nasal polyposis in the literature. Collins et al.24 reported similar findings in regard to gender in nasal polyposis. They suggested that males were more commonly smokers and had more occupational exposure to dust and chemicals in comparison to females. Busaba et al.25 demonstrated no statistically significant difference in regard to environmental allergy, asthma, psychiatric illness, or anatomical variance comparing the genders in chronic rhinosinusitis. However, chronic rhinosinusitis without polyposis was more commonly diagnosed among women, whereas chronic rhinosinusitis with polyposis was more commonly diagnosed among men (P < .05).
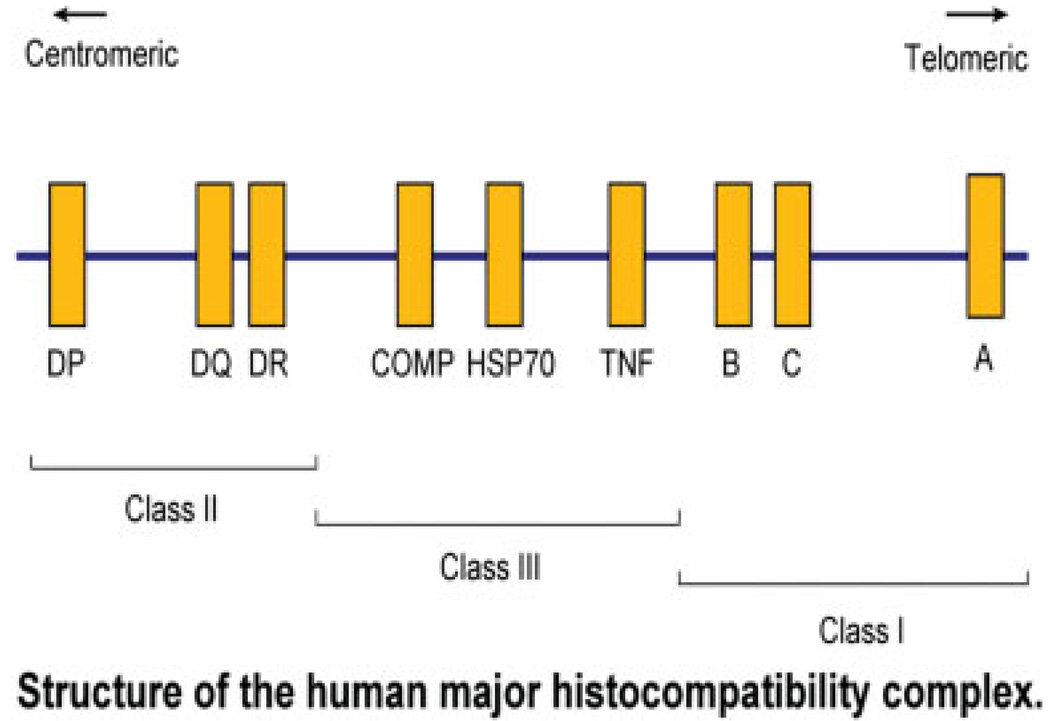
TNF-α is produced primarily but not exclusively by macrophages and T cells, respectively.26,27 Initially, identified for its cytotoxic and antitumor activity, it is now well-documented that this cytokine plays a critical role in a variety of immunological processes, including inflammation, secondary lymphoid organ development, and the control of intracellular pathogens.28 The TNF locus is located in the class 3 region of the human major histocompatibility complex on the short arm of chromosome 6 (Fig. 1). These genes include the HLA A, B, and C, HLA DR, DQ, and DP loci, which collectively have been called immune response genes. Because these genes are close together, they are usually transmitted from generation to generation in linkage disequilibrium; that is, they are almost always transferred from parent to child together. Whether or not the TNF-α polymorphisms themselves play a role in susceptibility to disease, or they are in linkage disequilibrium with other immune response genes close by, is not yet completely known.
Fig. 1.
Short arm of chromosome 6 demonstrating position of HLA antigens and some complement components and TNF-α and other inflammatory proteins. These genes are transmitted from generation to generation in genetic disequilibrium.
Previous investigations on genetic polymorphisms in nasal polyposis have been reported.11–15 Karjalainen and collaborators in Finland described the IL1A genotype as being associated with nasal polyposis in asthmatic adults.11 They analyzed the single G to T base exchange polymorphism in exon 5 at +4845 of the gene encoding IL1-α. The distribution of IL1-α genotype differed significantly between asthmatics with or without nasal polyps. The risk of nasal polyps was markedly increased in patients with the G homozygous allele subjects. Cheng et al.12 at National Taiwan University described the association of interleukin 1B and interleukin 1 receptor antagonists (IL-1Ra) gene polymorphisms and chronic sinusitis. The authors demonstrated an increased prevalence of IL1Ra polymorphism in patients with chronic rhinosinusitis suggesting that this polymorphism, or a polymorphism in linkage disequilibrium with it, may be involved in the susceptibility to chronic rhinosinusitis. Of the 88 patients in this group, almost 80% were diagnosed as having nasal polyposis. These investigators did not study TNF-α.
Erbeck and collaborators13 in Turkey reported IL-1A (+4845 GT and +4845 TT), IL-1B (−511 CC), and TNF-α (−238 AA and −308 GA) genotypes to be associated with nasal polyposis. They found that the GG homozygous haplotype was associated more with controls than with patients with nasal polyps. An unexpected finding in their report was that homozygous A genotype was much more common in controls than in cases. This is unexpected inasmuch as all reports in the literature thus far have suggested that the AA homozygous genotype is both very rare, and is always associated with the highest transcription of TNF-α, and therefore associated with chronic inflammatory disease in the great majority of patients. They do not explain this high incidence of homozygous AA in their control group for TNF-α −308.
Studies have described polymorphisms in genes involved with both leukotriene synthesis and remodeling in chronic hyperplastic sinusitis with nasal polyposis.14
Wang et al.15 studied the matrix metalloproteinase-2 (MMP2) gene polymorphisms in nasal polyps. They concluded the MMP2 gene does not play a crucial role in conferring risk for nasal polyps in a Taiwanese population.
One important aspect that was not investigated in all but the last two studies was HWE proportions. Broadly, significant deviation in the cases can actually buttress evidence of association, indicating possibly selective pressure in that genomic region under investigation; however, violations in HWE proportions in the control population indicate that the controls might actually not be an appropriate control population. Thus, both cases and controls should be tested separately for evidence of violation of HWE proportions. The impact of not considering HWE proportions in genetic association studies was investigated in a meta-analysis of almost 50 studies.19
Exclusion of the violating studies resulted in loss of statistical significance of this overall meta-analysis in three instances and more than a 10% change in the summary odds ratio in six. Thus, clearly for accuracy and particularly replication purposes, HWE proportions as well as nonsignificant deviations from the exact frequencies expected under HWE proportions in gene-disease association studies should be carefully considered. We reviewed the genotype distributions presented in the five studies previously mentioned and tested the controls for violations of HWE proportions. HWE proportions were violated in the significant SNPs in two studies.11,13 These violations should be carefully considered when evaluating the evidence for genetic association with CHSwNP.
The mechanism by which lymphocytes and especially eosinophils migrate into the epithelium and lamina propria of nasal polyps is now becoming unraveled. Our knowledge of the trafficking of these cells from the bloodstream to the nasal mucosa via the cell surface receptors on inflammatory cells and their counter receptors on the venules of nasal mucosal tissue has been established. TNF-α and IL1-β, two potent proinflammatory cytokines, known to be upregulated in nasal polyps, appear to be responsible for this trafficking and migration of inflammatory cells into the nasal polyps. The mechanisms by which this takes place have been elucidated.29,30
TNF-α upregulates VCAM-1 and NF-κB in fibroblasts from nasal polyps.29 Nasal fibroblasts produce VCAM-1, and that production is increased by TNF-α stimulation. VCAM-1 expression in nasal fibroblasts is induced through an NF-κB dependent pathway. These findings might provide a rationale for using NF-κB inhibitors as a treatment for nasal inflammation.
The receptor on the surface of lymphocytes and eosinophils, which migrate to these counter receptors on endothelial venules, have also been established to be very late antigen 4, either α-4 β-1 or α-4 β-7. Our laboratory has established that both VCAM-1 in the endothelium of nasal polyps, as well as VLA-4 on the surface of eosinophils, are present in nasal polyposis.4
In summary, the increased transcription of TNF-α in the cells of nasal polyps can increase integrins on the surface of eosinophils and lymphocytes in the polyp tissue, but also increase the counter receptor VCAM-1 on the endothelium, thus, driving the eosinophils into the polypoid tissue. The eosinophils that have accumulated in the tissue of the polyp can also synthesize and release TNF-α. By using southern blot analysis after a reverse transcription-PCR., Finotto and colleagues31 detected a signal specific for TNF-α mRNA in nasal polyp eosinophils. Thus an autocrine up-regulation of eosinophils may occur in the nasal polyp. Eosinophils that arrive in the nasal polyp tissue may then synthesize TNF-α, which subsequently recruit more eosinophils and, thus, a chronic inflammatory disorder, in which eosinophils and lymphocytes predominate emerges in CHSwNP.
Finally, TNF-α can also increase the secretion of chemokines that attract eosinophils into the nasal polyp mucosa. Not only does TNF-α increase the secretion of eotaxin,32 but it can also increase the expression of RANTES (regulated on activation, normal T cell expressed and secreted) from polyp fibroblasts.33 Thus, there is evidence that TNF-α increases the secretion of chemokines that attract eosinophils into the nasal polyp from constitutive cells that are present, such as fibroblasts.
CONCLUSION
We found that allelic frequencies at the promoter region of the cytokine gene TNF-α at the locus −308 were significantly different in patients with massive nasal polyposis compared to healthy controls, and that patients with the A allele at this locus showed an increased susceptibility to disease. This finding coupled with the fact that the presence of the A allele at this SNP is associated with higher secretion rates for this proinflammatory cytokine provides plausible evidence that this locus is in the casual disease pathway. The rational for this is as follows. If the levels of TNF-α are increased in the nasal polyp tissue (as with the presence of the A allele), a programmed sequence of events occurs that includes; 1) upregulation of the integrins on the surface of eosinophils; 2) increased synthesis of the counter receptor for these integrins on the endothelia of venules, namely, VCAM-1; the increased integrins on eosinophils and the counter receptor VCAM-1 allow eosinophils to migrate into the polyp lamina propria; 3) the eosinophils may then secrete more TNF-α, which recruits more eosinophils; and 4) TNF-α also may be responsible for increasing the synthesis and release of the chemokines RANTES and eotaxin from structural cells, namely fibroblasts, in the nasal polyp, which recruit even more eosinophils.
In conclusion, the A allele in the promoter region of the TNF-α gene at position −308 leads to an increase in the production of TNF-α and a cascade of events that yield increased susceptibility to developing CHSwNP.
BIBLIOGRAPHY
- 1.Feezor RJ, Moldawer LL. Genetic polymorphisms, functional genomics and the host inflammatory response to injury and inflammation. In: Cynober L, Moore FA, editors. Nutrition and Critical Care. Vol. 8. Basel, Switzerland: Karger AG; 2003. pp. 15–37. [DOI] [PubMed] [Google Scholar]
- 2.Bone RC. Toward a theory regarding the pathogenesis of the systemic inflammatory response syndrome: what we do and what we do not know about cytokine regulation. Crit Care Med. 1996;24:163–172. doi: 10.1097/00003246-199601000-00026. [DOI] [PubMed] [Google Scholar]
- 3.Hollaway AF, Rao S, Shannon MF. Regulation of cytokine gene transcription in the immune system. Mol Immunol. 2002;38:567–580. doi: 10.1016/s0161-5890(01)00094-3. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein JM. The molecular biology of nasal polyposis. Curr Allergy Asthma Rep. 2001;1:262–267. doi: 10.1007/s11882-001-0017-3. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Dong Z, Zhu DD, Guan B. Expression profile of immune-associated genes in nasal polyps. Ann Otol Rhinol Laryngol. 2006;115:450–456. doi: 10.1177/000348940611500609. [DOI] [PubMed] [Google Scholar]
- 6.Kratka Z, Bartoba J, Krejsa O, Otcenaskova M, Janatova T, Duskova J. Interleukin-1 gene polymorphisms as assessed in a 10-year study of patients with early-onset periodontitis. Folia Microbiol. 2007;52:183–188. doi: 10.1007/BF02932157. [DOI] [PubMed] [Google Scholar]
- 7.Sykora J, Subrit I, Didek P, et al. Cytokine tumor necrosis factor-α promoter gene polymorphism at position −308 G → A and pediatric inflammatory bowel disease: implications in ulcerative colitis and Crohn’s disease. J Pediatr Gastroenterol Nutr. 2006;42:479–487. doi: 10.1097/01.mpg.0000221917.80887.9e. [DOI] [PubMed] [Google Scholar]
- 8.Dong LM, Potter JD, White E, Ulrich CM, Cardon LR, Peters U. Genetic susceptibility to cancer: the role of polymorphisms in candidate genes. JAMA. 2008;299:2423–2436. doi: 10.1001/jama.299.20.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Texereau J, Chiche JD, Taylor W, Choukroun G, Comba B, Mira JP. The importance of Toll-like receptor 2 polymorphisms in severe infections. Clin Infect Dis. 2005;41 suppl 7:S408–S415. doi: 10.1086/431990. [DOI] [PubMed] [Google Scholar]
- 10.Joki-Erkkila VP, Puhakka H, Hurme M. Cytokine gene polymorphism in recurrent acute otitis media. Arch Otolaryngol Head Neck Surg. 2002;128:17–20. doi: 10.1001/archotol.128.1.17. [DOI] [PubMed] [Google Scholar]
- 11.Karjalainen J, Joki-Erkkila VP, Hulkkonen J, et al. The IL-1A genotype is associated with nasal polyposis in asthmatic adults. Allergy. 2003;58:393–396. doi: 10.1034/j.1398-9995.2003.00118.x. [DOI] [PubMed] [Google Scholar]
- 12.Cheng YK, Der Lin C, Chang WC, et al. Increased prevalence of interleukin-1 receptor antagonist gene polymorphism in patients with chronic rhinosinusitis. Arch Otolaryngol Head Neck Surg. 2006;132:285–290. doi: 10.1001/archotol.132.3.285. [DOI] [PubMed] [Google Scholar]
- 13.Erbeck SS, Yurtcu E, Erbek S, Belginatac F, Sahin FI, Cakmak O. Pro-inflammatory cytokine single nucleotide polymorphisms in nasal polyposis. Arch Otolaryngol Head Neck Surg. 2007;133:705–709. doi: 10.1001/archotol.133.7.705. [DOI] [PubMed] [Google Scholar]
- 14.de Alarcon A, Steinke JW, Caughey R, et al. Expression of leukotriene C4 synthase and plasminogen activator inhibitor 1 gene promoter polymorphisms in sinusitis. Am J Rhinol. 2006;20:545–549. doi: 10.2500/ajr.2006.20.2934. [DOI] [PubMed] [Google Scholar]
- 15.Wang LF, Chien CY, Kuo WR, Tai CF, Juo SH. Matrix metalloproteinase- 2 gene polymorphisms in nasal polyps. Arch Otolaryngol Head and Neck Surg. 2008;134:852–856. doi: 10.1001/archotol.134.8.852. [DOI] [PubMed] [Google Scholar]
- 16.Gaskins RE. A surgical staging system for chronic sinusitis. Am J Rhinol. 1992;6:5–12. [Google Scholar]
- 17.Lund VJ, MacKay IS. Staging in rhinosinusitis. Rhinology. 1993;31:183–184. [PubMed] [Google Scholar]
- 18.Kang K, Fu DJ, Julien D, Braun A, Cantor CR, Koster H. Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci USA. 1999;96:10016–10020. doi: 10.1073/pnas.96.18.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP. Impact of violations and deviations in Hardy-Weinberg Equilibrium on postulated gene-disease associations. Am J Epidemiol. 2006;163:300–309. doi: 10.1093/aje/kwj046. [DOI] [PubMed] [Google Scholar]
- 20.Armitage P. Test for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- 21.Cochran WG. Some methods for strengthening the common chi-squared tests. Biometrics. 1954;10:417–451. [Google Scholar]
- 22.Kroeger KM, Carville KS, Abraham LJ. The −308 tumor necrosis factor-α promoter polymorphism effects transcription. Mol Immunol. 1997;34:391–399. doi: 10.1016/s0161-5890(97)00052-7. [DOI] [PubMed] [Google Scholar]
- 23.Wilson AG, Symons J, McDowell TL, McDebitt HO. Effects of a polymorphism in the human tumor necrosis factor-α promoter on transcription activation. Proc Natl Acad Sci U S A. 1997;94:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins MM, Pang YT, Loughran S, Wilson JA. Environmental risk factors and gender in nasal polyposis. Clin Otolaryngol Allied Sci. 2002;27:314–317. doi: 10.1046/j.1365-2273.2002.00573.x. [DOI] [PubMed] [Google Scholar]
- 25.Busaba NY, Sin HJ, Salman SD. Impact of gender on clinical presentation of chronic rhinosinusitis with and without polyposis. J Laryngol Otol. 2008;122:1180–1184. doi: 10.1017/S0022215107001302. [DOI] [PubMed] [Google Scholar]
- 26.Lieu ZZ, Lock JG, Hammond LA, La Gruta NL, Stow JL, Gleeson PA. A trans-Golgi network golgin is required for the regulated secretion of TNF in activated macrophages in vivo. Proc Natl Acad Sci U S A. 2008;105:3351–3356. doi: 10.1073/pnas.0800137105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komorowska A, Komorowski J, Banasik M, Lewkowicz P, Tchorzewski H. Cytokines locally produced by lymphocytes removed from the hypertrophic nasopharyngeal and palatine tonsils. Int J Pediatr Otorhinolaryngol. 2005;69:937–941. doi: 10.1016/j.ijporl.2005.01.035. [DOI] [PubMed] [Google Scholar]
- 28.Tracey D, Klareskog L, Sasso EH, Salfeld GG, Gak PP. Tumor necrosis factor antagonist mechanisms of action: a comprehensive review. Pharmacol Ther. 2008;117:244–279. doi: 10.1016/j.pharmthera.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Ohori J, Ushikai M, Sun D, et al. TNF-α upregulates VCAM-1 and NF-κB in fibroblasts from nasal polyps. Auris Nasus Larynx. 2007;34:177–183. doi: 10.1016/j.anl.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Jahnsen FL, Brandtzaeg P, Haye R, Haraldsen G. Expression of functional VCAM-1 by cultured nasal polyp derived microvascular endothelium. Am J Pathol. 1997;150:2113–2123. [PMC free article] [PubMed] [Google Scholar]
- 31.Finotto S, Ohno I, Marshall JS, et al. TNF-α production by eosinophils in upper airways inflammation (nasal polyposis) J Immunol. 1994;153:2278–2289. [PubMed] [Google Scholar]
- 32.Yoshifuku K, Matsune S, Ohori J, Sagara Y, Fukuwa T, Kurono Y. IL-4 and TNF-α increased the secretion of eotaxin from cultured fibroblasts of nasal polyps with eosinophil infiltration. Rhinology. 2007;45:235–241. [PubMed] [Google Scholar]
- 33.Sajai F, Nonaka M, Pawankar R. Expression of RANTES by IL-1β and TNF-α stimulated nasal polyp fibroblasts. Auris Nasus Larynx. 2000;27:247–252. doi: 10.1016/s0385-8146(00)00052-3. [DOI] [PubMed] [Google Scholar]