Abstract
Assembly of MHC class I molecules with peptide in the endoplasmic reticulum requires the assistance of tapasin. Alternative splicing, which is known to regulate many genes, has been reported for tapasin only in the context of mutations. Here, we report on an alternate splice form of tapasin (tpsnΔEx3) derived from a human melanoma cell line that does not appear to be due to mutations. Excision of exon 3 results in deletion of amino acids 70 to 156 within the beta barrel region, but the membrane proximal Ig domain, the transmembrane domain and cytoplasmic tail of tapasin are intact. Introduction of tpsnΔEx3 into a tapasin deficient cell line does not restore MHC class I expression at the cell surface. Similar to a previously described tapasin mutant (tpsnΔN50), tpsnΔEx3 interacts with TAP. Therefore we utilized these altered forms of tapasin to test the importance of MHC class I interaction with TAP. In the presence of wild type tapasin, transfection of tpsnΔN50, but not tpsnΔEx3, reduced MHC class I expression at the cell surface likely due its ability to compete MHC class I molecules from TAP. Together these findings suggest that tumor cells may contain alternate splice forms of tapasin which may regulate MHC class I antigen presentation.
Keywords: antigen presentation, antigen processing
Introduction
MHC class I molecules consist of three components which must form a stable complex in the endoplasmic reticulum (ER) before transport to the cell surface; the highly polymorphic heavy chain, β2-microglobulin, (β2m) and a small peptide 8-10 amino acids in length. This trimeric complex is recognized by a specific clonotypic T cell receptor on the surface of a CD8+ T lymphocyte. Recognition of an immunogenic peptide associated with MHC class I molecules leads to the activation of a CD8+ T cell and subsequent killing of the target cell. Although in vitro studies suggest that a few hundred or possibly even one MHC-peptide complex is sufficient for activation of a T cell [1, 2], many reports have demonstrated that detectable, but reduced, MHC class I expression is associated with poor recognition by T cells [3-7]. Thus, T cell recognition is limited by the availability of surface MHC class I molecules bound to specific peptides.
Nascent MHC class I heavy chains first interact with calnexin upon their translocation into the ER [8]. Dissociation of heavy chains from calnexin is thought to precede their association with a preformed complex of TAP and tapasin [9-11]. These molecules, together with β2m, the chaperone calreticulin, the thiol-oxidoreductase ERp57 and B cell receptor associated protein 31 (BAP31) make up the ‘peptide loading complex’ which brings several proteins together to assist in the assembly of MHC class I molecules with their peptide ligands [10, 12-17]. Peptides, required for the stabilization of the immature heavy chain: β2m dimers, are derived from the cytosol by proteasome activity and trimming peptidases [18]. The peptides are transported into the ER by the TAP1 and TAP2 heterodimer [19, 20] where they are further trimmed by ERAAP [21, 22] to optimize their size for binding to the cleft of the MHC class I molecule. The loading of the processed peptides into the peptide binding site of MHC class I molecules is enhanced by the coordinated effort of tapasin and ERp57.
Tapasin was first identified as a TAP associated protein that was required to detect MHC class I molecules associated with TAP [10, 12, 13]; an interaction whose importance in MHC class I assembly and export was suggested earlier [23-25]. This bridging function of tapasin is thought to be important for efficient MHC class I assembly with peptide as the absence of tapasin results in poor MHC class I export from the ER and hence low expression at the surface [13, 25-27]. Although some MHC allelic products, such as HLA-B27, can achieve considerable surface expression in the absence of a TAP-MHC class I interaction (i.e. in the absence of tapasin), the MHC I-peptide complexes folded in the absence of tapasin are less stable arguing that the TAP-MHC class I interaction is important for optimal assembly [28]. Furthermore, single mutations in the transmembrane domain of tapasin diminishes interaction with TAP and compromises the conformation of surface MHC class I molecules [29, 30]. Together these data argue that the bridging function is important for efficient assembly, export and surface expression. However, soluble tapasin which does not detectably associate with TAP, but promotes MHC class I expression challenged the idea that tapasin acted simply as a bridge between TAP and MHC class I molecules [31]. While more recent studies have demonstrated that the tapasin promotes peptide loading by enhancing the binding of peptides with slow off-rates [33, 34], likely through the action of a tapasin-ERp57 disulfide intermediate [32, 35], the importance of TAP-MHC class I proximity remains unclear.
Much of our information on how tapasin functions has been derived from transfection studies in the human B-LCL 721.220, tapasin deficient mice or more recently a tapasin deficient melanoma cell line [10, 13, 26, 27, 36]. Although the complete lack of tapasin protein drastically affects MHC class I expression, lower levels of tapasin is associated with lower MHC class I expression [37]. Therefore, the regulation of tapasin is an important aspect of efficient MHC class I expression and subsequent CTL and/or NK responses. Tapasin is upregulated by cytokines and extracellular signals such as IFN, IFNβ, TNF, IL-4 and ligation of TLRs [38, 39]. Gene regulation also may occur by alternate splicing [40] however, alternate splicing of tapasin has only been reported in the context of genomic mutations [41, 42].
Here we report on an alternate splice form of tapasin (tpsnΔEx3) arising in a human melanoma cell line. This alternate splice form of tapasin lacks exon 3 but does not appear to be due to genomic mutation surrounding exon 3. TpsnΔEx3, like tpsnΔN50, interacts with TAP, but does not support MHC class I assembly and surface expression. Therefore, we tested if tapasinΔEx3 could act as a dominant negative by competing with wild type tapasin for TAP occupancy.
Materials and Methods
Cell lines
The human melanoma cell lines M1195 and M553 (HLA-A28, A2-haplotype loss, HLA-B*5001 and HLA-B*5701) and HeLaM cells have been described previously [11, 36, 43]. All cell lines used in this report were grown in RPMI-1640 medium (Life Technologies, Grand Island, NY) supplemented with 10% bovine calf serum (Hyclone Laboratories, Logan, UT) and 10mM HEPES.
Antibodies
The monoclonal antibody (mAb) W6/32 recognizes β2-microglobulin associated HLA-A, -B and -C molecules. The mAb SFR8.B6 recognizes the HLA-Bw6 epitope and is highly dependent upon the presence of residues Arg82 and Gly83 [44]. As we have previously shown in the M553 transfected with wild type tapasin cells, the SFR8.B6 supratypic antibody likely reacts with the HLA-B*5001 (Arg82, Gly83) product but not with HLA-B*5701 (Leu82, Arg83). The mAb TT4-A20 recognizes the HLA-Bw4 epitope, which is highly dependent upon residues 79–83 that are conserved in HLA-B*5701 expressed by M553 cells [36]. The following mAbs and antisera have been described previously: R.Ring4c antiserum (C-terminal peptide of TAP1) [23], mAbs NOB-2 (TAP2) and TO-3 (tapasin) [45, 46]; R.gp48C antiserum (C-terminal peptide of tapasin [47]; mAb 3B10.7 (heavy chain) [48], mAbs TP25.99 (heavy chain) and CR11.351 (HLA-Aw68) [49]. The rabbit polyclonal calreticulin antibody was purchased from Stressgen (Vancouver, Canada). (San Diego, CA). The mAb specific for human tubulin was purchased from Sigma-Aldrich (St. Louis, MO). The mAb PaSta1 recognize a lumenal epitope within tapasin [32].
RT-PCR, cloning and genomic PCR
Total mRNA from M1195 cells was isolated using TRIZOL (Invitrogen, Carlsbad, CA) as per the manufacturer’s recommendations. Two micrograms of mRNA were reverse transcribed to generate the first cDNA strand by using Superscript II (Invitrogen, Carlsbad, CA). cDNA was then used as template in a PCR reaction using the Accuprime enzyme (Invitrogen, Carlsbad, CA) to amplify tapasin cDNA using the primers 1F (5′-AGCGCCATGAAGTCCCTGTCTCTGCTC-3′) and 1R (5′-GTGCCCTCACTCTGCTTTCTTCTTTGA-3′). Amplicons were then gel purified and cloned into pDrive (Qiagen, Valencia, CA) before sequencing. The tapasinΔEx3 sequence obtained from pDrive has been submitted to genbank (EU693375). The cassette encoding wild type tapasin or tpsnΔEx3 cDNA was then excised and subcloned into pCDNA3.1(−) puro which was derived from pCDNA3.1(−)neo (Invitrogen, Carlsbad, CA) by replacement of the neomycin gene with the puromycin resistance gene. For analysis of genomic tapasin, genomic DNA was isolated from M1195 cells using TRIZOL according to manufacturers recommendations. PCR was performed using the primers FP_Ex3Genomic-225 (5′ AGTGTACACGGTGAGTCTCTAG 3′) and RP_Ex3Genomic+340 (5′ CATCACCTGACAAGTCTCTCAA 3′). The 826 bp product was cloned into pGEM-T-Easy (Promega Madison, WI) and single colonies were isolated and plasmid DNA purified for sequencing using the T7 and SP6 primers.
Transfection and cloning
For stable transfectants, tapasin expression constructs were transfected into M553, M553(R240)tpsn or HeLaM cells using Lipofectamine (Invitrogen, Carlsbad, CA) or Effectene (Qiagen, Valencia, CA) according to the manufacturer’s recommendations. Selection of clones containing the plasmid pCDNA3.1(−) PURO encoding a wild type tapasin cDNA or the tpsnΔEx cDNA was performed by limiting dilution and supplementing the media with puromycin (Sigma-Aldrich St Louis, MO) at a final concentration of 1 microgram per mL. For transient transfection experiments, 100 ng of the pmaxGFP plasmid which encodes an enhanced GFP protein driven by the CMV promoter (Amaxa, Gaithersburg, MD) and 2 micrograms of the tpsnΔEx3 cDNA in pCDNA3.1(−)PURO or empty pCDNA3.1(−)puro plasmid were transfected into 1 million cells using an Amaxa Nucleoporator II instrument (solution V, program T-030) according to manufacturers recommendations.
Western blotting
Cells were lysed using 1% TritonX-100 in 10mM Tris, 150mM NaCl, pH7.4 supplemented with protease inhibitors PMSF (0.5mM) and N-ethylmaleimide (5mM) and incubated at 4°C for 30 minutes. Protein concentration of post nuclear supernatants was determined by the Bradford assay (Bio Rad, Hercules, CA). After SDS-PAGE and transfer of proteins to Immobilon-P membrane (Millipore, Bedford, MA), membranes were incubated in 3% Bovine Serum Albumin (BSA). Proteins of interest were detected using the indicated primary antibodies overnight at 4°C followed by three washes in PBS/0.02% TWEEN 20. Secondary antibodies coupled to alkaline phosphatase were then added in 3% Bovine Serum Albumin and incubated at room temperature for 1 hour. After three washes in PBS/0.02% TWEEN 20, VistraECF substrate (Amersham Pharmacia, Piscataway, NJ) was applied and fluorescence was measured using a STORM860 instrument (Molecular Dynamics, Palo Alto, CA). Fluorescence intensity was acquired and analyzed using ImageQuant 5.2 software by integration of area under the histogram analysis.
Radiolabeling and Immunoprecipitation
Cells were washed in PBS and starved for 30 minutes in the absence of cysteine and methionine prior to labeling with 35S-methionine for 2.5 hours at 37°C. For immunoprecipitation cells were lysed in either 1% Digitonin (Calbiochem, Gibbstown, NJ) or 1% TritonX-100 in 10mM Tris, 150mM NaCl, (pH7.4) 0.5mM PMSF and 5mM NEM. After pelleting nuclei, post nuclear supernatants were diluted in 1% TritonX-100. The resultant supernatants were subjected to immunoprecipitation using the antibodies indicated and protein G Sepharose. After washing, the bound proteins were eluted with reducing Laemmli sample buffer, separated by SDS-PAGE and subjected to autoradiography. For immunoprecipitation of TAP, cells were lysed in digitonin containing lysis buffer indicated above (to preserve interactions between TAP and MHC class I molecules) without the addition of the 1% TritonX-100 dilution. After immunoprecipitation, bound proteins were eluted by boiling in 1% SDS, 10mM Tris-Cl, 150mM NaCl. The resulting supernatants were diluted 10 fold in 1% TritonX-100, 10mM Tris-Cl, 150mM NaCl (pH7.4) and subjected to a second round of immunoprecipitation.
Flow cytometric analysis
Cells were washed and incubated with saturating concentrations of W6/32, SFR8.B6 and TT4-A20 or control antibodies for 30 minutes at 4°C. After washing, half a million cells were resuspended in saturating concentrations (10 microgram/ml) of fluorescein isothiocyanate or phycoerythrin conjugated, goat anti-mouse immunoglobulin G antibodies. When cells transfected with GFP were used for flow cytometric analysis, the cells were stained with W6/32-PE directly conjugated antibody (BD biosciences, San Jose, CA) and analyzed without fixation. Otherwise, after removing the excess primary and secondary antibody by washing, cells were fixed in 2% paraformaldehyde and cells were analyzed on a FACScan instrument (Becton Dickinson, San Jose, CA). Data were analyzed with WINMDI software (Joe Trotter, The Scripps Research Institute, La Jolla, CA).
Results
M1195 cells encode a tapasin variant lacking the third exon in the absence of genomic mutations
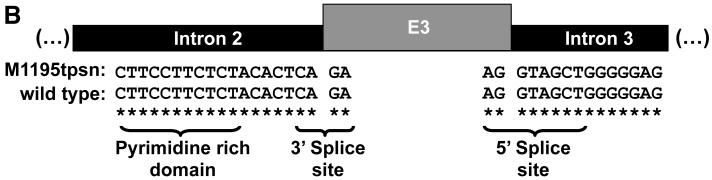
While characterizing antigen presentation genes in human melanoma cell lines we discovered an alternate splice form of tapasin lacking exon 3 in M1195 (tpsnΔEx3, Figure 1A). Tapasin cDNA was amplified by PCR using primers designed to anneal at the 5′ and 3′ ends of the coding region. Sequencing of independent colonies revealed a full length wild type tapasin gene encoding the R240 tapasin allele ((R240)tpsn) and tpsnΔEx3. To determine if a genomic mutation in the splice sites surrounding exon 3 may be the cause of tpsnΔEx3 we sequenced genomic DNA from M1195 cells (Figure 1B). Exon3 including 200bp 5′ and 200bp 3′ of the exon was amplified by PCR. In an attempt to isolate and sequence both genomic copies of tapasin, eight independent PCR reactions and 24 independent colonies from those eight PCR reactions were sequenced. All 32 sequences revealed identical wild type splice site sequences (Figure 1B). All sequences contained an intact exon 3 suggesting that deletion of exon 3 in the genomic DNA is unlikely. Furthermore, these wild type sequences indicate intact splice acceptor and donor sites on either end of exon3.
Figure 1. TpsnΔEx3 is likely not due to genomic mutations.
A. Sequence of wild type tapasin and a variant alternate splice form lacking exon 3. B. Exon 3 is not deleted from the genome and splice sites surrounding exon 3 are not mutated. Genomic sequence surrounding exon 3 in M1195 cells compared to wild type sequence (NT007592).
TpsnΔEx3 encodes a protein that can be expressed at appreciable levels within cells
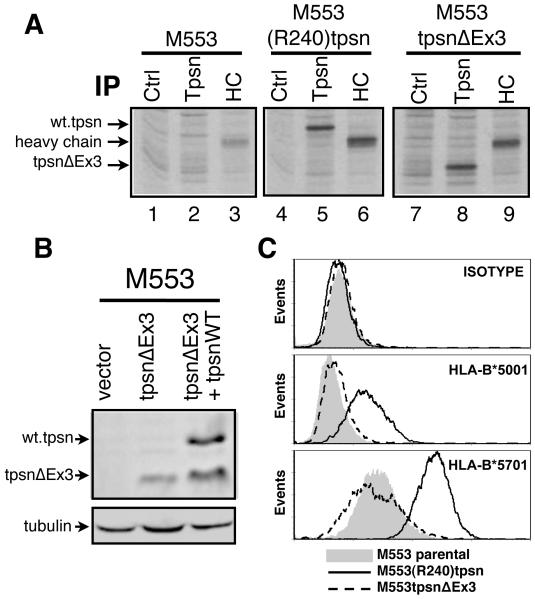
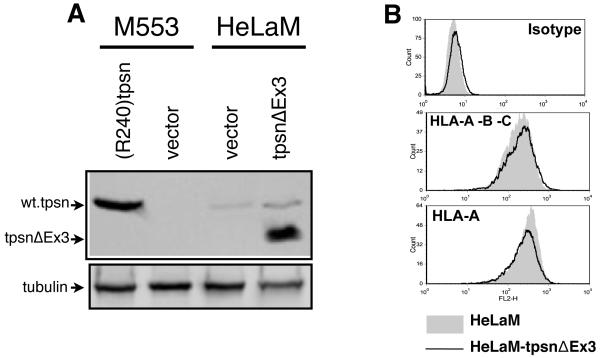
The tpsnΔEx3 sequence predicts an in frame truncated protein lacking 86 amino acids (from position 70 to 156) and an expected molecular weight of 38kD instead of the wild types 48kD. To test whether the alternate splice lacking exon 3 could be translated and expressed at appreciable levels, the tpsnΔEx3 cDNA isolated from M1195 was transfected into M553 cells where endogenous tapasin protein is undetectable [36]. To verify the expression of the truncated protein product we examined newly synthesized tpsnΔEx3 protein (Figure 2A) and steady state levels (Figure 2B). Control, tapasin or heavy chain specific antibodies were used to immunoprecipitate (IP) the respective proteins from 35S-methionine labeled M553, M553(R240)tpsn or M553tpsnΔEx3 cell lysates (Figure 2A). While class I heavy chains are detectable in all three cell types (lanes 3, 6 and 9), tapasin IP from M553 lysates confirmed the lack of detectable endogenous tapasin protein (compare control antibody, lane 1, and tapasin antibody, lane 2). Tapasin IP from M553(R240)tpsn cell lysates revealed a specific band corresponding to wild type tapasin (lane 5) that was not present in the M553 parental cell line (lane 2). In M553tpsnΔEx3 cells, the tapasin specific IP (lane 8) revealed a band of lower molecular weight (as predicted) than that of the transfected wild type tapasin (lane 5) and specific to the tapasin antibody (compare control and tapasin specific lanes 7 and 8 respectively). The presence of a detectable radiolabeled tpsnΔEx3 after 35S-methionine labeling argues that tpsnΔEx3 protein is synthesized. The tpsnΔEx3 protein from M553tpsnΔEx3 cells was also successfully detected by western blot (Figure 2B) suggesting that tpsnΔEx3 is not rapidly degraded following synthesis.
Figure 2. TpsnΔEx3 expression does not restore tapasin deficiency.
A. Tapasin deficient M553 cells or the indicated transfectants were metabolically labeled, lysed in TritonX100 containing lysis buffer and immunoprecipitations (IP) performed with control (Ctrl) antibody (L243), tapasin (Tpsn) specific antibody TO-3 or heavy chain (HC) specific antibody TP25.99. B. Lysates from M553 cells transfected with empty plasmid (vector) or tpsnEx3 were assessed for for tapasin by western blot using TO-3 antibody. Control lane shows relative position of wild type tapasin and tpsnΔEx3. C. Untransfected M553 cells (filled histograms) or stable transfectants containing wild type R240tpsn (solid line) or tpsnΔEx3 (dotted line) were examined for MHC class I expression by TT4-A20 staining (HLA-B*5701), SFR8.B6 (HLA-B*5001) staining, or isotype control staining. A representative figure of at least three independent experiments is shown.
TpsnΔEx3 does not restore MHC class I surface expression in M553 cells
We have previously reported that levels of either HLA-B allele, (HLA-B*5701 and – B*5001), on M553 cells are low to undetectable because this cell line does not express tapasin protein, and that this defect is restored by transfecting a wild type tapasin cDNA [36]. To determine if tpsnΔEx3 was sufficient to promote efficient class I expression at the cell surface (in the absence of wild type tapasin) tpsnΔEx3 cDNA was over-expressed in M553 cells and compared to wild type tapasin transfectants. As expected, transfection of wild type tapasin restores MHC class I expression at the cell surface (Figure 2C, MFI for M553 parental and (R240)tpsn transfected were 25 and 228 for HLA-B*5701, and 4 and 17 for HLA-B*5001, respectively). Surface MHC class I levels on MtpsnΔEx3 cells remained unaltered when compared to the parental M553 cells (Figure 2C, MFI values for M553 parental vs M553tpsnΔEx3 were 25 vs 22 for HLA-B*5701 and 4 vs 5 for HLA-B*5001), arguing that tpsnΔEx3 is not capable of restoring the tapasin deficiency of M553 cells.
TpsnΔEx3 interacts with TAP
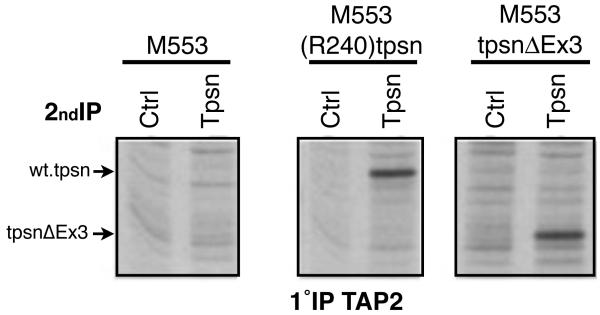
The transmembrane domain of tapasin is required for its interaction with TAP [29, 31] and this domain is present in tpsnΔEx3. We therefore hypothesized that tpsnΔEx3 would interact with the TAP heterodimer. M553, M553(R240)tpsn and M553tpsnΔEx3 cells were metabolically labeled as in Figure 2A, and an equivalent number of cells were lysed in digitonin containing buffer to preserve the interactions within the peptide loading complex. TAP2 IPs were eluted and re-precipitated using either a control or tapasin specific antibody (Figure 3). Parental M553 cells were used to distinguish between the specific interaction mediated by the transfected tapasin and the interactions that occur in the absence of tapasin (Figure 2, lanes 1 and 2). As previously shown, in M553(R240)tpsn cells, there is a specific band corresponding to the transfected wild type tapasin (compare tapasin IP, lane 4, to lane 3 control IP). In M553tpsnΔEx3 cells, a specific band in lane 6 corresponds to tpsnΔEx3, evidenced by a lower molecular weight band than the wild type tapasin (lane 4). Similar results were obtained using TAP1 specific antibody for the first immunoprecipitation (data not shown). Together, these results suggest that tpsnΔEx3 interacts with TAP despite the lack of exon 3.
Figure 3. TpsnΔEx3 interacts with TAP.
M553 cells were treated with IFN for 16-20 hours and metabolically labeled using 35S methionine prior to lysis in digitonin containing lysis buffer. Cell lysates were subjected to immunoprecipitation (1°IP) using a TAP2 specific antibody. After elution, re-IP (2°IP) with a control antibody (Ctrl) or TO-3 (Tpsn) was performed and gels subject to autoradiography.
TpsnΔN50, but not TpsnΔEx3 diminishes MHC class I surface expression in the presence of wild type tapasin
Previous studies have demonstrated that deletion of the N-terminal 300 amino acids (tpsnΔN300) or as little as the N-terminal 50 residues from tapasin renders it incapable of interacting with MHC class I molecules, yet able to interact with, and stabilize, TAP [47]. Therefore, if the tapasin bridging function is important, then tapasin mutants that interact with TAP but not with MHC class I molecules may compete for TAP binding resulting in fewer MHC class I molecules associated with TAP. We therefore tested if tpsnΔEx3 or tpsnΔN50 could compete for TAP and diminish MHC class I assembly and expression at the cell surface.
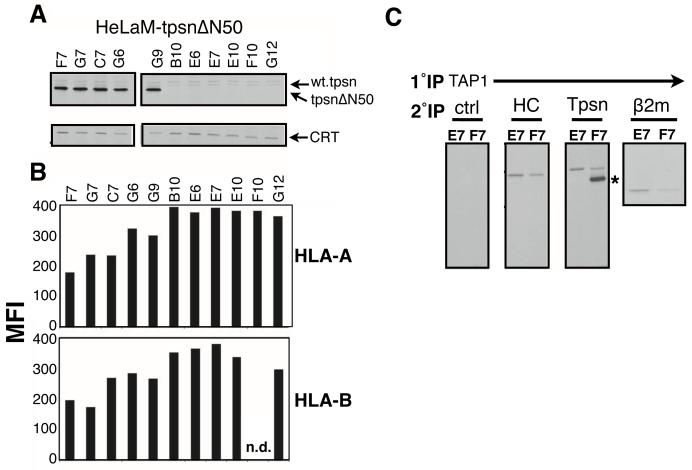
TpsnΔN50 cDNA was transfected into HeLaM cells, which have an intact peptide loading complex, and transfectants were selected for puromycin resistance. Of 11 isolated clones, five expressed abundant tpsnΔN50 at steady state (at a much greater level than the barely detectable endogenous tapasin in HeLaM cells), while six did not express detectable tpsnΔN50 (Figure 4A). Flow cytometric analysis of the HLA-A and HLA-B alleles revealed that tpsnΔN50 expression correlates with lower levels of MHC class I at the cell surface (Figure 4B). Two clones were chosen for further examination of interactions with TAP; clone F7, which expresses high levels of tpsnΔN50 and a clone that does not express tpsnΔN50, clone E7. In clone F7, the tpsnΔN50 interacts robustly with TAP (Figure 4C, marked by *) and the level of endogenous wild type tapasin associated with TAP1 was diminished by approximately fifty percent when compared to clone E7 (Figure 4C). Confirmation of the reduced ability of endogenous tapasin to function as a bridge between the TAP and MHC class I molecules is shown by the decreased level (~50%) of associated heavy chain and β2m (Figure 4C, HC panel and β2m panel). Together these data argue that tpsnΔN50 can act as a dominant negative by reducing MHC class I association with TAP and reducing MHC class I surface assembly and surface expression.
Figure 4. TpsnΔN50 downregulates surface MHC class I molecules by competing with wild type tapasin in HeLa cells.
A. The indicated puromycin resistant transfectants of HeLaM cells were labeled with 35S-methionine and immunoprecipitated for tpsnΔN50 (Rgp48C) or calreticulin. B. HLA-A (CR11.351) and HLA-B (SFR8.B6) expression on the surface of clones indicated. Mean Fluorescence Intensity (MFI) is plotted for each clone. C. Clones F7 (expressing tpsnN50) and E7 (not expressing tpsnN50), were lysed in digitonin and immunoprecipitated using a Ring4C (TAP1) specific antibody. After elution, re-immnoprecipitation was performed using control, heavy chain (HC, 3B10.7), tapasin (R.gp48C) or β2m specific antibodies. A representative figure of at least three independent experiments is shown.
Next we tested if tpsnΔEx3 could also act as a dominant negative. Stable transfection of tpsnΔEx3 into HeLaM cells also led to high expression relative to the endogenous wild type tapasin (Figure 5A). This ratio of mutant to wild type tapasin is comparable to that observed in HeLaM tpsnΔN50 cells (Figure 5A vs 4A). Flow cytometric analysis of staining with CR11-351 (recognizing HLA-Aw68 on HeLaM cells) or W6/32 (anti-HLA-A, -B, -C) showed no significant differences between HeLaM and HeLaMtpsnΔEx3; (Figure 4B the MFI for HeLaM and HeLaMtpsnΔEx3 were 266 and 235 for CR11-351 and 171 and 191 for W6/32, respectively). Therefore it appears that tpsnΔEx3 does not act as a dominant negative.
Figure 5. TpsnΔEx3 does not reduce MHC class I expression on HeLa cells.
A. Stable transfectants of HeLaM, or M553, cells containing empty vector or tpsnΔEx3 were analyzed for tapasin protein by Western blot using TO-3 antibody. B. The effect of stable tpsnΔEx3 expression on MHC class I expression on the surface of HeLaM cells was determined using W6/32 (HLA-A, -B, -C) or CR11-351 (HLA-Aw68).
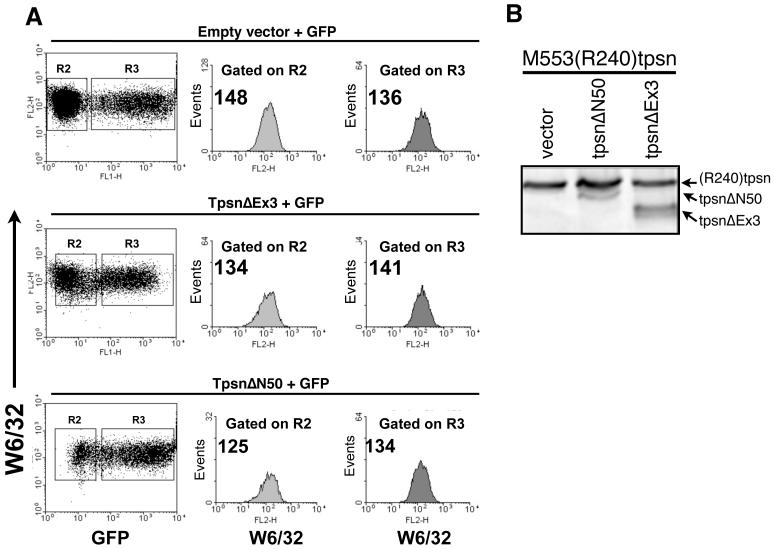
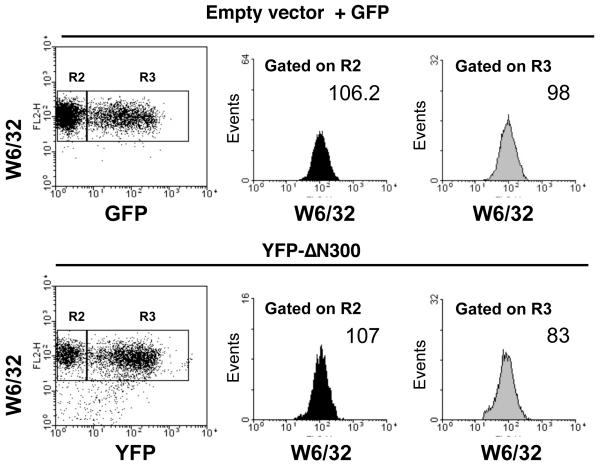
Everett and Edidin recently reported that transient (rather than stable) transfection of a tapasin truncation mutant reduced MHC class I expression at the cell surface [50]. Therefore, we attempted a similar transient transfection approach of M553(R240)tpsn cells with tpsnΔEx3 or tpsnΔN50 along with an EGFP encoding plasmid. Populations of cells successfully co-transfected were gated based on GFP fluorescence. Then, MHC class I levels were analyzed in cells transfected, gated on R3 (GFP positive) and untransfected, gated on R2 (GFP negative) using W6/32 directly conjugated to PE (FL-2). Examination of the mean fluorescent intensity (MFI) on FL-2 revealed no significant difference in the surface expression of MHC class I in M553(R240)tpsn cells transfected with tpsnΔN50 or tpsnΔEx3 when compared to those transfected with empty vector and GFP (Figure 6A). The lack of effect in the transient transfection may be due to the lower ratio of transfected tpsnΔN50 to wild type tapasin in the M553 cells compared to HeLaM cells (compare Figure 6B to Figure 4A). Transient transfection of YFP-ΔN300 into M553(R240)tpsn cells reduced surface MHC class I expression by approximately 20% suggesting that the transient transfection approach to demonstrate dominant negative tapasin activity is feasible (Figure 7). Collectively these data argue that tpsnΔN50 and YFP-tpsnΔN300 can have dominant negative activity when expressed at high enough levels. However, similar over-expression of tpsnΔEx3 (in HeLaM cells) does not result in dominant negative activity.
Figure 6. Transient expression of truncated tapasin molecules does not reduce MHC class I expression in M553(R240)Tpsn cells.
A. Transient co-transfection of M553(R240)Tpsn cells with pmaxGFP and the indicated tapasin variants followed by W6/32 staining 24 hours post transfection. W6/32 staining on populations of cells gated on forward and side scatter (R1) and for the negative or positive expression of GFP (R2 vs. R3, respectively). B. The transient transfectants from A. were analyzed by western blot for the ratio of wild type (R240)tpsn to tpsnΔEx3 or tpsnΔN50 using TO-3 antibody.
Figure 7. YFP-N300 downregulates MHC class I expression in M553(R240)tpsn cells.
M553(R240)Tpsn cells were transiently transfected with YFP-N300 or co-transfected with empty vector and pmaxGFP. Twenty-four hours post transfection, MHC class I expression was analyzed by flow cytometry using a PE directly conjugated W6/32 (MHC class I) antibody. Mean fluorescent intensity was determined for W6/32 staining on populations of cells gated on forward and side scatter (R1) and for the negative or positive expression of GFP/YFP (R2, GFP/YFP negative) and R3, (GFP/YFP positive).
Discussion
In the present study we have demonstrated the presence of a novel splice form of tapasin in a human melanoma cell line. This splice form, which lacks exon 3, encodes a protein that can be synthesized within cells and associate with TAP but does not promote MHC class I assembly and surface expression when expressed in a tapasin deficient cell line. Over-expression of tpsnΔEx3 in the presence of wild type tapasin (in HeLaM or M553(R240)tpsn cells) does not appear to diminish MHC class I assembly and surface expression. However, a deletion mutant of tapasin that lacks the N-terminal fifty amino acids does appear to compete with wild type tapasin and reduce MHC class I association with TAP and subsequent assembly and surface expression.
Two previous reports of tapasin alternate splice forms have demonstrated that their occurrence was due to genomic mutations. First, Copeman and colleagues identified the tapasin defect in the B-LCL .220; a cell line that was selected for low class I expression after γ-irradiation [41]. In .220 cells, a single nucleotide substitution abolishes the 5′ splice site of the second intron leading to the generation of a truncated tapasin protein that lacked a portion of the signal sequence and the N-terminal fifty amino acids. The protein product of this alternate splice form is barely detectable in .220 cells presumably due to the incomplete signal sequence that is predicted to impair its translocation into the ER. Second, Elliott and colleagues analyzed an alternate splice form of tapasin from the existing cDNA database [42]. This splice variant of tapasin failed to exclude introns 4 through 6 due to a mutation in the intron-exon boundary. However, when this mutated splice form of tapasin was transfected into .220 cells, it was processed to remove the 4th and the 6th intron but the 5th intron remained as part of the translated product, introducing an early stop codon generating a soluble tapasin protein. This truncated tapasin protein, although unable to interact with TAP (as the transmembrane domain was absent), was able to restore surface presentation of HLA-B5 expression of the .220 cells. However, the stability of the HLA-B5 allele was much lower when .220 cells were transfected with the alternate splice form of tapasin than with wild type tapasin suggesting suboptimal assembly with peptide [42]. We have identified a unique previously undescribed tapasin alternate splice form in which the third exon is deleted. Exon 3 ‘skipping’ is likely not caused by genomic mutations because sequencing of exon 3 and the surrounding nucleotides reveals intact splice acceptor and donor sites (Figure 1B).
Deletion of exon 3 (amino acid residues 70-156) eliminates the central beta sheet (S4, S5, S6 and S7) of the N-terminal three-tiered beta sheet sandwich. This structure of tapasin is known to be essential for interaction with ERp57 since Cys95 forms a disulfide intermediate with ERp57 (Cys57 on ERp57). The phenotype of tpsnΔEx3 is more severe than C95A mutation [32]. Mutation of Arg72 within this region leads to diminished peptide loading by the tapasin-ERp57 conjugate [35] supporting the idea that residues beyond Cys95 are critical for efficient assembly by the tapasin-ERp57 conjugate. Therefore, it is not surprising that tpsnΔEx3 does not support efficient MHC class I export and surface expression. However, its ability to interact with TAP prompted us to test whether tpsnΔEx3 may compete with wild type tapasin for TAP occupancy which would result in fewer MHC class I molecules associated with TAP. Although our studies demonstrate that tpsnΔEx3 does not reduce surface expression, we show that tpsnΔN50 can reduce MHC class I association with TAP and surface expression significantly. Everett and Edidin recently showed that a mutant tapasin lacking 300 amino acids at the N-terminus fused with yellow fluorescent protein (YFP) diminishes MHC class I expression possibly due to competition for TAP accessibility [50]. In our studies, we demonstrate a correlation between reduced MHC class I association with TAP and reduced surface expression of MHC class I molecules. It should be noted that we observe this dominant negative activity when the mutant tapasin is expressed at much higher levels than the wild type tapasin (in HeLaM cells) but could not reproduce this phenotype in M553(R240)tpsn cells likely due to the high expression of wild type tapasin.
Together these data provide evidence for the importance of the MHC class I – TAP interaction and also for the presence of alternate splice forms of tapasin which may represent a potential mechanism of regulation. Initial attempts to determine the prevalence of tapasinΔExon3 in cell lines that express wild type tapasin have been unsuccessful. Whether this is due to the low abundance of tapasinΔEx3 transcripts relative to wild type tapasin transcripts is unclear. Future studies into identifying the prevalence of tapasinΔEx3 and/or the signals that regulate the generation of tapasinΔEx3 may uncover novel aspects of regulation of antigen presentation in normal and tumor cells.
Acknowledgements
We thank Soldano Ferrone for his generosity in providing mAbs NOB-2, TO-3 and TP25.99; Michael Edidin for providing the YFP-BAP31 expression construct and the Roswell Park Cancer Institute Biopolymer Facility for DNA sequencing. This work was supported by Investigator startup funds (NB), the Roswell Park Alliance Foundation (NB), NIH PHS grants AI071183 (NB), CA55791 (SG) and CA98156 (SG) and the NCI Cancer Center Support Grant to Roswell Park Cancer Institute [CA016156].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Christinck ER, Luscher MA, Barber BH, Williams DB. Peptide binding to class I MHC on living cells and quantitation of complexes required for CTL lysis. Nature. 1991 Jul 04;352(6330):67–70. doi: 10.1038/352067a0. [DOI] [PubMed] [Google Scholar]
- [2].Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002 Oct 24;419(6909):845–9. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- [3].Ferrone S, Marincola FM. Loss of HLA class I antigens by melanoma cells: molecular mechanisms, functional significance and clinical relevance. ImmunolToday. 1995 Oct;16(10):487–94. doi: 10.1016/0167-5699(95)80033-6. [DOI] [PubMed] [Google Scholar]
- [4].Wolfel T, Klehmann E, Muller C, Schutt KH, Meyer zum Buschenfelde KH, Knuth A. Lysis of human melanoma cells by autologous cytolytic T cell clones. Identification of human histocompatibility leukocyte antigen A2 as a restriction element for three different antigens. The Journal of experimental medicine. 1989 Sep 1;170(3):797–810. doi: 10.1084/jem.170.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern PL. Natural history of HLA expression during tumour development. Immunology today. 1993 Oct;14(10):491–9. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- [6].Cormier JN, Panelli MC, Hackett JA, Bettinotti MP, Mixon A, Wunderlich J, et al. Natural variation of the expression of HLA and endogenous antigen modulates CTL recognition in an in vitro melanoma model. IntJCancer. 1999 Mar 01;80(5):781–90. doi: 10.1002/(sici)1097-0215(19990301)80:5<781::aid-ijc24>3.0.co;2-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kageshita T, Hirai S, Ono T, Hicklin DJ, Ferrone S. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: association with disease progression. AmJPathol. 1999 Mar;154(3):745–54. doi: 10.1016/S0002-9440(10)65321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Degen E, Williams DB. Participation of a novel 88-kD protein in the biogenesis of murine class I histocompatibility molecules. JCell Biol. 1991 Mar;112(6):1099–115. doi: 10.1083/jcb.112.6.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Degen E, Cohen-Doyle MF, Williams DB. Efficient dissociation of the p88 chaperone from major histocompatibility complex class I molecules requires both beta 2-microglobulin and peptide. JExpMed. 1992 Jun 01;175(6):1653–61. doi: 10.1084/jem.175.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sadasivan B, Lehner PJ, Ortmann B, Spies T, Cresswell P. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996 Aug;5(2):103–14. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- [11].Diedrich G, Bangia N, Pan M, Cresswell P. A role for calnexin in the assembly of the MHC class I loading complex in the endoplasmic reticulum. JImmunol. 2001 Feb 01;166(3):1703–9. doi: 10.4049/jimmunol.166.3.1703. [DOI] [PubMed] [Google Scholar]
- [12].Li S, Sjogren HO, Hellman U, Pettersson RF, Wang P. Cloning and functional characterization of a subunit of the transporter associated with antigen processing. Proc Natl Acad Sci U S A. 1997 Aug 5;94(16):8708–13. doi: 10.1073/pnas.94.16.8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ortmann B, Copeman J, Lehner PJ, Sadasivan B, Herberg JA, Grandea AG, et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997 Aug 29;277(5330):1306–9. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- [14].Solheim JC, Harris MR, Kindle CS, Hansen TH. Prominence of beta 2-microglobulin, class I heavy chain conformation, and tapasin in the interactions of class I heavy chain with calreticulin and the transporter associated with antigen processing. J Immunol. 1997 Mar 1;158(5):2236–41. [PubMed] [Google Scholar]
- [15].Hughes EA, Cresswell P. The thiol oxidoreductase ERp57 is a component of the MHC class I peptide-loading complex. CurrBiol. 1998 Jun 04;8(12):709–12. doi: 10.1016/s0960-9822(98)70278-7. [DOI] [PubMed] [Google Scholar]
- [16].Lindquist JA, Jensen ON, Mann M, Hammerling GJ. ER-60, a chaperone with thiol-dependent reductase activity involved in MHC class I assembly. EMBO J. 1998 Apr 15;17(8):2186–95. doi: 10.1093/emboj/17.8.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Paquet ME, Cohen-Doyle M, Shore GC, Williams DB. Bap29/31 influences the intracellular traffic of MHC class I molecules. JImmunol. 2004 Jun 15;172(12):7548–55. doi: 10.4049/jimmunol.172.12.7548. [DOI] [PubMed] [Google Scholar]
- [18].Seifert U, Maranon C, Shmueli A, Desoutter JF, Wesoloski L, Janek K, et al. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol. 2003 Apr;4(4):375–9. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- [19].Shepherd JC, Schumacher TN, Ashton-Rickardt PG, Imaeda S, Ploegh HL, Janeway CA, Jr., et al. TAP1-dependent peptide translocation in vitro is ATP dependent and peptide selective. Cell. 1993 Aug 13;74(3):577–84. doi: 10.1016/0092-8674(93)80058-m. [DOI] [PubMed] [Google Scholar]
- [20].Androlewicz MJ, Anderson KS, Cresswell P. Evidence that transporters associated with antigen processing translocate a major histocompatibility complex class I-binding peptide into the endoplasmic reticulum in an ATP-dependent manner. ProcNatlAcadSciUSA. 1993 Oct 01;90(19):9130–4. doi: 10.1073/pnas.90.19.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].York IA, Chang SC, Saric T, Keys JA, Favreau JM, Goldberg AL, et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002 Dec;3(12):1177–84. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- [22].Saric T, Beninga J, Graef CI, Akopian TN, Rock KL, Goldberg AL. Major histocompatibility complex class I-presented antigenic peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J Biol Chem. 2001 Sep 28;276(39):36474–81. doi: 10.1074/jbc.M105517200. [DOI] [PubMed] [Google Scholar]
- [23].Ortmann B, Androlewicz MJ, Cresswell P. MHC class I/beta 2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 1994 Apr 28;368(6474):864–7. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- [24].Suh WK, Cohen-Doyle MF, Fruh K, Wang K, Peterson PA, Williams DB. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science. 1994 May 27;264(5163):1322–6. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- [25].Grandea AG, III, Androlewicz MJ, Athwal RS, Geraghty DE, Spies T. Dependence of peptide binding by MHC class I molecules on their interaction with TAP. Science. 1995 Oct 06;270(5233):105–8. doi: 10.1126/science.270.5233.105. [DOI] [PubMed] [Google Scholar]
- [26].Garbi N, Tan P, Diehl AD, Chambers BJ, Ljunggren HG, Momburg F, et al. Impaired immune responses and altered peptide repertoire in tapasin-deficient mice. NatImmunol. 2000 Sep;1(3):234–8. doi: 10.1038/79775. [DOI] [PubMed] [Google Scholar]
- [27].Grandea AG, III, Golovina TN, Hamilton SE, Sriram V, Spies T, Brutkiewicz RR, et al. Impaired assembly yet normal trafficking of MHC class I molecules in Tapasin mutant mice. Immunity. 2000 Aug;13(2):213–22. doi: 10.1016/s1074-7613(00)00021-2. [DOI] [PubMed] [Google Scholar]
- [28].Peh CA, Burrows SR, Barnden M, Khanna R, Cresswell P, Moss DJ, et al. HLA-B27-restricted antigen presentation in the absence of tapasin reveals polymorphism in mechanisms of HLA class I peptide loading. Immunity. 1998 May;8(5):531–42. doi: 10.1016/s1074-7613(00)80558-0. [DOI] [PubMed] [Google Scholar]
- [29].Tan P, Kropshofer H, Mandelboim O, Bulbuc N, Hammerling GJ, Momburg F. Recruitment of MHC class I molecules by tapasin into the transporter associated with antigen processing-associated complex is essential for optimal peptide loading. JImmunol. 2002 Feb 15;168(4):1950–60. doi: 10.4049/jimmunol.168.4.1950. [DOI] [PubMed] [Google Scholar]
- [30].Petersen JL, Hickman-Miller HD, McIlhaney MM, Vargas SE, Purcell AW, Hildebrand WH, et al. A charged amino acid residue in the transmembrane/cytoplasmic region of tapasin influences MHC class I assembly and maturation. JImmunol. 2005 Jan 15;174(2):962–9. doi: 10.4049/jimmunol.174.2.962. [DOI] [PubMed] [Google Scholar]
- [31].Lehner PJ, Surman MJ, Cresswell P. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line .220. Immunity. 1998 Feb;8(2):221–31. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- [32].Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity. 2002 Jan;16(1):87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- [33].Zarling AL, Luckey CJ, Marto JA, White FM, Brame CJ, Evans AM, et al. Tapasin is a facilitator, not an editor, of class I MHC Peptide binding. JImmunol. 2003 Nov 15;171(10):5287–95. doi: 10.4049/jimmunol.171.10.5287. [DOI] [PubMed] [Google Scholar]
- [34].Howarth M, Williams A, Tolstrup AB, Elliott T. Tapasin enhances MHC class I peptide presentation according to peptide half-life. ProcNatlAcadSciUSA. 2004 Aug 10;101(32):11737–42. doi: 10.1073/pnas.0306294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol. 2007 Aug;8(8):873–81. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- [36].Belicha-Villanueva A, McEvoy S, Cycon K, Ferrone S, Gollnick SO, Bangia N. Differential contribution of TAP and tapasin to HLA class I antigen expression. Immunology. 2008 Jan 11;124:112–20. doi: 10.1111/j.1365-2567.2007.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ogino T, Bandoh N, Hayashi T, Miyokawa N, Harabuchi Y, Ferrone S. Association of tapasin and HLA class I antigen down-regulation in primary maxillary sinus squamous cell carcinoma lesions with reduced survival of patients. Clin Cancer Res. 2003 Sep 15;9(11):4043–51. [PubMed] [Google Scholar]
- [38].Seliger B, Schreiber K, Delp K, Meissner M, Hammers S, Reichert T, et al. Downregulation of the constitutive tapasin expression in human tumor cells of distinct origin and its transcriptional upregulation by cytokines. Tissue Antigens. 2001 Jan;57(1):39–45. doi: 10.1034/j.1399-0039.2001.057001039.x. [DOI] [PubMed] [Google Scholar]
- [39].Abarca-Heidemann K, Friederichs S, Klamp T, Boehm U, Guethlein LA, Ortmann B. Regulation of the expression of mouse TAP-associated glycoprotein (tapasin) by cytokines. ImmunolLett. 2002 Oct 01;83(3):197–207. doi: 10.1016/s0165-2478(02)00104-9. [DOI] [PubMed] [Google Scholar]
- [40].Wechsler-Reya R, Sakamuro D, Zhang J, Duhadaway J, Prendergast GC. Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J Biol Chem. 1997 Dec 12;272(50):31453–8. doi: 10.1074/jbc.272.50.31453. [DOI] [PubMed] [Google Scholar]
- [41].Copeman J, Bangia N, Cross JC, Cresswell P. Elucidation of the genetic basis of the antigen presentation defects in the mutant cell line .220 reveals polymorphism and alternative splicing of the tapasin gene. EurJImmunol. 1998 Nov;28(11):3783–91. doi: 10.1002/(SICI)1521-4141(199811)28:11<3783::AID-IMMU3783>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- [42].Gao B, Williams A, Sewell A, Elliott T. Generation of a functional, soluble tapasin protein from an alternatively spliced mRNA. Genes Immun. 2003 Dec 11; doi: 10.1038/sj.gene.6364043. [DOI] [PubMed] [Google Scholar]
- [43].Marincola FM, Shamamian P, Alexander RB, Gnarra JR, Turetskaya RL, Nedospasov SA, et al. Loss of HLA haplotype and B locus down-regulation in melanoma cell lines. The Journal of Immunology. 1994 Aug 01;153(3):1225–37. [PubMed] [Google Scholar]
- [44].Radka SF, Kostyu DD, Amos DB. A monoclonal antibody directed against the HLA-Bw6 epitope. JImmunol. 1982 Jun;128(6):2804–6. [PubMed] [Google Scholar]
- [45].Ogino T, Wang X, Kato S, Miyokawa N, Harabuchi Y, Ferrone S. Endoplasmic reticulum chaperone-specific monoclonal antibodies for flow cytometry and immunohistochemical staining. Tissue Antigens. 2003 Nov;62(5):385–93. doi: 10.1034/j.1399-0039.2003.00114.x. [DOI] [PubMed] [Google Scholar]
- [46].Wang X, Campoli M, Cho HS, Ogino T, Bandoh N, Shen J, et al. A method to generate antigen-specific mAb capable of staining formalin-fixed, paraffin-embedded tissue sections. JImmunolMethods. 2005 Apr;299(1-2):139–51. doi: 10.1016/j.jim.2005.02.006. [DOI] [PubMed] [Google Scholar]
- [47].Bangia N, Lehner PJ, Hughes EA, Surman M, Cresswell P. The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. EurJImmunol. 1999 Jun;29(6):1858–70. doi: 10.1002/(SICI)1521-4141(199906)29:06<1858::AID-IMMU1858>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- [48].Lutz PM, Cresswell P. An epitope common to HLA class I and class II antigens, Ig light chains, and beta 2-microglobulin. Immunogenetics. 1987;25(4):228–33. doi: 10.1007/BF00404692. [DOI] [PubMed] [Google Scholar]
- [49].Russo C, Ng AK, Pellegrino MA, Ferrone S. The monoclonal antibody CR11-351 discriminates HLA-A2 variants identified by T cells. Immunogenetics. 1983;18(1):23–35. doi: 10.1007/BF00401353. [DOI] [PubMed] [Google Scholar]
- [50].Everett MW, Edidin M. Tapasin increases efficiency of MHC I assembly in the endoplasmic reticulum but does not affect MHC I stability at the cell surface. J Immunol. 2007 Dec 1;179(11):7646–52. doi: 10.4049/jimmunol.179.11.7646. [DOI] [PubMed] [Google Scholar]