Abstract
Genetic capsid labeling of conditionally replicative adenoviruses (CRAds) with fluorescent tags offers a potentially more accurate monitoring of those virotherapy agents in vivo. The capsid of an infectivity-enhanced CRAd, Ad5/3, delta 24, was genetically labeled with monomeric red fluorescent protein 1 (mRFP1) or its advanced derivative, “mCherry,” to evaluate the utility of each red fluorescent reporter and the benefit of CRAd capsid labeling for noninvasive virus tracking in animal tumor models by a new spectral imaging approach. Either reporter was incorporated into the CRAd particles by genetic fusion to the viral capsid protein IX. Following intratumoral injection, localization and replication of each virus in orthotopic breast cancer xenografts were analyzed by spectral imaging and verified by quantitative polymerase chain reaction. Fluorescence in tumors increased up to 2,000-fold by day 4 and persisted for 5 to 7 weeks, showing oscillatory dynamics reflective of CRAd replication cycles. Capsid labeling in conjunction with spectral imaging thus enables direct visualization and quantification of CRAd particles in tumors prior to the reporter transgene expression. This allows for noninvasive control of CRAd delivery and distribution in tumors and facilitates quantitative assessment of viral replication. Although mCherry appeared to be superior to mRFP1 as an imaging tag, both reporters showed utility for CRAd imaging applications.
Virotherapy with conditionally replicative adenoviruses (CRAds) represents a promising approach to treatment of cancer. Genetic manipulations with human adenovirus (Ad) have enabled cancer selectivity of viral replication as well as modification of the Ad’s natural tropism to enhance its infectivity for cancer cells.1 Despite exciting preclinical data, adenoviral cancer gene therapy approaches have yet to display therapeutic efficacy in human clinical trials.2 Insufficient transduction of cancer cells by Ad and the lack of noninvasive methods of monitoring CRAd localization and replication in vivo represent major obstacles that hamper clinical improvement of the CRAd-based virotherapy approach.2–4 In that regard, noninvasive imaging technologies offer a means to better understanding in vivo biology of CRAds and improving their therapeutic efficacy.
The first vector imaging applications, based on reporter transgene expression from a replication-defective viral genome, allowed visualization of viral infection in cells and proved feasibility of viral targeting to a specific cell population.5–8 Subsequently, expression of imaging transgenes from replicative virus genomes offered a tool for tracking viral replication and spread within the infected tissues.9 However, the conventional vector imaging strategies, critically dependent on the viability of infected cells to generate a transgene expression–dependent imaging signal, are incompatible with the cell-killing function of oncolytic agents and, therefore, inadequate for assessment of CRAd accumulation and spread in tumors.
An alternate strategy to engineering of an imaging reporter into Ad vectors involves its genetic fusion with the viral minor capsid protein IX (pIX). This approach not only allows for coordinated expression of a reporter transgene with this virus-specific protein to mirror the onset of Ad replication 10 but also results in direct incorporation of an imaging modality in the viral capsid.11–14 This strategy holds a potential utility for detection and quantification of viral particles in tumors prior to the onset of the virus-specific gene expression and replication as well as direct tracking of the replicating viral mass in vivo. 11,15
Red fluorescent reporters of the “mFruit” family with far-red emission spectra, such as mCherry, hold potential for live imaging in animal models of human diseases owing to deeper tissue penetration and higher photostability of their fluorescence. 16 Among the monomeric red fluorescent proteins, derivatives of Discosoma sp protein DsRed, mCherry offers the longest wavelength fluorescence (only 3 nm longer than monomeric red fluorescent protein 1 [mRFP1]), higher photostability (t0.5 to bleach = 68 seconds vs 6.2 seconds for mRFP1), and brightness of the signal (27% of DsRed vs 21% for mRFP1), as well as faster maturation time (15 minutes versus < 1 hour for mRFP1) and higher tolerance to N-terminal fusions compared to mRFP1. Despite a slightly lower quantum yield (0.22 vs 0.25 for mRFP1), these characteristics altogether make mCherry a superior imaging modality to mRFP1 owing to a more robust predicted fluorescence yield. 17,18
One of the obstacles for in vivo imaging of the red fluorescent reporters is a background autofluorescence, which is an intrinsic property of mammalian cells, tissues, and diet and is indistinguishable from the red fluorescence of protein reporters by the traditional fluorescent imaging technique. 19 A new spectral imaging technology enables specific detection of fluorescent reporters in animal cells and tissues by their distinct spectral profiles. Special image processing software can then “unmix” spectral profiles of fluorescent reporters from those of autofluorescence and “extract” a specific signal component from a digital image. 20
The technical feasibility of genetic capsid labeling of Ad vectors has been established, 11–13, 15,21–23 and replication-competent Ads with red fluorescent tags on the capsid, including mRFP1, have recently been generated 15,23 and validated in vivo by a conventional fluorescent imaging approach. 15 However, the benefits of Ad capsid labeling for in vivo imaging applications have not been demonstrated directly. On the other hand, to our knowledge, no capsid-labeled CRAd agent has been reported to date, except the ones generated in our most recent study. 23 Moreover, no capsid-labeled CRAd has ever been evaluated in vivo for imaging potential.
In the present study, we sought to develop a novel CRAd, fluorescently labeled with a new-generation red fluorescent reporter mCherry, and to evaluate mCherry as a CRAd imaging modality relative to its ancestor mRFP1 in a mouse model of breast cancer using a new spectral imaging technique. The capsid labeling modifications were introduced in the genome of the infectivity-enhanced CRAd, known as Ad5/3, delta 24, with previously demonstrated strong oncolytic potential in animal models of cancer. 24
The present study for the first time demonstrates the utility of Ad capsid labeling for direct visualization of the CRAd in tumors immediately after its intratumoral (IT) administration as well as the utility of spectral imaging as a tool for distinguishing the virus-associated fluorescence from background autofluorescence and noninvasive tracking of CRAd replication in tumors. Potential implications of the spectral imaging technology for clinical and basic research are discussed.
Materials and Methods
Cells
Breast cancer cells DY36T2 and 2LMP were maintained in modified improved minimal essential medium (MEM), (GIBCO, Gaithersburg, MD), supplemented with 10% fetal bovine serum (FBS). Human fibroblasts VH-10 were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, HyClone Laboratories, Logan, UT); MCF7 and LCC6 breast cancer cell lines were maintained in an alpha-modified MEM (Mediatech, Manassas, VA); MDA-MB-231, MDA-MB-468, and MDA-MB-435* 25 breast cancer cells were grown in low-glucose DMEM (HyClone Laboratories), supplemented with MEM vitamins, MEM nonessential amino acids, and 1 mM sodium pyruvate; BT-474 breast cancer cells were maintained in RPMI-1640 medium (Mediatech) supplemented with 10 μg/mL insulin, 4.5 g/L glucose, 10 mM HEPES, and 1 mM sodium pyruvate; human embryonic kidney 293 (HEK 293) cells (Microbix, Toronto, ON) and A549 lung adenocarcinoma cells were cultured in DMEM/F12 (50:50) (Mediatech). All media were supplemented with 10% FBS (HyClone Laboratories), 2 mM L-glutamine, 100 IU/mL penicillin, and 100 μg/mL streptomycin (all Mediatech). All cells were grown in monolayers in plastic flasks at 37°C in a humidified atmosphere with 5% CO2.
Recombinant Ad Construction
The genome of the CRAd with mCherry red fluorescent protein on the capsid was constructed by homologous recombination in Escherichia coli (BJ5183 strain) 26,27 using a fiber gene–modified AdEasy-1 backbone vector AdEz-F5/3 (Ad5ΔE1/ΔE3-F5/3)14 and a modified pShuttle vector pSlΔ24-pIX-mCherry. This shuttle vector contained the mCherry coding sequence, inserted downstream of the Ad5 minor capsid pIX gene to generate a C-terminal pIX fusion and a 24-basepair deletion in the Ad5 E1A gene coding sequence (delta 24). 28 The plasmid with mCherry reporter coding sequence, kindly provided by Dr. Roger Tsien (Howard Hughes Medical Institute, University of California, San Diego, La Jolla, CA), was used to amplify mCherry open reading frame by polymerase chain reaction (PCR) using the following sense and antisense primers: 5′-ATTAGCTAGCCGGAATGGTGA-3′ and 5′-TATGACTA GTCGACTTACTTGTACAGCTCGTCCATGCCG-3′ (Nhe I site is underlined, Sal I site is shown in bold, Spe I is shown in italics). The resulting product was digested with Nhe I and Spe I (generates Nhe I–compatible cohesive ends) and subcloned into the unique Nhe I site of the pSlΔ24, pIX-tk 23 by replacing the HSV-tk (tk) gene coding sequence. Construction of the control virus with mRFP1 instead of mCherry and the corresponding shuttle vector pSlΔ24-pIX-mRFP1 derived from the previously described pScsΔ2429 was recently reported elsewhere. 23 Both shuttle vectors carried a linker sequence, encoding a 10–amino acid peptide (SADYKDDDDK) with an octapeptide flag motif (shown in bold), inserted downstream of the polypeptide IX coding sequence in place of the stop codon, followed by Nhe I (pSlΔ24-pIX-mCherry) or Bam H1 (pSlΔ24-pIX-mRFP1) restriction sites. Following homologous recombination and colony selection on kanamycin, the recombinants were screened by a recombinant-specific PCR assay of our own design (unpublished data, 2007) and verified by partial sequencing. Following a restriction test (Hind III, Nhe I), DNA isolated from positive clones was linearized with Pac I and transfected into the HEK 293 helper cells with lipofectamine LTX (Invitrogen, Carlsbad, CA). The expression of pIX fusions was under the control of the native pIX promoter. The deletion of the E3 genomic region, including nucleotides 28,133–30,817, was described previously. 14
Generation and Molecular Validation of CRAds
All viruses were generated and plaque-purified in E1-complementing retinoblast 911 cells according to standard procedures. 30 The CRAds were propagated in A549 adenocarcinoma cells, whereas the replication-defective virus was amplified in helper HEK 293 cells. All viruses were purified by double cesium chloride (CsCl) ultracentrifugation (AdEasy system, Qbiogene, Carlsbad, CA) and dialyzed against phosphate-buffered saline (PBS) containing 0.5 mM Mg 2+, 0.9 mM Ca 2+, and 10% glycerol. Purified preparations of each virus were analyzed for viral particle titer, determined by absorbance at 260 nm (A260) using a conversion factor 1 A260 = 1.1 × 1012 vp/mL, as described earlier. 31 The genomic identity of each virus was verified by conventional PCR assays and sequencing of some PCR products. (The PCR primer sequences used in those assays are available on request.) The absence of contamination with the wild-type E1A sequence–containing genomes (replication-competent adenovirus [RCA]) in virus preparations was confirmed by an in-house developed PCR assay of our own design (unpublished data, 2007). Incorporation of pIX-mRFP1 and pIX-mCherry fusion proteins in the viral capsids and their molecular integrity were confirmed by Western blot. Equal amounts (1 × 1010 vp) of each virus were subjected to SDS-PAGE gel electrophoresis and transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked and hybridized with a 1:1,000 dilution of mouse monoclonal ANTI-FLAG M2 primary antibody (Sigma, St. Louis, MO) for 1 hour. The membrane was washed in Tris-buffered saline–Tween and incubated with a secondary horseradish peroxidase–conjugated sheep antimouse antibody diluted 1:1,000 (Amersham Biosciences, Piscataway, NJ) for 1 hour. For normalization, the same PVDF filters were rehybridized with monoclonal (4D2) antibodies against an Ad5 fiber tail at 1:5,000 dilution. Antibody binding was detected using an electrogenerated chemiluminescence (ECL) detection system (GE Healthcare, Piscataway, NJ). Hybridization procedures were performed using a Western breeze kit (Invitrogen, Carlsbad, CA). For molecular-weight reference, prestained protein standards (Bio-Rad, Hercules, CA) were used.
Infectious titers were determined in HEK 293 helper cells by the 50% tissue culture infectious dose (TCID50) method. The limiting dilution end point and infectious titer (TCID50/mL) were determined by the Kärber equation: TCID50/mL = 10[1 + D(S-0.5)]/V, where D is the logarithm of the dilution factor (eg, D = 1 for the 10-fold serial dilution), S is the logarithm of the initial dilution plus the sum of ratios of infection-positive wells per total wells at each subsequent dilution, and V is the volume of the diluted vector (in mL) used for inoculation.
In Vitro Screening of Breast Cancer Cells for CRAd Infection and Replication
Nine different breast cancer cell lines, mentioned above, were infected with the mCherry-labeled CRAd at the multiplicity of infection (MOI) of 1 TCID50/cell and subjected to fluorescent microscopy analysis 48 hours later. To assess the cytotoxic effect of the oncolytic agents on each cell line, the same CRAd-infected cells were further incubated for 2 weeks and stained with 0.2% crystal violet dye in buffered formalin for 20 minutes followed by multiple washes with water.
In Vitro Analysis of CRAd Cell Binding and Replication by Real-Time PCR
DY36T2 or MDA-MB-361 cells were incubated with the labeled viral particles at 4°C at an MOI of 1.5 × 104 vp/cell to synchronize the infection process at the receptor-binding step. Virus binding to cell membranes was visualized by fluorescence microscopy using an inverted IX-70 microscope (Olympus, Melville, NY), equipped with a Magnifire digital charge-coupled device camera (Optronics, Golets, CA) 5 to 10 hours postinfection. Images were taken with 40× objective using red (mCherry or mRFP1) and blue (Hoechst) excitation filters. Images were overlaid by Adobe Photoshop C2 software. CRAd internalization and trafficking through the cytoplasm to the nuclei were observed 2 to 5 hours after changing cell incubation temperature from 4° to 37°C.
Cell binding of mRFP1- and mCherry-CRAds was further compared by quantification of Ad genomic DNA recovered from the infected cells. Specifically, DY36T2 or MDA-MB-361 cells were plated in 12-well plates in the recommended media and grown to 90% confluency. Each cell line was infected with the mCherry-CRAd or the mRFP1-CRAd at an MOI of 500 vp/cell. After 1 hour of infection at 4°C, the cells were washed three times with PBS to remove the unbound virus. Next, total DNA was isolated from infected cells using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). Quantitative real-time polymerase chain reaction (qPCR) for the E4 region of the Ad5 genome was then performed to determine the number of bound infectious particles. The following primers were used for amplification of the Ad5 E4 region: 5′-GGAGTGCGCCGAGACAAC-3′ (forward) and 5′-ACTACGTCCGGCGTTCCAT-3′ (reverse). Detection was performed with the following probe: 6FAM-TGG CATGACACTACGACCAACAC-TAMRA. A housekeeping β-actin gene was used for normalization. For replication analysis, DY36T2 or MDA-MB-361 cells were infected at an MOI of 100 vp/cell and harvested 12, 24, and 60 hours postinfection. All other experimental details were exactly as described above for the cell binding experiment.
Generation of Orthotopic Xenografts in Nude Mice
Female athymic mice, 6 weeks of age (Frederick Animal Production Program, Frederick, MD), were allowed to acclimate at the University of Alabama for 2 weeks before use. Strict adherence to the University of Alabama at Birmingham Institutional Animal Care and Use Committee–approved protocol was maintained throughout the study. MDA-MB-361 or DY36T2 breast cancer cells (5 × 107) in sterile PBS (100 μL) were injected into the mammary fat pads of 3% isoflurane-anesthetized mice to orthotopically implant xenografts. Tumors were allowed to grow for 2 weeks to become approximately 5 mm in diameter before they were used for in vivo experiments.
Spectral Imaging of CRAds
Orthotopic breast carcinoma tumors, resulting from orthotopic implantation of DY36T2 or MDA-MB-361 cells, were directly injected with 3 × 1010 vp of either mCherry-CRAd or mRFP1-CRAd. Prior to IT virus injection, background autofluorescence images of each tumor were collected. Tumors were imaged 5 minutes and 1 hour after virus injection using a Leica MZFLIII fluorescent stereomicroscope (Meyer Instruments, Inc., Houston, TX), equipped with a HQ560/55xHQ excitation filter and a 600LP emission filter and a “Nuance” spectral imaging camera N-MS1-500-FL(CRI, Inc., Woburn, MA). A liquid-crystal tunable wavelength filter in the camera was set for collection of emission images from 600 to 720 nm in 5 nm increments. To enable comparison across time points and avoid potential signal saturation, the spectral imaging data sets (600–720 nm) were collected at 100-, 400- and 5,000-millisecond exposures per image, using 4 × 4 binning for fluorescence and 1 × 1 binning for bright field images. All tumors were imaged twice weekly for the duration of the experiment, after the initial data collections.
Composite images (unmixed composites) were generated for each tumor by unmixing the spectral signature of the red fluorescent reporter fusion proteins (pIX-mCherry or pIX-mRFP1) from those of background autofluorescence using a spectral library that was compiled from numerous spectral profiles collected from uninjected control tumors under the same conditions.
The original image of each tumor at a given time point was analyzed using the Nuance 2.4.2 image quantification software (CRI, Inc) after extraction of its 630 nm component (excitation maximum, shifted in the pIX fusions from the 607 to 610 nm excitation maximums of the red fluorescent proteins) and normalizing signals as photons per millisecond of exposure. Background autofluorescence of each 630 nm wavelength image was quantified within three different locations outside the tumor area of the same image, and the mean value was subtracted from the tumor fluorescence. Of note, data for one mouse from each group with the DY36T2 tumors were excluded from the charts, presented in Figure 6, for technical reasons.
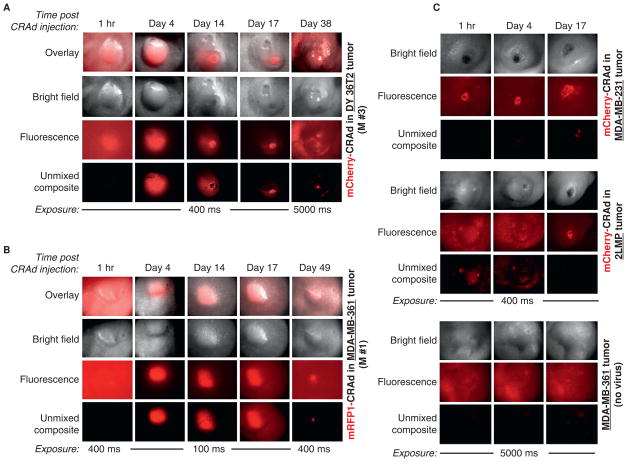
Figure 6.
Noninvasive tracking of CRAd replication in tumors by spectral imaging. Arrays of images of representative mammary fat pad DY36T2 (A) or MDA-MB-361 (B) tumors showing dynamic changes in the tag-specific red fluorescence in the course of intratumoral replication of the labeled CRAds. Repetitive spectral imaging of the same tumors (corresponding to the M #3 and M #1 charts on Figure 7, A and D, respectively) was performed before (1 hour) and after (days 4–49) the onset of viral replication. All types of tumors received intratumoral injection of the same dosage (viral particles) of mCherry- (A, C) or mRFP1-CRAd (B). Overlay (bright field plus fluorescence) images were made by using Adobe Photoshop C2 software. Other details are as in Figure 5. C, Representative images of other types of breast cancer tumors MDA-MB-231 and 2LMP with a substantially lower permissiveness to the CRAd infection and weaker fluorescence (top and middle panels), following injection with the same dose of mCherry-CRAd as in A and B, at indicated time points. Representative images of a single MDA-MB-361 tumor that received no CRAd are shown as control for autofluorescence. Bright foci on original images represent autofluorescence of ulcerated (wounded) areas on the surface of some tumors.
To take into account lower infectious titer of the mCherry CRAd compared to mRFP1-CRAd, all fluorescence intensity values obtained in tumors with replicating CRAds (from day 4 until the last day of the experiment) were normalized to equal the number (1 × 109) of infectious (TCID50) units per tumor.
The efficiency of CRAd capsid labeling (pIX incorporation) was assessed by quantification of fluorescent signals, emitted by 1010 CsCl-purified particles of each capsid-labeled virus in 10% glycerol containing dialysis buffer (10 mM Tris-HCl; pH 8.0; 50 mM NaCl; 2 mM MgCl2) sampled on a glass slide, using the same approach as above. Unlabeled pIX-WT CRAd was used as a nonfluorescent virus/background control.
Statistical Analysis
All values were compared between groups (mCherry vs mRFP1) or (DY36T2 vs MDA-MB-361) by nonparametric Mann-Whitney test using GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA). The differences were assumed to be statistically significant when the p values were ≤ .05.
Results
Construction and Molecular Validation of the mCherry-Labeled CRAd
To achieve capsid labeling of an infectivity-enhanced CRAd, Ad5/3-delta 24, with a fluorescent tag, optimal for in vivo imaging applications, a red fluorescent protein mCherry was genetically fused to the C-terminus of the Ad minor capsid pIX. To facilitate transduction of tumors, this virus also carried a fiber modification, genetically replacing the C-terminal “knob” domain of the Ad5 fiber with its counterpart from the Ad serotype 3 (Ad3) fiber. 24,30 In addition, the entire E3 region of the Ad genome was deleted to prevent genomic instability of the pIX-modified virus (see Discussion). 32,33 A previously reported CRAd with mRFP1 on the capsid 23 was evaluated in this study in parallel as an isogenic variant with an alternate imaging modality.
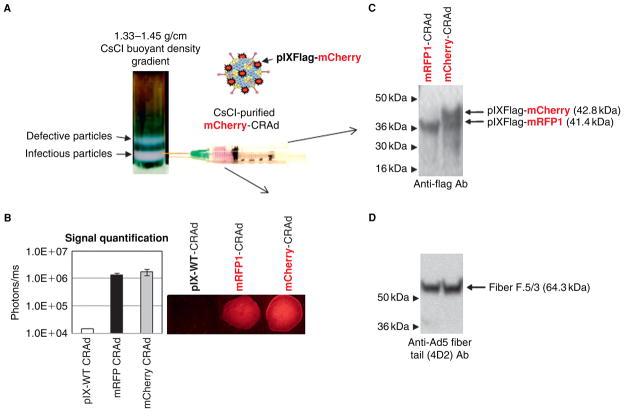
Infectious viral particles of either the mCherry- (Figure 1A) or the mRFP1-labeled CRAd (data not shown) preparations represented a predominant fraction (lower band) on CsCl-density gradients. Fluorescent labeling of the viral particles was indicated by the pink color of the purified virus bands, visible even without fluorescence imaging. Capsid labeling of each CRAd was further evidenced by strong fluorescence of the purified virus solutions, sampled on a glass slide, whereas a CRAd with wild-type pIX (pIX-WT CRAd) showed no fluorescence (Figure 1B). The efficiency of capsid incorporation of the pIX fusions was estimated by quantification of viral particle–associated fluorescence (see Figure 1B, chart), as well as by Western blot analysis (Figure 1C). Subsequent detection of the modified fiber protein on the same blots verified that an equal number of viral particles were used for comparison (Figure 1D). These experiments demonstrated similar capsid incorporation efficiencies of the pIX-mCherry and pIX-mRFP1 fusions. Furthermore, Coomassie gel staining showed comparable amounts of other Ad capsid proteins, suggesting that capsid labeling did not affect incorporation of other structural proteins into the viral particles and the CRAd capsid assembly (data not shown).
Figure 1.
Molecular validation of CRAd capsid labeling. A, Capsid incorporation of mCherry-pIX fusion was evidenced by the color of the purified CRAd solution pooled from the CsCl gradient. A wild-type pIX-containing virus (pIX-WT-CRAd) was used as an unlabeled control. B, An equal number (1010) of purified viral particles of control (white bar), mRFP1- (black bar), and mCherry- (gray bar) CRAds were sampled on a glass slide and the red fluorescence was quantified by spectral imaging (bar chart). C, Incorporation of the pIX fusions with imaging reporters into the CRAd capsids was verified by Western blot using antiflag antibodies detecting a flag sequence at the C-terminus of pIX in the purified CRAd particles. Arrowheads indicate the positions of molecular-weight markers. D, The same blot (as C) was reprobed with the Ad5 fiber tail–specific antibodies (4D2) to confirm equal loading of each CRAd on the gel.
In Vitro Selection of Breast Cancer Cells for Establishing In Vivo Xenograft Tumor Models
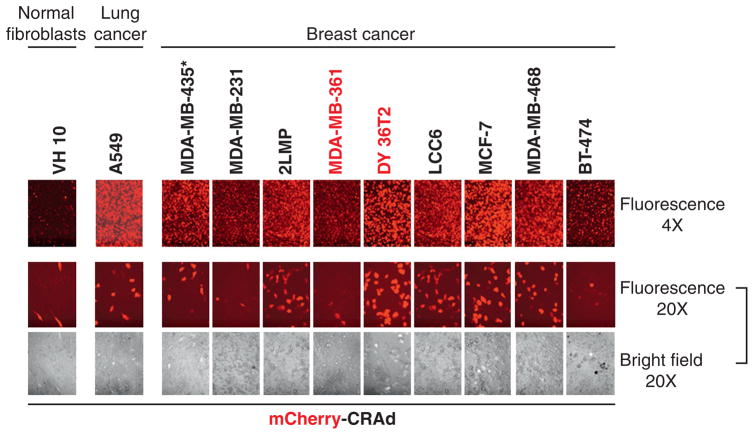
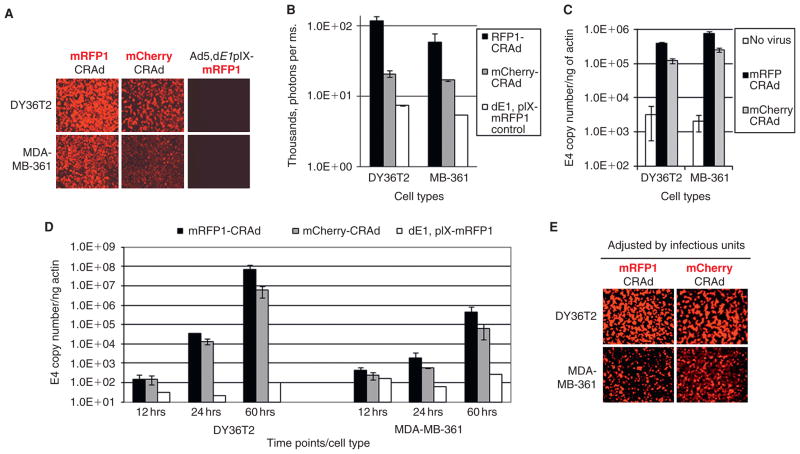
To establish an optimal animal model of human breast cancer for in vivo CRAd imaging studies, a panel of available breast cancer cell lines were analyzed for their ability to be infected with and support replication of the capsid-labeled infectivity-enhanced CRAd Ad5/3-delta 24-pIX-mCherry (mCherry-CRAd) using fluorescence microscopy. Cell lines DY36T2 and MCF7 displayed the highest levels of fluorescent labeling (Figure 2). The strong fluorescence in DY36T2 and MCF7 cells, indicative of efficient CRAd infection and replication, correlated with high susceptibility of those cells to the CRAd-mediated oncolysis. The latter was evidenced by early development of cytopathic effect (see Figure 2, bright field images) followed by an efficient killing of the infected cells 2 weeks later, as revealed by crystal violet assay (data not shown). Conversely, a relatively weak fluorescence observed in BT-474, MDA-MB-231, and MDA-MB-361 cell lines was consistent with the absence of cell-killing effect or its dramatic delay compared to the DY36T2-infected cells (see Figure 2 and data not shown). On the basis of these observations, DY36T2 and MDA-MB-361 cells lines were chosen as highly susceptible for (positive) and resistant to (negative) the Ad5/3-delta-24 CRAd transduction and/or replication, respectively. The choice was also based on the known ability of these cells to grow in vivo and generate orthotopic mammary fat pad tumors in athymic nude mice.
Figure 2.
Susceptibility of various breast cancer cells to Ad5/3, delta24-pIX-mCherry virus infection in vitro. A panel of breast cancer cell lines were infected with Ad5/3, delta24-pIX-mCherry (mCherry-CRAd) particles at a multiplicity of infection of 1 TCID50/cell, and fluorescence was visualized 48 hours later by fluorescence microscopy at different magnifications using 4× or 20× objectives. Bright field and fluorescent images of the same views were taken in parallel to assess the early cytopathic effect of the replicating CRAd. Normal human fibroblasts VH-10 and lung carcinoma A549 cells were used as negative and positive controls for the cancer-selective virus replication, respectively. DY36T2 and MDA-MB-361 cell lines (highlighted in red) were chosen for implantation in mice to generate tumor xenografts. The asterisk indicates that the origin of the MDA-MB-435 cell line is highly controversial and was recently suggested to be a melanoma rather than a breast cancer line. However, the controversy has not been fully resolved.25
Biologic Characterization and Comparison of the Capsid-Labeled CRAds In Vitro
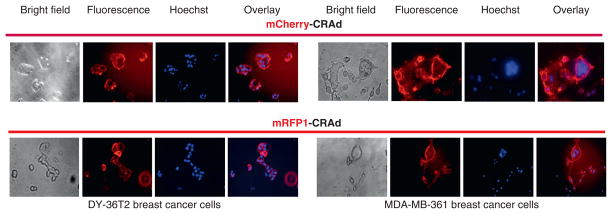
The labeled CRAd particles demonstrated the ability to bind to cell surface receptors, as revealed by fluorescent labeling of the chosen target cells, following several hours of incubation with an excess of purified viruses at 4°C (Figure 3). These conditions allow receptor binding but not internalization step of the Ad infection. Furthermore, gradual translocation of the cell surface–associated fluorescence first to the cytoplasm and then to the nuclei of the infected cells was observed on changing the cell incubation temperature from 4° to 37°C (data not shown). The ability of the fluorescent CRAds to bind cell surface receptors with subsequent internalization and intracellular trafficking confirmed the full biologic activity of the capsid-modified particles. Of note, the mRFP1-generated cell surface fluorescence bleached out detectably faster than that produced by the mCherry tag on prolonged exposure of the culture cells to the excitation light (data not shown).
Figure 3.
Validation of biologic activity of labeled CRAd particles in vitro. DY36T2 and MDA-MB-361 cells were infected by mCherry- or mRFP1-CRAds at 4°C at the multiplicity of infection of 1.5 × 104 vp/cell to block internalization but allow binding of labeled virions to their cognate (Ad3) receptors. Cell surface labeling by fluorescent viral particles was visualized 5 hours later by fluorescence microscopy (Fluorescence images). Prior to viral infection, cells were stained with Hoechst reagent for 10 minutes to label nuclear DNA with blue fluorescence (Hoechst images). Hoechst and fluorescence images were overlaid (Overlay images) using Photoshop C2 software. Bright field, fluorescence, and Hoechst images were obtained from the same view field by using the corresponding optical filters.
The physical (viral particle) titers of the mCherry- and the mRFP1-CRAds were similar (3.2 × 1012 and 4.3 × 1012 vp/mL, respectively) and only slightly lower than that of unmodified control pIX-WT CRAd (6 × 1012 vp/mL), suggesting only a minor, if any, effect of the pIX modifications on the CRAd production. In contrast, the infectious titer of the mCherry-CRAd (1.66 × 1010TCID50/mL) was sixfold lower than that of the mRFP1 virus (1 × 1011TCID50/mL) (p = .017). Lower infectivity was observed for several independent preparations of the mCherry-CRAd. In line with this finding, the intensity of red fluorescence in mCherry-CRAd-infected cells was significantly lower (p = .008) compared to the mRFP1-CRAd-infected cells (Figure 4, A and B).
Figure 4.
Biologic characterization of capsid-labeled CRAds in vitro by imaging and molecular methods. A, Fluorescence in DY36T2 and MDA-MB-361 cells was visualized by fluorescent microscopy (with 4× objective) 48 hours postinfection with either mCherry- or mRFP1-CRAd and incubation at 37°C. A capsid-labeled replication-defective virus Ad5, dE1, pIX-mRFP1 was used as a negative control for Ad replication. B, Quantification of fluorescence intensity in CRAd-infected cells from images shown on A by using Nuance 2.4.2 spectral imaging software. Fluorescent signals are presented as photons per millisecond (ms) of image exposure. C, Comparison of the labeled CRAds for their cell binding ability. An equal number of mRFP1- and mCherry-CRAd particles were used to infect either DY36T2 or MDA-MB-361 breast cancer cells in vitro for 1 hour at 4°C (multiplicity of infection [MOI] = 500 vp/cell). Total DNA was isolated from the cells for quantitative PCR analysis of the Ad5 genomic DNA (E4) copy number and the housekeeping β-actin gene for normalization. CRAd-infected cell samples were analyzed in triplicates. D, Comparison of the CRAd DNA replication in vitro. Each cell line was infected with an equal number of viral particles of either mRFP1- or mCherry-CRAds at an MOI of ≈100 vp/cell, and total DNA was harvested 12, 24, and 60 hours later. The rest of the analysis was the same as described for C. CRAd-infected cell samples were analyzed in triplicates. Black and gray bars correspond to mRFP1- and mCherry-CRAds, respectively. White bars correspond to replication-defective control virus dE1, pIX-mRFP1. Standard deviation is shown by brackets on all charts. All graphs are presented in log scale. E, Fluorescence microscopy analysis of intracellular fluorescence, similar to the one shown in A, except equal infectious units (TCID50 units) of each CRAd were used. Specifically, six times more viral particles of the mCherry-CRAd was used for infection to correlate infectious particles with produced cell fluorescence during the linear phase of CRAd replication in vitro. Images were taken between 48 and 60 hours postinfection.
To verify the difference in infectivity by molecular methods, equal numbers of the mCherry- and the mRFP1-CRAd particles were compared for their ability to bind to the selected breast cancer culture cells using a real-time qPCR approach. The results demonstrated 3.35-fold (p = .002) and 3-fold (p = .01) more particles of the mRFP1-CRAd bound to DY36T2 and MDA-MB-361 cells compared to the mCherry-CRAd particles, respectively (Figure 4C). On the other hand, approximately twice as many particles of the mRFP1- (p = .021) and the mCherry-CRAds (p = .01) were bound to MDA-MB-361 cells than to DY36T2 cells (see Figure 4C), suggesting that the former cells express approximately twice as much Ad3 receptor.
To further verify the difference in infectivity between the viruses, DNA replication of the mCherry- and the mRFP1-CRAds was compared in each cell type at various times following infection with an equal number of particles of each virus. Both CRAds showed a robust amplification of genomic DNA between 24 and 72 hours postinfection, which, according to our pilot study data (not shown), corresponded to the linear portion of the CRAd growth curves. In line with the observed difference in infectious titers, the mRFP1-CRAd produced increasingly more genomic DNA in DY36T2 cells over time compared to the mCherry-CRAd (Figure 4D). The calculated differences between the amplified mRFP1- and the mCherry-CRAd genomes in DY36T2 cells at 24 and 60 hours postinfection were 2.4-fold (p = .014) and 12.2-fold (p = .132), respectively. Replication of both CRAds in MDA-MB-361 cells was substantially attenuated as a detectable increase in the genomic DNA occurred only 24 hours postinfection and the amount of both CRAd genomes at 60 hours postinfection was over 100-fold lower (p = .01) than in the DY36T2 cells (see Figure 4D). Thus, the lower number of particles bound to each cell type and fewer replicated CRAd genomes, resulting in reduced reporter gene expression and fluorescence in the infected cells (see Figure 4A), were consistent with the lower infectious titer of the mCherry-CRAd observed by the TCID50 method.
As predicted, adjusting the number of the mCherry-CRAd infectious particles by a factor of 6 (the CRAd infectious titer ratio) resulted in nearly equal levels of fluorescence in the CRAd-infected cells between 48 and 60 hours postinfection (Figure 4E ). This experiment suggested that during the linear phase of the CRAd replication in vitro, the level of intracellular fluorescence is almost proportional to the number of infectious viral particles used for infection.
Direct Visualization of Tumor-Injected Labeled Viral Particles by Spectral Imaging
This part of the study was aimed at evaluation of CRAd capsid labeling as a tool for direct visualization and quantification of the oncolytic agents in tumors shortly after their direct IT administration as a means to control the accuracy of virus delivery and IT distribution. In this regard, we employed spectral imaging as a unique technology, capable of distinguishing a specific fluorescent signal associated with the labeled viral particles from autofluorescence background.
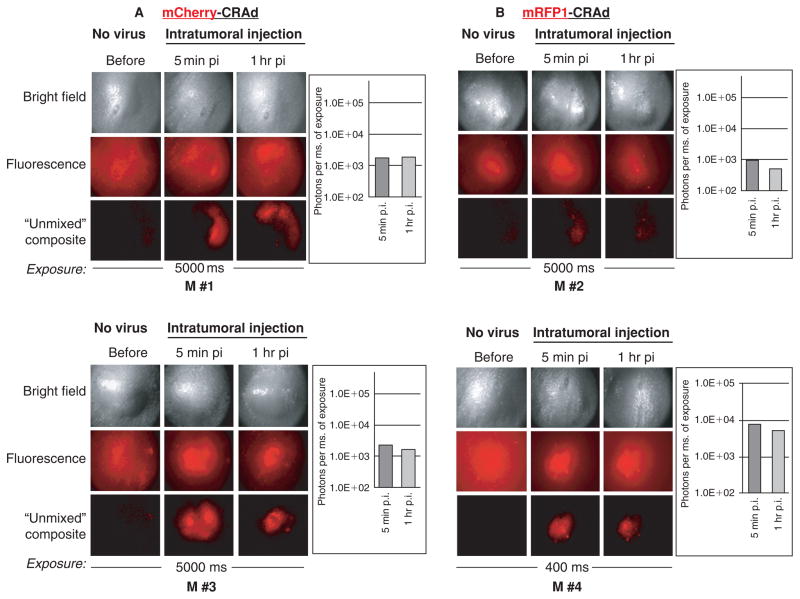
A specific red fluorescence could be detected in orthotopic mammary fat pad tumors (~5–7 mm in diameter) following IT injection of 3 × 1010 fluorescently labeled viral particles. These imaging experiments also revealed high tumor-to-tumor variability in the input signal intensity (Figure 5).
Figure 5.
Direct visualization of labeled viral particles in xenograft tumors. An equal number of purified viral particles (3 × 1010 vp in 10 μL of saline) of capsid-labeled mCherry-CRAd (A) or mRFP1-CRAd (B) were delivered into mammary fat pad tumors orthotopically implanted in athymic nude mice by a single intratumoral injection. Sample images of four representative tumors (M #1–M #4) are shown to reflect signal variability. Images were captured before and 5 minutes or 1 hour after injection of labeled viruses at 5,000 or 400 milliseconds (M #4) exposure. Signals on original unsaturated fluorescent images were quantified (bar graphs) using Nuance 2.4.2 spectral imaging software, as described in Materials and Methods. Unmixed composite images are the result of digital processing of the original (fluorescence) images by subtracting fluorescence components with spectral profiles, distinct from those of fluorescent proteins (mRFP1/mCherry). Bright field panels represent phase contrast images of the same tumors. Exposure times in milliseconds (ms) are indicated below each figure panel. M #1 to M #4 indicate names of the tumors/mice within each experimental tumor group, whose replication profiles are depicted in Figure 6. M #1 and M #3 belong to DY36T2 tumor groups; M #2 and M #4 belong to the MDA-MB-361 tumor groups; M #1 and M #3 received injection of the mCherry-CRAd; M #2 and M #4 were injected with the mRFP1-CRAd.
Although the intensity of the virus-associated fluorescence in most injected tumors was high enough to be clearly visualized without the spectral “unmixing” procedure (see Figure 5A, panel M #3; see Figure 5B, panel M #4), in some tumors, this image-processing step was absolutely critical for distinguishing the virus-associated fluorescence from background autofluorescence (see Figure 5A, panel M #1; see Figure 5B, panel M #2). Quantification of fluorescence emitted by the tumor-delivered CRAds revealed only a subtle, up to twofold decrease in signal intensity within 1 hour following virus administration (see Figure 5, panels M #2, M #3, M #4, bar graphs) in most tumors, suggesting diffusion of the CRAds from or redistribution within the tumor via tumor vasculature and/or owing to viral particle disintegration.
Live Tracking of CRAd Replication in Orthotopic Breast Cancer Mouse Models
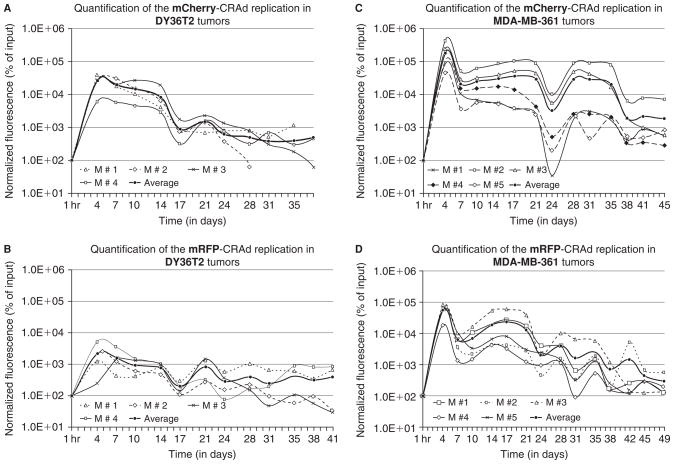
The replication dynamics of each CRAd in orthotopic DY36T2 or MDA-MB-361 xenografts was analyzed by repetitive noninvasive spectral fluorescence imaging of the mRFP1 or mCherry reporters and signal quantification for each time point in each individual tumor. We observed a marked increase in red fluorescence in DY36T2 tumors after injection of either mCherry (Figure 6A, Figure 7A) or mRFP1 (Figure 7B) CRAds. Despite some variations in signal intensity among tumors of each experimental group, the major peaks of fluorescence were consistently observed in all injected DY36T2 tumors between days 4 and 7 as well as between days 10 and 14 after virus administration. These were invariably followed by a sharp signal decrease on day 17. Another minor peak of intensity in the DY36T2 tumors was consistently observed around day 21 (see Figure 7, A and B).
Figure 7.
Fluorescence intensity profiles of capsid-labeled CRAd replication in breast cancer xenografts. Fluorescence profiles were obtained for each tumor by plotting integrated density of each fluorescent signal measured at each indicated time point (days after injection) as a percentage of the input signal (fluorescence intensity of injected labeled particles 1 hour after injection) following normalization of signals for each time point, starting from day 4, to 109 infectious (TCID50) units of each CRAd. The “Average” curves represent mean values-based charts for each experimental group. Four mice/tumors were analyzed in experimental groups A and B (DY36T2 tumors) and five mice/tumors in groups C and D (MDA-MB361 tumors). Fluorescence signals were quantified as described in Materials and Methods.
Unexpectedly, tumors derived from the MDA-MB-361 cells that were less permissive for the CRAd replication in vitro (see Figure 4, A–D) produced a substantially stronger fluorescence in vivo than the DY36T2-derived tumors (see Figure 6B and Figure 7, C and D). The CRAd fluorescence profiles in the MDA-MB-361 tumors were discernibly different from those in the DY36T2 tumors. Although the position of the major peak was the same as in DY36T2 tumors (day 4), this peak was sharper in the MDA-MB-361-derived tumors. The second fluorescence peak in the MDA-MB-361 tumors occurred between days 15 and 20, followed by a sharp signal decrease on day 24 (see Figure 7, C and D). Despite the strong variability in signal intensity, the relative position of both peaks was consistent among all tumors within each experimental group. Three additional peaks were observed on days 28, 35, and 42, indicating productive CRAd replication even at later time points. A specific fluorescent signal could still be detected in each tumor type for as long as 38 to 49 days after a single dose injection of each virus (see Figure 6 and Figure 7). In contrast to MDA-MB-361 tumors, the level of fluorescence in tumors derived from other breast cancer cell lines (MDA-MB-231 and 2LMP) with low in vitro permissiveness for the CRAds was dramatically lower (see Figure 6C) and in line with the in vitro results.
Owing to the fact that the mCherry-CRAd showed a sixfold lower infectivity than the mRFP1-CRAd but equal intensity of capsid labeling, a direct comparison of fluorescent yields of the two in vivo replicating CRAds would be inadequate. In this regard, to estimate relative fluorescence yields of each CRAd during its replication in tumors, we reasoned that signal intensities, measured for each time point of CRAd replication (on day 4 and thereafter), could be normalized to an equal number of infectious CRAd particles per tumor. The validity of such adjustment was rationalized by the following two assumptions: (1) the fluorescence level in infected cells/tumors is proportional to imaging reporter gene expression, which, in turn, is proportional to the number of replicated viral genomes, and (2) the fluorescence profiles observed in vivo for each CRAd and tumor type between 24 and 96 hours (4 days) following CRAd infection correspond to the linear phase of the CRAd growth curves in vitro and, therefore, are directly proportional to the number of infectious viral particles used for tumor transduction. These assumptions were based on the results of fluorescence microscopy and the CRAd DNA replication in vitro analyses presented in Figure 4.
Comparison of the infectious units–normalized signals revealed 11-fold (p = .028) and a 3.6-fold (p = .032) higher fluorescence yields for the mCherry-CRAd compared to the mRFP1-CRAd in DY36T2 (see Figure 7, A and B) and MDA-MB-361 tumors (see Figure 7, C and D), respectively. On the other hand, whereas the mCherry-CRAd produced, on average, only 7.6-fold (p = .016) stronger signals in the MDA-MB-361 tumors compared to the DY36T2 tumors (see Figure 7, A and C), the mean intensity of the major peaks for the mRFP1-CRAd replication profiles was 24-fold higher (p = .016) in MDA-MB-361 tumors (see Figure 7, B and D).
Whereas maximal amplification of the signal in the DY36T2 tumors injected with the mCherry-CRAd was, on average, 263-fold (see Figure 7A), the mRFP1-CRAd showed only a 23-fold signal increase (see Figure 7B). In contrast, the average fluorescence increase in MDA-MB-361 tumors was approximately 2,000-fold (see Figure 7C) and 546-fold (see Figure 7D) for the mCherry- and the mRFP1-CRAds, respectively.
Ex Vivo Analysis of Isolated and Sliced Tumors by Spectral Imaging
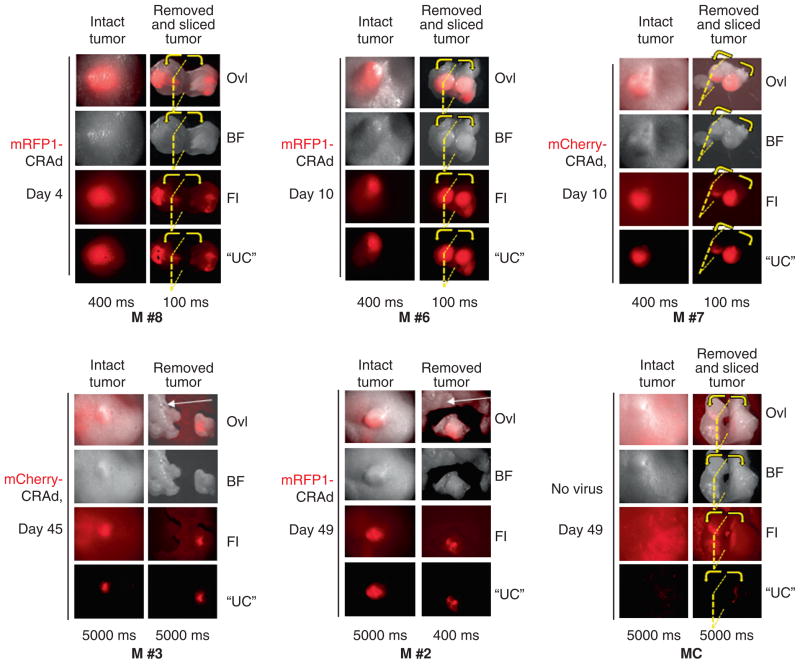
Imaging of surgically removed tumors revealed a high level of tissue-associated autofluorescence background (Figure 8C). Spectral imaging was also found critical for identification and localization of the specific, virus-associated red fluorescence in isolated (see Figure 8, panels M #2, M #3) or isolated and sliced (see Figure 8, panels M #6, M #7, M #8) tumors as well as signal quantification (data not shown). No statistically significant difference in signal intensity was observed between the tumors before (“intact tumor”) and after their surgical removal (“removed tumor”) (see Figure 8, panels M #2, M #3). In contrast, substantially stronger signals were measured in the same tumors after slicing (“removed and sliced tumor” on Figure 8, panels M #6, M #7, M #8), suggesting that the intensity of in vivo detected signals is largely dependent on the location of the signal (virus-infected cells) relative to the tumor surface. Importantly, ex vivo imaging of the sliced tumors revealed a substantial signal distribution across the tumor mass. Nevertheless, no tumors showed viral spread throughout the entire tumor volume (see Figure 8, panels M #6, M #7, M #8), suggesting limited CRAd lateralization.
Figure 8.
Ex vivo analysis of isolated and sliced tumors by spectral imaging. Fluorescent signals in tumors were analyzed by spectral imaging before and after surgical removal and slicing of the tumors. Images taken noninvasively (“intact tumor”) are shown on the left-side columns of each image panel. Images of the same tumors after their surgical removal (panels M #2 and M #3) or both removal and slicing (panels M #6, M #7, M #8, MC) are shown on the right-side columns (“removed tumor” or “removed and sliced tumor”) for signal comparison. Images of six representative tumors (M #2, M #3, M #6, M #7, M #8, MC) surgically removed at different time points (day 4, day 10, and day 45/49) are displayed. Tumor cross-section plane is shown by a yellow dotted line, and the direction of the slice separation is indicated by yellow arrows. All shown tumors are of MDA-MB-361 cell origin. A tumor that did not receive any virus was used as autofluorescence control (panel “MC”). Exposure times in milliseconds are indicated below each image column. Shorter exposures are shown for cross-sections or some isolated tumor images (M #2) to avoid signal saturation. Tissues of nontumor origin shown for autofluorescence level comparison are indicated by white arrows (panels M #2 and M #3). BF = bright field images; Fl= fluorescence images; Ovl = overlay; “UC” = “unmixed composites”. Other details as in Figure 5 and Figure 6. Panels M #6, M #7, and M #8 display images of additional tumors of the MDA-MB-361 experimental groups dissected early in the experiment (day 4 or day 10 time point). Tumors M #2, M #6 and M #8 received mRFP1-CRAd; tumor M #3 and M #7 received mCherry-CRAd.
Quantification of CRAd Genomes in Tumors at Various Times Post–Virus Administration
To obtain direct evidence for CRAd replication in tumors on the molecular level, the viral genomic DNA was specifically detected and quantified by real-time qPCR in the whole-tumor lysates prepared on the last day of the experiment (days 38/41 for the DY36T2 tumor groups and days 45/49 for the MDA-MB-361 tumor groups) as well as from the tumors of each experimental group on day 4 and day 10 post–virus administration. The tumors representing these earlier time points of the experiment were part of each experimental group from the beginning of the study but removed on days 4 and 10 post–virus administration for ex vivo tumor imaging (tumors M #6, M #7, and M #8 on Figure 8 ) and subsequent CRAd genome quantification. Comparison of the CRAd genomic DNA content in tumors at three different time points (day 4, day 10, and the last day) demonstrated a close correlation between the intensity of fluorescent signal and the amount of the tumor-contained virus (data not shown), thereby supporting CRAd replication in the xenografts.
Discussion
The aim of this study was to develop a conditionally replicative adenovirus (CRAd) carrying an advanced-generation red fluorescent reporter mCherry on its capsid and to evaluate the utility of this reporter for in vivo imaging of oncolytic agents by using a new spectral imaging technique as well as the benefits of the CRAd capsid labeling strategy for in vivo CRAd imaging applications. Use of mCherry as an imaging tag on the CRAd capsid was rationalized by the advanced optical characteristics of this fluorescent reporter, established by previous studies. 18 Although mCherry was recently employed for genetic labeling of human immunodeficiency virus 1 (HIV-1)34, it has not been used in the context of Ad capsid labeling or any Ad-based vectors.
The new oncolytic agents generated in this study on the basis of the Ad5/3-delta 24 CRAd carried either mRFP1 or mCherry fluorescent proteins, genetically fused to the Ad structural pIX, represented by 240 copies per Ad5 capsid. To achieve cancer targeting of viral replication, both CRAds carried a 24-basepair deletion in the E1A gene that abolishes its binding to tumor suppressor protein retinoblastoma (Rb). This deletion prevents the E1A-mediated induction of the cell proliferation mechanism, necessary for the onset of Ad replication during the Ad5 infection of normal cells. As a result, the delta 24 mutation renders Ad replication selective for cancer cells with the defective Rb/p16 signaling pathway, where Rb blocking is no longer required to induce the Ad replication-permissive state. 28
In addition, both capsid-labeled CRAds carried a “serotype chimera” fiber modification (F5/3), providing one of the strongest known infectivity enhancement effects in most types of cancer cells both in vitro and in vivo. 24,31,35 Replacement of the fiber knob domain with its counterpart from the Ad3 results in ablation of the natural coxsackie and adenovirus receptor (CAR) tropism and retargeting of the virus to a different set of receptors, currently believed to be CD46 and/or CD80/86. 36,37
Finally, both CRAd genomes had a fully deleted E3 region to avoid viral DNA packaging constraints, known to result from the lack or underincorporation of pIX in the Ad capsids, and to prevent possible genomic instability of the pIX-modified virions. 32,33
Although capsid-labeled Ads with replication-defective and replication-competent genomes have been described, 11–13,15,21,22 the viruses generated in this and another recent study of ours 23 are, to the best of our knowledge, the first reported CRAd agents with imaging tags on the capsids. This study also provides the first imaging validation of a capsid-labeled CRAd in an orthotopic animal model of cancer.
The biologic properties of the labeled CRAd particles have been characterized, and no discernible defects in their assembly were found as a result of pIX-mCherry or pIX-mRFP1 incorporation in the CRAd capsids. Importantly, the efficiency of incorporation of each fusion protein into the viral capsids and the resulting efficiency of capsid-associated red fluorescence were comparable between the two CRAds (see Figure 1, B–D). Besides, the modified CRAd particles were biologically active, as evidenced by their ability to fluorescently label cell surface receptors (see Figure 3) with subsequent internalization and intracellular trafficking of the labeled capsids or their fluorescent components.
However, for unknown reasons, the infectious titers of the mCherry CRAd preparations were lower than those of the mRFP1 virus. Owing to the high degree of similarity in size and structure between mCherry and mRFP1 protein tags, the presence of mCherry per se was unlikely to account for the observed loss of infectivity. In fact, the pIX-mCherry fusion is slightly longer than pIX-mRFP1 partially owing to a slightly longer linker sequence between the flag and the fluorescent protein. Although the linker size difference is negligible, the sequence of the linker peptide is different from that in the mRFP1 fusion and could possibly impose some adverse effect on the pIX-mCherry fusion structure and/or stability. Some evidence for its effect on stability comes from the smeary appearance of the pIX-mCherry band on the Western blot (see Figure 1C). Degradation of the fusion could potentially result in its underincorporation or partial loss from the particles, which would reduce fluorescence of the mCherry-CRAd particles. Although pIX-free Ad particles are known to be noninfectious, 32,33 this explanation is not consistent with our results of quantification of capsid-associated fluorescence (see Figure 1B). After all, it is not clear if the lower infectious titers have any genetic determination at all.
Whereas the fluorescence microscopy approach failed to detect any difference between mRFP1- and mCherry-CRAds in their cell-binding ability at 4°C (see Figure 3), a more sensitive qPCR-based approach revealed approximately three times more particles of the mRFP1-CRAd bound to both types of breast cancer cells (see Figure 4C) at the same MOI (viral particle/cell). This finding agreed with higher infectivity of the mRFP1-CRAd preparation, which was also consistent with the higher number of replicated viral genomes (see Figure 4D) and a discernibly stronger fluorescence observed in the mRFP1-CRAd-infected cells 48 hours postinfection (see Figure 4, A and B). A barely detectable fluorescence in the cells infected with a nonreplicative mRFP1-labeled control virus Ad5 dE1pIX-mRFP1 (see Figure 4, A and B) supported replication dependence of the fluorescent signal in the CRAd-infected cells. This observation was consistent with an earlier study demonstrating a correlation between mRFP1 fluorescence and replication of a capsid-labeled Ad carrying wild-type E1 genes.15
In contrast to the cell surface–labeling assay, a PCR-based cell-binding analysis was able to demonstrate that MDA-MB-361 breast cancer cells can bind twice as many CRAd particles compared to DY36T2 cells (see Figure 4C). This suggested that lower fluorescence intensity produced in the CRAd-infected MDA-MB-361 cells 48 hours postinfection in vitro (Figure 4, A and B) was rather due to the lower rate of the delta 24 CRAd replication in these cells than to the lack of the Ad3 receptor and, hence, poor transduction. This conclusion was further supported by a direct comparison of the CRAd DNA replication dynamics in vitro (see Figure 4D). Wheras MDA-MB-361 cells appeared to be less permissive for the delta 24 CRAd replication, also evidenced by poor sensitivity to CRAd-mediated oncolysis in vitro (data not shown), xenograft tumors derived from MDA-MB-361 cells unexpectedly showed up to 24-fold stronger fluorescence compared to the DY36T2 tumors following infection with either of the delta 24 CRAds (see Figure 6 and Figure 7). This finding supports the notion that some biologic properties of cancer cells can differ dramatically in vitro and in vivo.
Although an in vivo imaging study of a capsid-labeled adenovirus has been reported, 15 the feasibility of direct visualization of labeled Ad particles in tumors without contribution of the reporter gene expression and viral replication has not been addressed or demonstrated. Moreover, a conventional fluorescent imaging was used for signal detection as opposed to more sensitive spectral imaging. Our CRAd imaging approach uniquely allowed detection of subtle changes in intensity and distribution of the CRAd particle–associated fluorescence within the first hour after IT CRAd administration. The observed reduction in signal intensity probably reflects virus diffusion from and/or within the tumor mass via tumor vasculature or possibly even physical disintegration of the CRAds. Detection of CRAds shortly after administration and quantification of the specific, viral particle–associated fluorescence provides a valuable noninvasive tool for controlling CRAd delivery and redistribution in tumor tissues prior to the onset of viral replication. Importantly, this also offers a basis for quantitative assessment of changes in IT fluorescence levels, allowing normalization of the total tissue fluorescence at a given time point to the input signal. Calculation of relative signal amplification can be used as a means to quantitatively assess and compare CRAd replication rates in vivo.
Owing to the strong variability in the measured intensity of the tumor-injected (input) fluorescence, we were unable to observe any significant difference in fluorescent yields between the mRFP1- and the mCherry-CRAd particles in the tumors. On the other hand, a substantial difference in infectious titers between the mCherry- and mRFP1-CRAd preparations did not allow for an accurate quantitative comparison of fluorescent yields produced by replicating mRFP1- and mCherry-CRAds in the animal tumor model either. On the other hand, if equal infectious units of each CRAd were to be used, it would result in an unequal number of tumor-injected particles (unequal input signal) and, given the signal calculation approach (as a percentage of the input fluorescence), would lead to underestimation of the relative fluorescence increase for the replicating mCherry-CRAd. Thus, a formally accurate comparison of the CRAd fluorescent yields in tumors would require that both the number of replicating viral genomes and the level of input fluorescence (number of injected labeled virions) be the same. Unfortunately, such a condition could not be met in this study.
Therefore, the only conceivable way of estimating the relative fluorescent yields of the two CRAds in tumors was to make an approximation by normalizing all fluorescence values obtained for the replicating viruses in tumors to equal CRAd infectious (TCID50) units, assuming proportionality of the fluorescent signals to the number of replicated viral genomes and, correspondingly, to the number of the input infectious CRAd units during the linear phase of CRAd replication. The validity of those assumptions was supported by our in vitro “simulation” experiments (see Figure 4E).
The maximal fluorescent yields for the mCherry-CRAd in tumors estimated with such approximation were severalfold (from 3.6- to 11-fold) higher than those for the mRFP1-CRAd depending on the tumor type (see Figure 7, C and D, and Figure 7, A and B, respectively). Although the mCherry reporter appeared to provide some advantage over mRFP1, both fluorescent reporters showed utility as imaging tags for noninvasive monitoring of CRAds in mouse models of breast cancer.
A continuous noninvasive tracking of the CRAds in breast cancer xenografts by spectral fluorescent imaging showed a conspicuous, up to 2,000-fold increase in fluorescence intensity relative to the input signal (see Figure 7). Together with the oscillating nature of each tumor’s fluorescence profile and remarkable longevity of the signal persistence (for nearly 2 months), this observation suggested that both CRAds underwent continuous replication cycles in the tumors. CRAd replication in the xenografts was also evidenced by direct correlation of the red fluorescence with the number of CRAd genomes, recovered from tumors at various time points of the experiment (data not shown). Thus, red fluorescence can be effectively used to noninvasively and quantitatively monitor replication of the capsid-labeled CRAds in tumors. This finding is consistent with an earlier study using mRFP1-labeled Ad particles. 15
A high degree of similarity was observed between replication profiles of the mCherry- and the mRFP1-CRAds within each type of tumor (see Figure 7, A and B, and Figure 7, C and D), whereas substantially more difference in the profiles was noted between different tumor types (see Figure 7, A and C, and Figure 7, B and D). This might reflect biochemical differences between the two types of cancer cells, resulting in drastically different ability of MDA-MB-361 cells to support the Ad5/3, delta-24 CRAd replication in vitro and in vivo.
Although the timing of the major fluorescence peak in the CRAd replication profiles observed in this study was consistent with that observed in the previous study, 15 the longevity of the fluorescence (viral replication) in tumors, observed in our study, was substantially longer and the oscillatory dynamics of viral replication was much more conspicuous (see Figure 7).
Finally, imaging of dissected tumors demonstrated an efficient penetration of both mCherry and mRFP1 fluorescence through the mouse skin, with only a slight signal attenuation. Much stronger signal attenuation and scatter were observed for the tumor tissue, as revealed by imaging of cross sections of tumors after slicing (see Figure 8). This suggests that the depth of imaging signal localization in a solid tumor is a critical factor, determining the intensity of detected fluorescent signal. Spectral imaging of sliced tumors also provided important information regarding the signal distribution across the tumor mass (see Figure 8), suggesting that the viral spread in the tumor mass following IT injection was limited (see Figure 8, panels M #8, M #6, M #7). This was consistent with the earlier observations of limited lateralization of CRAds in vivo. 15 Given that the focus of this study was primarily CRAd imaging capabilities, the oncolytic potential of the viruses in tumors was not analyzed.
Owing to the high levels of fluorescence maintained in the tumors throughout the CRAd replication cycles from day 4 to day 17, the spectral definition (unmixing) capacity of the spectral imaging method was not crucial for identification of the mRFP1- or mCherry-specific fluorescence during this initial period of the CRAd in vivo life cycle (see Figure 6, days 4–17). However, the spectral signature–based signal identification, uniquely provided by this method, became increasingly important at later time points when signal levels approached tissue autofluorescence levels. The spectral unmixing function also demonstrated utility for ex vivo imaging of isolated and sliced tumors owing to high levels of background fluorescence (see Figure 8C).
In the aggregate, this study represents the first imaging validation of a tumor-targeted CRAd agent with fluorescently labeled capsid in an animal tumor model and for the first time demonstrates the feasibility of direct visualization and quantification of fluorescent viral particles in orthotopic breast carcinoma tumors immediately or shortly after their intratumoral administration with the help of a new spectral imaging technology. This method demonstrated important utility for sensitive monitoring of fluorescent CRAd agents and signal quantification in tumors by means of circumventing the problem of tissue autofluorescence.
Spectral imaging holds another important potential for separating the virus particle–associated fluorescence from that of the free form of a reporter fusion owing to tentative changes in the spectral profile of the signal that could possibly be induced by capsid incorporation of the reporter. This could allow highly specific monitoring of the replicating viral mass, critical for accurate tracking of replicating CRAds. This capacity could not be achieved in the present study (our unpublished observations, 2009) owing to technical limitations and would require improved resolution of the method as well as a more advanced instrumentation, which may become available in the years to come. Spectral imaging in conjunction with fluorescent labeling of CRAd agents thus holds promise for facilitating real-time analysis of the tagged CRAds in cancer models and thereby improving the clinical efficacy of the future virotherapy agents. This study represents the first step on this path.
Acknowledgments
This work was supported by University of Alabama School of Dentistry Institute of Oral Health Research and Dean’s Faculty Development Awards to AVB, the UAB Small Animal Imaging Shared Facility NIH Research Core Grant (P30CA013148) and the Department of Defense (BC050034) award to KRZ and the NIH Research Grant (5R01CA121187-03) to DTC. We thank Dr. Maria-Veronica Lopez for critical reading of the manuscript and Dr. Roger Y. Tsien for providing the mCherry gene-containing plasmid.
Footnotes
Financial disclosure of reviewers: None reported.
References
- 1.Nettelbeck DM. Virotherapeutics: conditionally replicative adenoviruses for viral oncolysis. Anticancer Drugs. 2003;14:577–84. doi: 10.1097/00001813-200309000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Makower D, Rozenblit A, Kaufman H, et al. Phase II clinical trial of intralesional administration of the oncolytic adenovirus ONYX-015 in patients with hepatobiliary tumors with correlative p53 studies. Clin Cancer Res. 2003;9:693–702. [PubMed] [Google Scholar]
- 3.Ahn M, Lee SJ, Li X, et al. Enhanced combined tumor-specific oncolysis and suicide gene therapy for prostate cancer using M6 promoter. Cancer Gene Ther. 2009;16:73–82. doi: 10.1038/cgt.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ranki T, Kanerva A, Ristimäki A, et al. A heparan sulfate-targeted conditionally replicative adenovirus, Ad5. pk7-Delta24, for the treatment of advanced breast cancer. Gene Ther. 2007;14:58–67. doi: 10.1038/sj.gt.3302830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Martin R, Raidl M, Hofer E, et al. Adenovirus-mediated expression of green fluorescent protein. Gene Ther. 1997;4:493–5. doi: 10.1038/sj.gt.3300408. [DOI] [PubMed] [Google Scholar]
- 6.Chen BE, Lendvai B, Nimchinsky EA, et al. Imaging high-resolution structure of GFP-expressing neurons in neocortex in vivo. Learn Mem. 2000;7:433–41. doi: 10.1101/lm.32700. [DOI] [PubMed] [Google Scholar]
- 7.MacLaren DC, Gambhir SS, Satyamurthy N, et al. Repetitive, non-invasive imaging of the dopamine D2 receptor as a reporter gene in living animals. Gene Ther. 1999;6:785–91. doi: 10.1038/sj.gt.3300877. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri TR, Cao Z, Rodríguez-Burford C, et al. A non-invasive approach for monitoring breast tumor cells during therapeutic intervention. Cancer Biother Radiopharm. 2007;17:205–12. doi: 10.1089/108497802753773829. [DOI] [PubMed] [Google Scholar]
- 9.Mittal SK, McDermott MR, Johnson DC, et al. Monitoring foreign gene expression by a human adenovirus-based vector using the firefly luciferase gene as a reporter. Virus Res. 1993;28:67–90. doi: 10.1016/0168-1702(93)90090-a. [DOI] [PubMed] [Google Scholar]
- 10.Vales LD, Darnell JE. Promoter occlusion prevents transcription of adenovirus polypeptide IX mRNA until after DNA replication. Genes Dev. 1989;3:49–59. doi: 10.1101/gad.3.1.49. [DOI] [PubMed] [Google Scholar]
- 11.Le LP, Everts M, Dmitriev IP, et al. Fluorescently-labeled adenovirus with pIX-EGFP for vector detection. Mol Imaging. 2004;3:105–16. doi: 10.1162/15353500200404100. [DOI] [PubMed] [Google Scholar]
- 12.Meulenbroek RA, Sargent KL, Lunde J, et al. Use of adenovirus protein IX (pIX) to display large polypeptides on the virion—generation of fluorescent virus through the incorporation of pIX-GFP. Mol Ther. 2004;9:617–24. doi: 10.1016/j.ymthe.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Tang Y, Le LP, Matthews QL, et al. Derivation of a triple mosaic adenovirus based on modification of the minor capsid protein IX. Virology. 2008;377:391–400. doi: 10.1016/j.virol.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 14.San Martín C, Glasgow JN, Borovjagin A, et al. Localization of the N-terminus of minor coat protein IIIa in the adenovirus capsid. J Mol Biol. 2008;383:923–34. doi: 10.1016/j.jmb.2008.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le LP, Le HN, Dmitriev IP, et al. Dynamic monitoring of oncolytic adenovirus in vivo by genetic capsid labeling. J Natl Cancer Inst. 2006;98:203–14. doi: 10.1093/jnci/djj022. [DOI] [PubMed] [Google Scholar]
- 16.Shu X, Shaner NC, Yarbrough CA, et al. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–47. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 17.Campbell RE, Tour O, Palmer AE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–82. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaner NC, Campbell RE, Steinbach PA, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 19.Troy T, Jekic-McMullen D, Sambucetti L, et al. Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models. Mol Imaging. 2004;3:9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 20.Tam JM, Upadhyay R, Pittet MJ, et al. Improved in vivo whole-animal detection limits of green fluorescent protein-expressing tumor lines by spectral fluorescence imaging. Mol Imaging. 2007;6:269–76. [PubMed] [Google Scholar]
- 21.Li J, Le L, Sibley DA, et al. Genetic incorporation of HSV-1 thymidine kinase into the adenovirus protein IX for functional display on the virion. Virology. 2005;338:247–58. doi: 10.1016/j.virol.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Matthews QL, Sibley DA, Wu H, et al. Genetic incorporation of a herpes simplex virus type 1 thymidine kinase and firefly luciferase fusion into the adenovirus protein IX for functional display on the virion. Mol Imaging. 2006;5:510–9. [PMC free article] [PubMed] [Google Scholar]
- 23.Kimball KJ, Rivera AA, Icyuz M, et al. A novel infectivity-enhanced oncolytic adenovirus with a capsid-incorporated dual imaging modality for monitoring virotherapy in ovarian cancer. Mol Imaging. 2009;5:264–77. [PMC free article] [PubMed] [Google Scholar]
- 24.Kanerva A, Zinn KR, Chaudhuri TR, et al. Enhanced therapeutic efficacy for ovarian cancer with a serotype 3 receptor-targeted oncolytic adenovirus. Mol Ther. 2003;8:449–58. doi: 10.1016/s1525-0016(03)00200-4. [DOI] [PubMed] [Google Scholar]
- 25.Chambers A. MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res. 2009;69:13. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 26.Chartier C, Degryse E, Gantzer M, et al. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–10. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He TC, Zhou S, da Costa LT, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–14. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heise C, Hermiston T, Johnson L, et al. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy [published erratum appears in Nat Med 2000;6:412] Nat Med. 2000;6:1134–9. doi: 10.1038/80474. [DOI] [PubMed] [Google Scholar]
- 29.Nettelbeck DM, Rivera AA, Balagué C, et al. Novel oncolytic adenoviruses targeted to melanoma: specific viral replication andcytolysisbyexpressionofE1Amutantsfromthetyrosinase enhancer/promoter. Cancer Res. 2002;62:4663–70. [PubMed] [Google Scholar]
- 30.Maizel JV, Jr, White DO, Scharff MD. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–25. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 31.Ulasov IV, Rivera AA, Han Y, et al. Targeting adenovirus to CD80 and CD86 receptors increases gene transfer efficiency to malignant glioma cells. J Neurosurg. 2007;107:617–27. doi: 10.3171/JNS-07/09/0617. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh-Choudhury G, Haj-Ahmad Y, Graham FL. Protein IX a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 1987;6:1733–9. doi: 10.1002/j.1460-2075.1987.tb02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sargent KL, Ng P, Evelegh C, et al. Development of a size-restricted pIX-deleted helper virus for amplification of helper-dependent adenovirus vectors. Gene Ther. 2004;11:504–11. doi: 10.1038/sj.gt.3302107. [DOI] [PubMed] [Google Scholar]
- 34.Campbell EM, Perez O, Melar M, et al. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 2007;360:286–93. doi: 10.1016/j.virol.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu ZB, Lu B, Park M, et al. Development of an optimized conditionally replicative adenoviral agent for ovarian cancer. Int J Oncol. 2008;32:1179–88. doi: 10.3892/ijo_32_6_1179. [DOI] [PubMed] [Google Scholar]
- 36.Sirena D, Lilienfeld B, Eisenhut M, et al. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J Virol. 2004;78:4454–62. doi: 10.1128/JVI.78.9.4454-4462.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Short JJ, Vasu C, Holterman MJ, et al. Members of adenovirus species B utilize CD80 and CD86 as cellular attachment receptors. Virus Res. 2006;122:144–53. doi: 10.1016/j.virusres.2006.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]