Abstract
TNF-R1 signal transduction is mediated through the assembly of scaffolding proteins, adaptors and kinases. TNF-receptor ubiquitous signaling and scaffolding protein (TRUSS), a 90.1 kDa TNF-R1-associated scaffolding protein, also interacts with TRAF2 and IKK and contributes to TNF-α-induced nuclear factor-κB (NF-κB) and c-Jun-NH2-terminal kinase (JNK) activation. Little is known about the mechanism of interaction between TRUSS, TNF-R1 and TRAF2. To address this issue, we used deletional and site-directed mutagenesis approaches to systematically investigate: (i) the regions of TRUSS that interact with TNF-R1 and TRAF2 and (ii) the ability of TRUSS to self-associate to form higher order complexes. Here we show that sequences located in the N-terminal (residues 1-248) and central regions (residues 249-440) of TRUSS are required to form a docking interface that supports binding to both TNF-R1 and TRAF2. While the C-terminal region (residues 441-797) did not directly interact with TNF-R1 or TRAF2, sequences located in this region were capable of self-association. Collectively, these data suggest that: (i) the interaction between TNF-R1 and TRAF2 requires sequences located in the entire N-terminal half (residues 1-440) of TRUSS, (ii) the binding interface for TNF-R1 is closed linked with the TRAF2 binding interface, and (iii) the assembly of homomeric TRUSS complexes may contribute to its role in TNF-R1 signaling.
Keywords: TRUSS, TNF-R1, TRAF2, signaling, homo-oligomerization
The TNF-α receptor, TNF-R1 (p55, CD120a), plays a key role in the initiation of inflammation, host defense, apoptosis, and cell survival through its ability to activate NF-κB, mitogen-activated protein kinases, caspase-8 and other signaling responses (1, 2). TNF-R1-dependent NF-κB activation is initiated by ligand-induced receptor oligomerization which facilitates the recruitment of TNF receptor-associated death domain protein (TRADD) to the cytoplasmic region of the receptor (3). TRADD serves as a platform for the recruitment of TNF receptor associated factor-2 (TRAF2) and receptor-interacting protein (RIP), which in turn recruit and activate the IκB kinase (IKK) complex (4, 5). The receptor-associated IKK complex then phosphorylates IκB and following its ubiquitination and degradation sets in motion the nuclear translocation of NF-κB and downstream transcriptional activation of NF-κB-dependent pro-inflammatory and pro-survival genes (as reviewed by Chen and Goeddel(6)). Subsequently, the IKK complex and associated adaptor proteins (the so-called complex I) dissociate from TNF-R1 and a new complex (complex II) capable of recruiting FADD and either caspase-8 or c-FLIPL forms in the cytosol and promotes apoptosis, provided that complex I fails to activate NF-κB and upregulate c-FLIPL expression (7). Other studies have also suggested that pro-apoptotic TNF-R1 signaling complexes are assembled on the cytosolic surfaces of endosomes (8). Together, these studies suggest that spatial and temporal elements contribute to the diversity of TNF-R1-induced signaling responses. However, while the assembly of these signaling complexes has been comprehensively studied, little is known about the mechanisms that regulate their composition or localization.
In an attempt to further understand the mechanisms of assembly and dissociation of TNF-R1 signaling complexes, we conducted yeast 2-hybrid screens using the membrane proximal region of TNF-R1 as bait and cloned TNF-Receptor Ubiquitous Scaffolding and Signaling protein (TRUSS) (9). In addition to interacting with TNF-R1, TRUSS was found to associate with TRAF2 and members of the IKK complex and activated NF-κB and JNK signaling pathways when overexpressed in cell lines (9, 10). Furthermore, in studies aimed at generating a map of the human protein interactome, Rual et al. (11) found that out of ~8000 human open reading frames included in their study, TRUSS (gene name TRPC4AP) only interacted with TNF-R1, TRAF2 and IKKγ. Together, these data suggest that TRUSS may contribute to the regulation of TNF-R1 signaling, possibly by facilitating the assembly and/or dissociation of TNF-R1-signaling complexes.
Little is known about how TRUSS interacts with TNF-R1 or TRAF2, though primary sequence analysis points to the presence of several protein-protein interaction motifs which include consensus TRAF2 binding motifs and a leucine zipper motif, consistent with TRUSS’ proposed function as a scaffolding protein. Furthermore, computational analysis suggests that TRUSS exists as a globular protein rich in α-helices. To gain insight into the question of how TRUSS interacts with TNF-R1 and TRAF2, we used a mutagenic approach to systematically investigate the region(s) of TRUSS that interact with these molecules. In addition, based on the known ability of TNF-R1 and TNF-R1 signaling adaptors and associated molecules to assembly into homomeric complexes, we addressed the question of whether TRUSS was also capable of self-association.
EXPERIMENTAL PROCEDURES
Materials
Western blot antibodies: anti-HA (Covance), anti-Flag (Sigma-Aldrich), anti-TRAF2 (Santa Cruz), anti-GFP (Clontech), and anti-myc (Clontech). Anti-TRUSS N-terminal and C-terminal antisera were prepared in rabbits by Alpha Diagnostics and affinity purified. Immunoprecipitation antibodies: anti-HA mAb (Roche), goat anti-mTNF-R1 (R&D Systems), and anti-TRAF2 (Santa Cruz Biotechnology). Vectors pcDNA3.1, pEGFP, pRSET, and pGEX were from Invitrogen, pCMV-myc was from Clontech, and all PCR primers were purchased from Genelink.
Constructs/Mutagenesis/Cloning
All HA-tagged TRUSS deletion mutants were cloned into pcDNA3.1 as described previously (9). Mutagenesis of TRAF2 binding sites was performed with the QuikChange® Site-directed Mutagenesis Kit (Stratagene). HA-TRUSS249-440 was cloned using a TOPO-TA cloning kit (Stratagene). His-TRUSS (pRSET), GFP-TRUSS (pEGFP-C1) and myc-TRUSS (pCMV-myc) constructs were cloned by excision of TRUSS from HA-TRUSS constructs with KpnI and NotI followed by gel purification and ligation into new vectors. The fidelity of all constructs and mutants were verified by sequencing.
GST bead and recombinant His-TRUSS preparation
GST and GST-TNF-R1207-425 in pGEX vectors were prepared in DH5-α cells (Stratagene) as previously described (12). BL-21 cells (Stratagene), transformed with pRSET-His-TRUSS, were grown at 30°C until an OD600 ≈ 0.3-0.4, induced for 3 hrs with 1 mM IPTG, and supernatant prepared as described above. NTA-Ni+ beads (Qiagen) were incubated with the supernatant, the beads were washed, and the recombinant His-TRUSS was eluted from the beads with PBS containing 250 mM imidazole.
Cell culture and Transfections
HEK293 cells (ATTC) were maintained in DMEM with 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% (v/v) FBS and plated at 5 × 105 cells per well in poly-D-lysine treated 6-well plates the day before transfection. HEK293 cells were transfected for 18-24 hr using the Lipofectamine 2000 standard transfection protocol (Invitrogen).
Pull-downs and co-immunoprecipitations
For GST-pull-downs, HEK293 cells were transfected with 1 μg of each HA-TRUSS deletion construct and lysed in an NP-40 lysis buffer (50 mM HEPES, pH 7.6, containing 150 mM NaCl, 5 mM EDTA, 1% (v/v) NP-40, 1 mM PMSF, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 1 mM NaF, and 1 mM Na3VO4). Equal amounts of post-nuclear supernatant (PNS) were incubated overnight with glutathione bead coupled with 15 μg of either GST or GST-TNF-R1207-425. After 3 washes with lysis buffer containing 500 mM NaCl, the PNS and bead-associated proteins were separated by SDS-PAGE on a 10% gel under reducing conditions, Western blotted, developed with ECL and exposed on Hyperfilm (GE Healthcare). The blots were then incubated in Ponceau S solution (0.1% (w/v) Ponceau S dissolved in 5% acetic acid) for detection of GST fusion proteins. For co-immunoprecipitations, HEK293 cells were co-transfected with up to 1 μg of each construct (see “Figure Legends” for the amount of each construct). The following day, the cells were lysed in the same lysis buffer that was used for the GST pull-downs for the anti-TNF-R1, anti-HA, and anti-GFP immunoprecipitations. A glycerol-based lysis buffer was used for the anti-TRAF2 immunoprecipitations (50 mM Tris (pH 7.6), 0.1% (v/v) NP-40, 250 mM NaCl, and 10% glycerol), as described (13). After protein quantification, equal amounts of lysate were incubated with 1 μg of the immunoprecipitating antibody (or non-immune IgG as a control) together with 30 μl of Protein-G coated beads (Santa Cruz Botechnology). The beads were washed 3 times with lysis buffer containing 500 mM NaCl, then PNS and bead-associated proteins were separated by SDS-PAGE through a 10% gel under reducing conditions and visualized by Western blotting as described above. For all pull-down and TNF-R1 and TRAF2 co-immunoprecipitation experiments, Western blot band intensities were quantified by densitometry using ImageJ64 software. The densities of pulled-down or immunoprecipitated bands were then normalized to the densities of the WCLs. For pull-down experiments, the density ratio (DR) = (GST-TNF-R1207-425 band − GST band)/PNS band, then % of 1-797 = DRmutant/DR1-797 * 100. When co-expression of another protein was involved (i.e. TNF-R1, TRAF2), the ratios were normalized to the density of the immunoprecipitating protein band to account for differences in co-expression between each mutant. For co-immunoprecipitation experiments, Density ratio (DR) = (co-IP band − n.i IgG band)/PNS band, density ratio normalization (DRNorm) = DR/IP band, then % = DRNormmutant/DRNorm1-797 * 100. TRUSS mutant interaction ratios were expressed as a percentage of the full-length TRUSS1-797 interaction ratio from each experiment. Data were compiled for 3-8 independent experiments, and the means and standard errors of the means were calculated. To distinguish signal from noise, we arbitrarily defined the binding threshold as 15% of the binding seen with full-length TRUSS.
FPLC gel filtration
A Superdex 200 16/60 prep-grade FPLC column (Amersham) and a BioRad control and collection system was used for gel filtration experiments. The column was equilibrated and calibrated with NP-40 lysis buffer or PBS (for recombinant His-TRUSS). Equal amounts of protein were loaded in a 2 ml sample loop, the column was eluted at an isocratic flow rate of 1 ml/min and 2 ml fractions were collected. Two hundred μl of each fraction were mixed with Laemelli/DTT sample buffer and boiled for 5 min before separation by SDS-PAGE through a 10% gel under reducing conditions. Individual proteins were visualized by Western blot analysis. For TRUSS self-association experiments, HEK293 cell lysate from cells transfected with HA-TRUSS or pcDNA3.1, or recombinant His-TRUSS eluted from nickel beads were fractionated by FPLC as described above.
RESULTS
TRUSS interacts with TNF-R1 through an N-terminal region containing residues 249-440
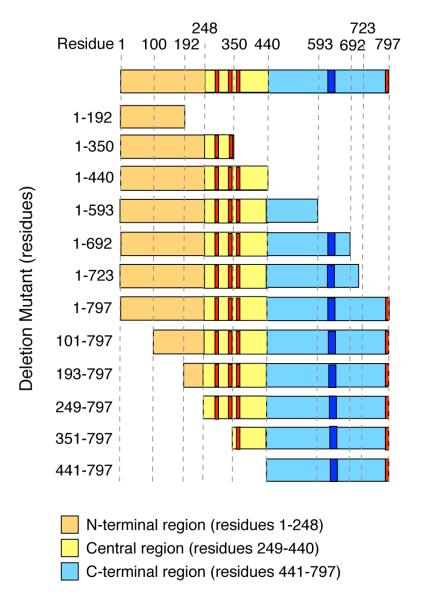
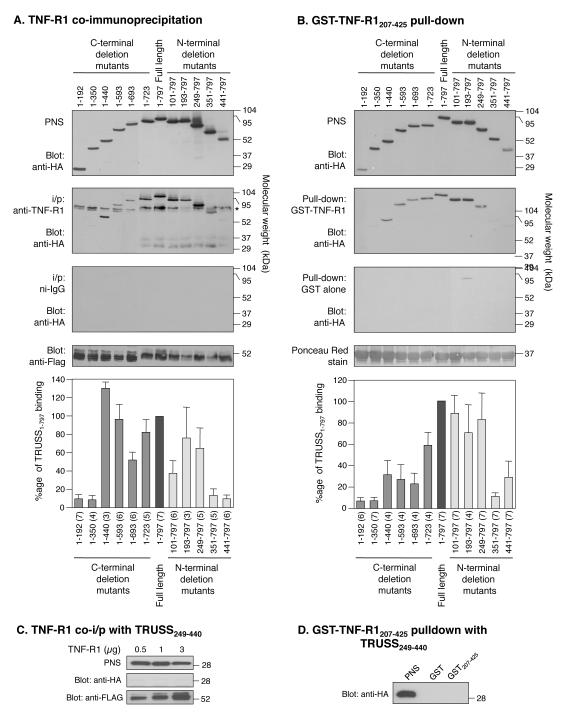
To investigate the region of TRUSS that interacts with TNF-R1, we created a panel of HA-epitope-tagged C-terminal and N-terminal TRUSS deletion mutants (Figure 1). Each mutant was tested for its ability to interact with full-length TNF-R1 following co-expression in HEK293 cells, immunoprecipitation with anti-TNF-R1 antibody, detection of co-immunoprecipitating TRUSS and TNF-R1 by immunoblotting with anti-HA and anti-Flag antibodies, respectively, and densitometric analysis as described in the “Methods”. Figure 2A shows that TRUSS C-terminal deletion mutants, 1-192 and 1-350, failed to interact with TNF-R1, while TRUSS mutants 1-440, 1-593, 1-693 and 1-723 all interacted with TNF-R1. Sequential deletion of the N-terminal region revealed that mutants 101-797, 193-797 and 248-797 interacted with TNF-R1, while mutants 351-797 and 441-797 did not (Figure 2A). None of the TRUSS deletion mutants were detected in co-immunoprecipitates conducted with isotype-matched non-immune IgG (Figure 2A). These findings suggest that the entire N-terminal region of TRUSS (residues 1-440) is involved in its interaction with TNF-R1 and that within this region, residues 249-440 contribute an important role.
Figure 1.
Cartoon depicting HA-tagged TRUSS deletion mutants. Orange rectangle = N-terminus; yellow = central region; blue = C-terminus; red bars = putative TRAF2 binding site motifs; dark blue bars = putative leucine zipper motif.
Figure 2.
TRUSS interacts with TNF-R1 via the N-terminal region (residues 249 to 440). A) TNF-R1 co-immunoprecipitation of TRUSS deletion mutants. HEK293 cells, transfected with 1 μg of each HA-tagged TRUSS construct plus 0.5 μg Flag-tagged TNF-R1 construct, were lysed and immunoprecipitated with anti-TNF-R1 antibody or non-immune IgG as a control. Western blots were probed with anti-HA then with anti-Flag to detect TNF-R1. * denotes a background band seen in TNF-R1 co-immunoprecipitations. B) GST-TNF-R1207-425 pull-down of TRUSS deletion mutants. HEK293 cells, transfected with 1 μg of each HA-tagged TRUSS deletion mutant construct, were lysed and incubated with GST-coated beads or GST-TNF-R1207-425-coated beads. Western blots were probed with anti-HA antibody and the blots were stained with PonceauS to detect of recombinant GST-TNF-R1207-425 protein. Error bars in panels A and B are the mean ± SEM calculated from between 3 and 7 independent experiments. The number of experiments for each construct is shown in parentheses. C. TRUSS249-440 does not co-immunoprecipitate with TNF-R1. HEK293 cells were transfected with 1 μg TRUSS249-440 plus increasing amounts of Flag-TNF-R1, lysed, and lysates immunoprecipitated with anti-TNF-R1. Western blots were probed with anti-HA then anti-Flag to detect TNF-R1. Representative blot of 10 independent experiments. D) TRUSS249-440 is not pulled down by GST-TNF-R1207-425. HEK293 cells were transfected with TRUSS249-440, lysed, and lysates were incubated with GST coated beads or GST-TNF-R1207-425 coated beads. Western blots were probed with an anti-HA antibody. Representative blot of 10 independent experiments.
To confirm these data we transfected HEK293 cells with full length TRUSS and each of the TRUSS deletion mutants shown in Figure 1. Cell lysates were then tested for their interaction with GST-TNF-R1 cytoplasmic domain fusion protein (GST-TNF-R1207-425) coupled to glutathione-conjugated Sepharose beads. Co-precipitating TRUSS was detected by immunoblotting with anti-HA antibody followed by densitometric analysis. Figure 2B shows that a qualitatively similar pattern of interactions between the TRUSS deletion mutants and TNF-R1 was detected using this approach. Neither full-length TRUSS nor any of the TRUSS deletion mutants interacted with GST-coupled beads, and equivalent levels of GST-TNF-R1207-425 were loaded onto each gel (Figure 2B). Thus, these data confirm the co-immunoprecipitation data and suggest that the TNF-R1 binding interface resides between residues 1-440 and that residues 249-440 play a prominent role in binding.
To determine if the sequence encompassed by residues 249 and 440 was sufficient for the interaction between TRUSS and the cytoplasmic domain of TNF-R1, an HA-tagged deletion mutant lacking the N-and C-terminal region (TRUSS249-440) was created and tested for its ability to interact with TNF-R1 by co-expression in HEK293 cells and co-immunoprecipitation. Figure 2C shows that TRUSS249-440 did not immunoprecipitate with TNF-R1 at any concentration of TNF-R1 transfected. Similarly, TRUSS249-440 did not interact with GST-TNF-R1207-425-coupled beads in pulldown experiments using lysates from TRUSS249-440 transfected HEK293 cells (Figure 2D). Taken together with the results obtained with the N-terminal deletion mutants, these findings suggest that while residues located in TRUSS249-440 are important for the interaction with TNF-R1 in the context of the full length molecule, additional residues located in the N-terminal region (1-248) also contribute to this interaction. Similar to the region encompassed by residues 249-440, these additional N-terminal residues are not sufficient to promote this interaction.
TRUSS interacts with TRAF2 through an N-terminal region containing residues 249-440
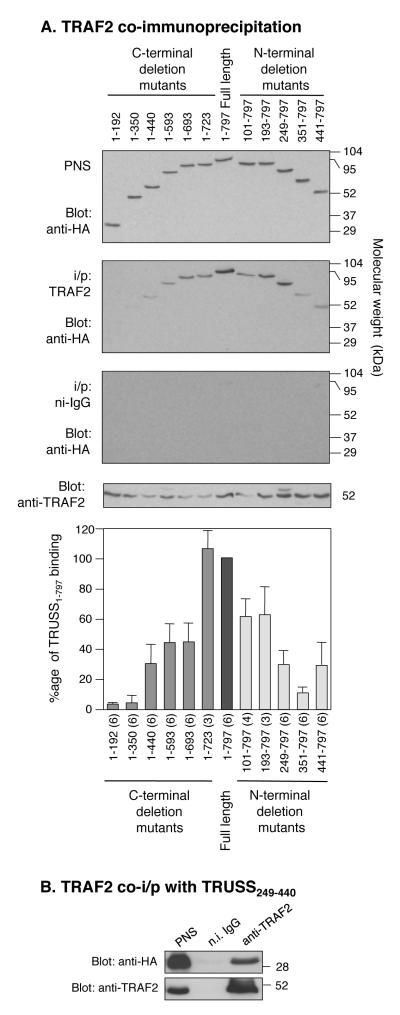
Previous studies have shown that TRUSS also interacts with TRAF2 (9, 11). To investigate the region of TRUSS involved in its interaction with TRAF2, HEK293 cells were co-transfected with full-length Flag-tagged TRAF2 and each N-terminal and C-terminal TRUSS deletion mutant shown in Figure 1. After lysis, TRAF2 was immunoprecipitated and co-immunoprecipitating TRUSS was detected by Western blotting with anti-HA antibody followed by anti-TRAF2 antibody and densitometric analysis. Figure 3A shows that the C-terminal TRUSS deletion mutants (1-192 and 1-350) failed to interact with TRAF2. Interactions were detected with the 1-440, 1-593 and 1-692 mutants, though at reduced levels compared to full-length TRUSS. The 1-723 mutant exhibited a similar degree of binding to TRAF2 as full-length TRUSS. The N-terminal deletion mutants exhibited progressively reduced interactions with TRAF2 as the N-terminal region was incrementally removed, as seen in mutants 101-797, 193-797 and 248-797. Mutant 351-797 did not interact with TRAF2, while mutant 441-797 exhibited weak binding to TRAF2. None of the TRUSS deletion mutants co-immunoprecipitated with isotype-matched non-immune IgG (Figure 3A). Collectively, these data suggest that the region between amino acids 249 and 440 is also important for the interaction between TRUSS with TRAF2.
Figure 3.
TRUSS interacts with TRAF2 via amino acids 249 to 440. A) TRAF2 co-immunoprecipitation of TRUSS deletion mutants. HEK293 cells, transfected with 1 μg of each HA-tagged TRUSS deletion mutant construct plus 0.5 μg Flag-tagged TRAF2 construct, were lysed and immunoprecipitated with anti-TRAF2 antibody or non-immune IgG as a control. Western blots were probed with anti-HA antibody and then with anti-TRAF2. Error bars in panel A are the mean ± SEM calculated from between 3 and 6 independent experiments. The number of experiments for each construct is shown in parentheses. B) TRAF2 co-immunoprecipitates with TRUSS249-440. HEK293 cells were transfected with 1 μg of TRUSS249-440 plus 0.5 μg Flag-tagged TRAF2 construct, lysed, and lysates immunoprecipitated with anti-TRAF2 antibody or non-immume IgG. Western blots were probed with an anti-HA antibody then with anti-TRAF2. Representative blot of 4 independent experiments.
Next, we used a co-immunoprecipitation approach to determine if the sequence between residues 249 and 440 was sufficient to support the interaction between TRUSS and TRAF2. HEK293 cells were co-transfected with Flag-tagged TRAF2 and TRUSS249-440 and complex formation was analyzed by immunoprecipitation with anti-TRAF2 antibody. Figure 3B shows that HA-tagged TRUSS249-440 co-immunoprecipitated with TRAF2 indicating the sufficiency of this sequence for binding to TRAF2.
The region of TRUSS that nominally interacts with TRAF2 (residues 249-440) contains three consensus TRAF2 binding motifs conforming to the consensus sequences (P/S/A/T)X(Q/E)E (14) and SXXE (15, 16). To determine if these motifs were required for the interaction between TRUSS and TRAF2, we performed alanine-scanning mutagenesis to systematically negate these putative TRAF2 binding motifs. All three TRAF2 binding motifs in the region between amino acids 249 and 440, were mutated to Ala residues, either alone or in combination. Because TRUSS441-797 demonstrated limited but detectable TRAF2 binding, we also mutated the extreme C-terminal consensus TRAF2 binding motif alone. As shown in Figure 3A, several of the TRUSS deletion mutants lacking the fourth putative TRAF2 binding motif were found to interact with TRAF2 (e.g. 1-440, 1-723). Each TRUSS Ala-point mutant was co-transfected with TRAF2 into HEK293 cells and cell lysates were subjected to TRAF2 immunoprecipitation. All the TRUSS-TRAF2 binding motif mutants co-immunoprecipitated with TRAF2 to the same extent as wild type TRAF2 (data not shown) signifying that TRUSS binds to TRAF2 independently of these putative TRAF2 binding motifs.
Self-interactions between the N-terminal, central and C-terminal regions of TRUSS
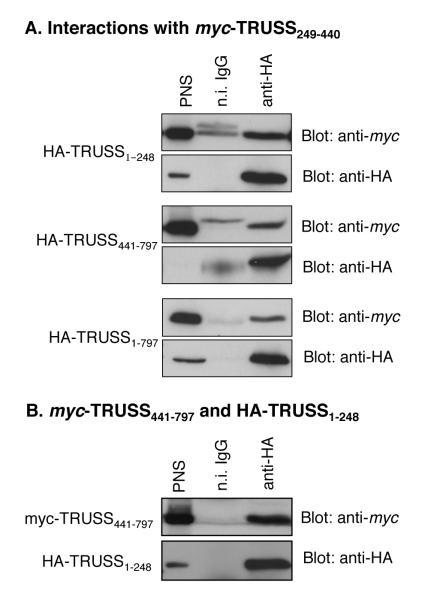
To gain insight into how TRUSS might fold to expose the TNF-R1 and TRAF2 binding interface, we investigated the ability of TRUSS-deletion mutants to interact with one another. Since the region encompassed by residues 249-440 was found to be important in the interaction between TRUSS and both TNF-R1 and TRAF2, we first determined if TRUSS249-440 interacted with the flanking N- and C-terminal regions. HEK293 cells were co-transfected with myc-tagged TRUSS249-440 and either HA-TRUSS1-248, HA-TRUSS441-797 or full length HA-TRUSS1-797 as a positive control. Cell lysates were immunoprecipitated with anti-HA antibody and co-immunoprecipitating myc-tagged TRUSS249-440 was detected by Western blotting with anti-myc antibody. Figures 4A shows that TRUSS249-440 interacted with both TRUSS1-248 and TRUSS441-797 as well as with full length TRUSS1-797 indicating that the central region (residues 249-440) was capable of interacting with sequences located in both the N-terminal and C-terminal regions.
Figure 4.
Intramolecular interactions of TRUSS. A) Interactions of TRUSS249-440. HEK293 cells were co-transfected with 1 μg of myc-TRUSS249-440 and either 1 μg of HA-TRUSS1-248, HA-TRUSS441-797, or HA-TRUSS1-797. The cells were lysed and immunoprecipitated with anti-HA antibody. Interacting proteins were detected by Western blot with anti-myc then anti-HA antibodies. Representative of at least 3 experiments; B) TRUSS N- and C-terminal interaction. HEK293 cells were co-transfected with 1 μg of myc-TRUSS441-797 and 1 μg of HA-TRUSS1-248, lysed, and lysates immunoprecipitated with anti-HA. Subsequent Westerns were blotted with anti-myc then anti-HA antibodies. Representative of at least 3 experiments.
Since the binding activity of the central region is dependent on sequences located in the N-terminal and part of the flanking C-terminal, we next determined if the N-terminal region could interact with the C-terminal region. HEK293 cells were co-transfected with HA-tagged TRUSS1-248 and myc-tagged TRUSS441-797, immunoprecipitated anti-HA antibody and co-immunoprecipitating myc-tagged TRUSS441-797 detected by immunoblotting. Figure 4B shows that the N-terminal region (residues 1-248) robustly interacted with the C-terminal region (residues 441-797). Together, these data suggest that intramolecular interactions between the N-terminal, central and C-terminal regions contribute to the tertiary structure of the full-length molecule.
TRUSS forms homomeric complexes
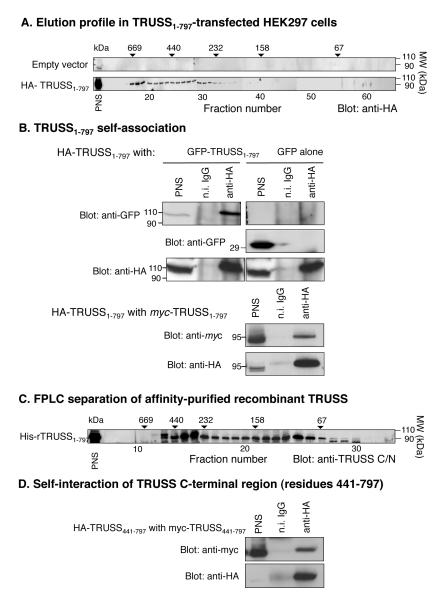
Given the fact that many proteins involved in TNF-R1 signaling function by forming homomeric and heteromeric signaling complexes, together with data showing that enforced expression of TRUSS is sufficient to activate NF-κB and AP-1 (9, 10), we next questioned whether TRUSS may also function by forming homomeric complexes. To address this question, we fractionated cell lysates from HEK293 cells transfected with full length HA-tagged TRUSS1-797 or empty vector as a control by gel filtration through a calibrated Superdex 200 column. Each fraction was then analyzed by SDS-PAGE and western blotting with anti-HA-antibody. Figure 5A shows that HA-TRUSS1-797 eluted as a broad peak ranging in molecular weight from ~700-200 kDa indicating the presence of homomeric and/or heteromeric complexes. To determine if TRUSS could specifically interact with itself, HEK293 cells were co-transfected with vectors encoding HA-tagged TRUSS1-797 together with either GFP-tagged TRUSS1-797 or GFP alone as a control. After lysis, the cells were immunoprecipitated with anti-HA antibody and co-immunoprecipitating GFP was detected by Western blotting. Figure 5B (upper panels) shows that GFP-TRUSS1-797 co-immunoprecipitated with HA-TRUSS1-797, whereas GFP did not. To verify that the GFP sequence did not contribute to the observed interaction, myc-tagged TRUSS1-797 and HA-TRUSS1-797 were co-transfected into HEK293 cells and tested for co-immunoprecipitation as described above. Figure 5B (lower panel) shows that myc-TRUSS1-797 also specifically co-immunoprecipitated with HA-TRUSS1-797. Together, these findings suggest that TRUSS molecules are capable of self-association to form homomeric complexes. To investigate the size distribution of homomeric TRUSS complexes, we determined the elution profile of bacterially-expressed, affinity-purified, recombinant His-tagged TRUSS1-797 by gel filtration as described above. Figure 5C shows that rHis-tagged TRUSS1-797 also eluted as a broad range of molecular weight species from approximately 500 to 70 kDa. However, the majority of the eluted TRUSS protein was detected in two peaks corresponding to molecular weights of ~300 kDa (centered in fractions 14-15) and ~100 kDa (centered in fractions 24-25). Taken together, these data suggest that TRUSS may exist both as a monomeric species (~90 kDa) and as homomeric complexes, including a trimer (~270 kDa).
Figure 5.
TRUSS is capable of self-interaction. A) Size fractionation of TRUSS-transfected lysate. HEK293 cells were transfected with either pcDNA3.1 empty vector or HA-tagged full-length TRUSS and lysed. Lysates were loaded onto a Superdex200 FPLC column and 1 ml fractions were collected. 20% of each fraction was subjected to SDS-PAGE and Western blotted for anti-HA. Molecular weight (MW) markers are located above the fraction where the each standard protein peak eluted. B) TRUSS/TRUSS co-immunoprecipitations. (Top panel) HEK293 cells, transfected with 1 μg HA-TRUSS1-797 plus 1 μg of either a GFP-TRUSS1-797 or GFP empty vector construct, were lysed and lysates immunoprecipitated with anti-HA antibody or non-immume IgG. Western blots were probed with anti-GFP antibody followed by anti-HA antibody. (Bottom panel) HEK293 cells were transfected with 1 μg each of myc-TRUSS1-797 and HA-TRUSS1-797, lysed and lysates immunoprecipitated with anti-HA antibody or non-immume IgG. Western blots were probed with anti-myc antibody followed by anti-HA antibody. C) Size fractionation of recombinant His-TRUSS. Recombinant His-TRUSS was prepared in BL-21 bacteria and purified with Ni+-beads. Purified His-TRUSS was loaded onto a Superdex 200 FPLC column and 1 ml fractions were collected. Twenty % of each fraction was subjected to SDS-PAGE and blotted with anti-TRUSS/C antibody. D) TRUSS C-terminal self-interactions. HEK293 cells were co-transfected with 1 μg each of myc-TRUSS441-797 and HA-TRUSS441-797, lysed and immunoprecipitated with anti-HA antibody. Western blots were probed with anti-myc followed by anti-HA antibody. Data shown are representative of at least 3 experiments.
Inspection of the primary sequence of TRUSS reveals an abundance of Leu and other hydrophobic residues in the C-terminal region (residues 441-797). Since the C-terminal region (residues 1-440) was not involved in the interaction with TNF-R1 and TRAF2, we questioned if one of the functions of the C-terminal region may be to promote TRUSS-self-association. To investigate this possibility, we co-transfected HEK293 cells with myc-tagged TRUSS441-797 and HA-tagged-TRUSS441-797. Cell lysates were immunoprecipitated with anti-HA antibody and co-immunoprecipitating myc-tagged TRUSS441-797 was detected by immunoblotting with anti-myc antibody. Figure 5D shows that the C-terminal region (region 441-797) was able to robustly self-associate and suggest that this region may contribute to TRUSS self-association and homomeric complex assembly.
DISCUSSION
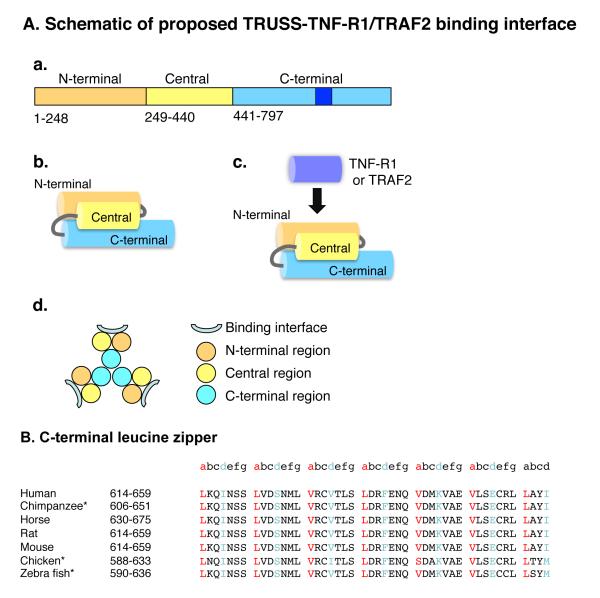
TNF-R1-dependent signaling is fundamentally based on the assembly of multimeric protein signaling complexes. Seeking to further understand the mechanisms of assembly of TNF-R1-induced signaling complexes, we cloned TRUSS, a TNF-R1-associated signaling and scaffolding molecule that also interacts with TRAF2 and the IKK complex. Based on the finding that a series of N-terminal and C-terminal TRUSS deletion mutants inhibited TNF-α-induced NF-κB and AP-1 activation, we concluded that TRUSS contributes to TNF-R1-induced NF-κB and JNK activation (9-11). However, further characterization of the role of TRUSS in TNF-R1-induced signaling was complicated by the fact that all TRUSS mutants tested inhibited TNF-R1 signaling to varying degrees (9, 10), consistent with the notion that to function as a scaffolding protein, TRUSS may need to expose multiple non-linear docking sites in different regions of the molecule. Primary sequence analysis has also provided little insight into the question of how TRUSS associates with its interaction partners, or whether it conforms to the trimeric paradigm expressed by some TNF-R1-associated signaling molecules. The goals of this study therefore, were to investigate: (i) the region(s) of TRUSS that facilitate its interaction with TNF-R1 and TRAF2, and (ii) the ability of TRUSS to form homomeric complexes through self-association. Using a systematic deletional mutagenesis approach, our findings suggest that the interaction of TRUSS with both TNF-R1 and TRAF2 requires sequences located in the N-terminal portion of TRUSS defined by residues 1-440 and that within this region, residues 249-440 appear to substantially contribute to these interactions. Our results also suggest that TRUSS exists as both a monomer and as a continuum of higher molecular weight homomeric complexes, including a trimer, and that the C-terminal region may contribute to TRUSS oligomerization. Based on these and other findings, we propose a structural model of TRUSS to explain how the TNF-R1 and TRAF2 binding interface is exposed (Figure 6A).
Figure 6.
A) Proposed structural organization and assembly of TRUSS. a) Primary structure organization. The putative leucine zipper is dark blue. b) Proposed tertiary organization of the N-terminal, central and C-terminal regions. c) Proposed position of the TNF-R1 and TRAF2 binding interface. d) Proposed organization of the TRUSS homotrimer. B. Alignment and conservation of the putative C-terminal leucine zipper sequences of vertebrate TRUSS. *The Entrez gene entries for these sequences only show isoform “b” which lacks an 8 residue sequence that arises by alternative splicing. Consequently, for comparison to the position of the second leucine like zipper, the numeric positions of the sequences are 8 residues shorter than those for the longer “a” isoform.
Our finding that the interaction between TRUSS and TNF-R1 was only revealed in the setting of larger TRUSS deletion mutants is consistent with the notion that the interaction occurs independently of a short linear docking motif, as, for example, has been suggested for the interaction between TRAF2 and CD40 (15, 17). Rather, our results suggest a model akin to the interaction between TRADD and the death domain of TNF-R1, in which a docking interface is created by individual amino acid residues that cluster together in the setting of the correctly folded molecules (18, 19). In these situations, binding activity is dependent on the functional integrity of long stretches of primary sequence that are required for correct folding and exposure of the binding interfaces (18, 19). In the case of TRUSS, we propose that residues distributed between positions 249-440, possibly just a few, cluster to form the TNF-R1 docking interface. Furthermore, based on the finding that the TRUSS249-440 mutant was unable to directly interact with TNF-R1, we propose that the flanking N-terminal region contributes to the correct folding and/or exposure of the TNF-R1 docking interface, as shown schematically in Figure 6A. Indeed, it is conceivable that the N-terminal region also contains residues that contribute to the binding interface (Figure 6A).
A similar deletion mutagenesis approach revealed that the region of TRUSS required for its interaction with TNF-R1 was also involved in its interaction with TRAF2. In contrast to its interaction with TNF-R1, the interaction between TRUSS and TRAF2 was detectable in the absence of the flanking N-terminal region. Systematic site-directed mutagenesis of the three consensus TRAF2 binding motifs located between residues 249-440 indicated that the TRUSS-TRAF2 interaction occurred independently of these TRAF2 consensus motifs. Furthermore, the low level of binding exhibited by TRUSS441-797 was not affected when residues comprising the extreme SXXE (residues 781-784) TRAF2 binding consensus motif were mutated to Ala. Taken together, these findings suggest that none of the consensus TRAF2 binding motifs participate in the interaction between TRUSS and TRAF2. It should be noted that the TRAF2 interaction motifs have been largely modeled on the interaction between the TRAF-C domain of TRAF2 and the non-death domain TNFRSF members, TNF-R2 and CD40 (15, 17). Since a wide array of non-TNFRSF molecules have also been shown to interact with TRAF2, e.g. filamin, cIAP1/2, ASK1, β-arrestin and TRADD (20-23), other binding motifs must exist to faciliate their interaction with TRAF2. Indeed, although TRADD interacts with TRAF2 via its TRAF-C domains (though TRADD binds to a different region of TRAF2 than that utilized in the interaction of TRAF2 with TNF-R1 and CD40 (24)), filamin interacts with the RING domain (20) and cIAP1/2 interacts with the TRAF-N domain (21). Thus, it might be anticipated that a series of different TRAF2 interaction motifs may exist to enable these proteins to interact with different regions of TRAF2. While future studies will be required to address the region(s) of TRAF2 that interact with TRUSS, our current findings suggest that the TRUSS N-terminal TRAF2 binding interface is closely linked to the TNF-R1 binding interface. Indeed, it is conceivable that TNF-R1 and TRAF2 compete for same binding interface on TRUSS.
Computational primary sequence analysis predicts TRUSS to be a hydrophobic, globular protein with ~50% α-helical character (25, 26). Approximately 25% of the amino acids comprising TRUSS are Leu, Iso or Val residues with ~15% being Leu residues alone. These findings raised the possibility that intramolecular hydrophobic interactions between the N-terminal, central and C-terminal regions may contribute to the correct exposure of the TNF-R1 and TRAF2 binding interface. To gain insights into this issue, we investigated potential interactions between the N-terminal (residues 1-248) region, the central region (residues 249-440) and the C-terminal region (residues 441-797). These studies suggested complexity in tertiary structure but were consistent with the single model depicted in Figure 6A(panel b) in which the N-terminal, central and C-terminal regions are aligned in an anti-parallel fashion to expose the TNF-R1 and TRAF2 interaction interface(s). Indeed, based on these findings, we speculate that interactions between the N- and C-terminal regions may contribute to the proper folding and exposure of the TNF-R1 and TRAF2 interface located in the central region (Figure 6A (panel c)).
As discussed earlier, TNF-R1 signaling is initiated by ligand-induced receptor clustering, which then initiates the formation of large, multimeric signaling complexes. Enforced overexpression of TNF-R1 and its associated signaling molecules, including TRUSS, can recapitulate many aspects of TNF-R1 signaling in a ligand-independent fashion (9, 27-31). Seeking to further investigate how enforced expression of TRUSS may affect TNF-R1 signaling, we determined that TRUSS self-associates to form homomeric complexes. Using a combination of co-immunoprecipitation approaches with epitope-tagged TRUSS constructs and gel filtration analysis of lysates from TRUSS-expressing cells together with bacterially-expressed, purified recombinant TRUSS, our results show that TRUSS exists both as a monomer and as a continuum of higher order complexes, including a trimer. In this respect, TRUSS resembles TNF-R1 and TRAF2, which, based on X-ray crystallographic approaches, have also been shown to exist as homotrimers (17, 32). Though future studies will be required to fully comprehend the mechanism of homo-oligomer assembly, our findings suggest that the C-terminal region may play an important role. Furthermore, theoretical modeling based on the data presented herein suggests a model (Figure 6A (panel d)) in which the C-terminal region of TRUSS contributes to homotrimer formation. In this model, the C-terminal region is predicted to be buried within the central region of the trimer, and the TNF-R1 and TRAF2 binding interfaces are exposed at the periphery. Interestingly, computational analysis identified a 46 amino acid sequence in the C-terminal region of TRUSS (encompassed by residues 614-659 of human and mouse TRUSS) that conforms to the heptad repeat structure of a leucine zipper (Figure 6B). Leucine zippers generally contain Leu, Iso or Val residues at the “a” and “d” positions, with charged amino acids frequently being present at the “e” and “g” positions (33). These residues are also important in trimer formation by leucine zippers (34, 35). The putative leucine zipper in the C-terminal region exhibits almost exact conservation across vertebrate species from man to zebra fish further suggesting that it plays an important role in the function(s) of TRUSS (Figure 6B). Leucine zipper sequences have been shown to be important in other protein-protein interactions including IKKγ, c-Cbl and c-Jun (36-38), and in membrane localization of the signaling adaptor, SLP-65 (39). Thus, it is conceivable that the C-terminal leucine zipper of TRUSS may contribute to the ability of TRUSS to self-associate. Clearly, X-ray crystallography or NMR studies are warranted to shed further light on this issue.
In summary, our results suggest that binding interface for TNF-R1 and TRAF2 is located in the N-terminal portion of TRUSS (residues 1-440). In addition, our findings indicate that TRUSS is capable of forming homomeric complexes. Additional information about the functions of TRUSS should be forthcoming with the development of TRUSS-deficient mice.
ACKNOWLEDGEMENTS
The authors wish to thank Linda Remigio and Ben Edelman for their outstanding technical assistance in these studies. We also wish to acknowledge Dr. Surinder Soond for his initial work on the cloning and characterization of TRUSS and Dr. Murry Wynes for helpful discussions.
This work was supported by Public Health Service grants HL055549, HL068628, AI070941 (DWHR) and GM080719 (GZ) from the National Institutes of Health. Dr. Terry Powers was supported by Institutional T-32 pulmonary training grant HL00048 from the National Institutes of Health and by a Predoctoral Emphasis Pathway in Tumor Immunology training grant from the Cancer Research Institute.
Abbreviations
- TRUSS
TNF-Receptor Ubiquitous Scaffolding and Signaling protein
- TRPC4AP
Transient Receptor Potential Canonical 4 Associated Protein
- TNF-R1
Tumor Necrosis Factor Receptor-1
- TNF-α
Tumor Necrosis Factor-α
- IKK
IκB Kinase
- FADD
Fas Associated Death Domain protein
- NF-κB
Nuclear Factor-κB
- RIP
Receptor Interacting Protein
- TRADD
TNF-R1 Associated Death Domain protein
- TRAF2
TNF Receptor Associated Factor 2
- FPLC
Fast Protein Liquid Chromatography
REFERENCES
- 1.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Wajant H, Henkler F, Scheurich P. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal. 2001;13:389–400. doi: 10.1016/s0898-6568(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 3.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 4.Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12:419–429. doi: 10.1016/s1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang SQ, Kovalenko A, Cantarella G, Wallach D. Recruitment of the IKK signalosome to the p55 TNF receptor: RIP and A20 bind to NEMO (IKKgamma) upon receptor stimulation. Immunity. 2000;12:301–311. doi: 10.1016/s1074-7613(00)80183-1. [DOI] [PubMed] [Google Scholar]
- 6.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 7.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 8.Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, Heinrich M, Merkel O, Ehrenschwender M, Adam D, Mentlein R, Kabelitz D, Schutze S. Compartmentalization of TNF Receptor 1 Signaling; Internalized TNF Receptosomes as Death Signaling Vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 9.Soond SM, Terry JL, Colbert JD, Riches DW. TRUSS, a novel tumor necrosis factor receptor 1 scaffolding protein that mediates activation of the transcription factor NF-kappaB. Mol Cell Biol. 2003;23:8334–8344. doi: 10.1128/MCB.23.22.8334-8344.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soond SM, Terry JL, Riches DW. TRUSS, a tumor necrosis factor receptor-1-interacting protein, activates c-Jun NH(2)-terminal kinase and transcription factor AP-1. FEBS Lett. 2006;580:4591–4596. doi: 10.1016/j.febslet.2006.06.098. [DOI] [PubMed] [Google Scholar]
- 11.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 12.Van Linden AA, Cottin V, Leu C, Riches DW. Phosphorylation of the membrane proximal region of tumor necrosis factor receptor CD120a (p55) at ERK consensus sites. J Biol Chem. 2000;275:6996–7003. doi: 10.1074/jbc.275.10.6996. [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi M, Rothe M, Goeddel DV. Anatomy of TRAF2. Distinct domains for nuclear factor-kappaB activation and association with tumor necrosis factor signaling proteins. J Biol Chem. 1996;271:19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- 14.Ye H, Park YC, Kreishman M, Kieff E, Wu H. The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol Cell. 1999;4:321–330. doi: 10.1016/s1097-2765(00)80334-2. [DOI] [PubMed] [Google Scholar]
- 15.McWhirter SM, Pullen SS, Holton JM, Crute JJ, Kehry MR, Alber T. Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc Natl Acad Sci U S A. 1999;96:8408–8413. doi: 10.1073/pnas.96.15.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothe M, Sarma V, Dixit VM, Goeddel DV. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 17.Park YC, Burkitt V, Villa AR, Tong L, Wu H. Structural basis for self-association and receptor recognition of human TRAF2. Nature. 1999;398:533–538. doi: 10.1038/19110. [DOI] [PubMed] [Google Scholar]
- 18.Tartaglia LA, Ayres TM, Wong GH, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 19.Telliez JB, Xu GY, Woronicz JD, Hsu S, Wu JL, Lin L, Sukits SF, Powers R, Lin LL. Mutational analysis and NMR studies of the death domain of the tumor necrosis factor receptor-1. J Mol Biol. 2000;300:1323–1333. doi: 10.1006/jmbi.2000.3899. [DOI] [PubMed] [Google Scholar]
- 20.Leonardi A, Ellinger-Ziegelbauer H, Franzoso G, Brown K, Siebenlist U. Physical and functional interaction of filamin (actin-binding protein-280) and tumor necrosis factor receptor-associated factor 2. J Biol Chem. 2000;275:271–278. doi: 10.1074/jbc.275.1.271. [DOI] [PubMed] [Google Scholar]
- 21.Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, Schmukle AC, Davidson AJ, Callus BA, Wong WW, Gentle IE, Carter H, Lee EF, Walczak H, Day CL, Vaux DL, Silke J. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (tnf) to efficiently activate nf-{kappa}b and to prevent tnf-induced apoptosis. J Biol Chem. 2009;284:35906–35915. doi: 10.1074/jbc.M109.072256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawamata Y, Imamura T, Babendure JL, Lu JC, Yoshizaki T, Olefsky JM. Tumor necrosis factor receptor-1 can function through a G alpha q/11-beta-arrestin-1 signaling complex. J Biol Chem. 2007;282:28549–28556. doi: 10.1074/jbc.M705869200. [DOI] [PubMed] [Google Scholar]
- 23.Fujino G, Noguchi T, Matsuzawa A, Yamauchi S, Saitoh M, Takeda K, Ichijo H. Thioredoxin and TRAF family proteins regulate reactive oxygen species-dependent activation of ASK1 through reciprocal modulation of the N-terminal homophilic interaction of ASK1. Mol Cell Biol. 2007;27:8152–8163. doi: 10.1128/MCB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsao DH, McDonagh T, Telliez JB, Hsu S, Malakian K, Xu GY, Lin LL. Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway. Mol Cell. 2000;5:1051–1057. doi: 10.1016/s1097-2765(00)80270-1. [DOI] [PubMed] [Google Scholar]
- 25.Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–235. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 26.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 27.Hsu H, Huang J, Shu HB, Baichwal V, Goeddel DV. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 28.Hu WH, Johnson H, Shu HB. Activation of NF-kappaB by FADD, Casper, and caspase-8. J Biol Chem. 2000;275:10838–10844. doi: 10.1074/jbc.275.15.10838. [DOI] [PubMed] [Google Scholar]
- 29.Rothe M, Wong SC, Henzel WJ, Goeddel DV. A novel family of putative signal transducers associated with the cytoplasmic domain of the 75 kDa tumor necrosis factor receptor. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 30.Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. Embo J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- 31.Baud V, Liu ZG, Bennett B, Suzuki N, Xia Y, Karin M. Signaling by proinflammatory cytokines: oligomerization of TRAF2 and TRAF6 is sufficient for JNK and IKK activation and target gene induction via an amino-terminal effector domain. Genes Dev. 1999;13:1297–1308. doi: 10.1101/gad.13.10.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 33.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckert DM, Malashkevich VN, Kim PS. Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues. J Mol Biol. 1998;284:859–865. doi: 10.1006/jmbi.1998.2214. [DOI] [PubMed] [Google Scholar]
- 35.Harbury PB, Kim PS, Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994;371:80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- 36.Agou F, Traincard F, Vinolo E, Courtois G, Yamaoka S, Israel A, Veron M. The trimerization domain of NEMO is composed of the interacting C-terminal CC2 and LZ coiled-coil subdomains. J Biol Chem. 2004;279:27861–27869. doi: 10.1074/jbc.M314278200. [DOI] [PubMed] [Google Scholar]
- 37.Schuermann M, Neuberg M, Hunter JB, Jenuwein T, Ryseck RP, Bravo R, Muller R. The leucine repeat motif in Fos protein mediates complex formation with Jun/AP-1 and is required for transformation. Cell. 1989;56:507–516. doi: 10.1016/0092-8674(89)90253-5. [DOI] [PubMed] [Google Scholar]
- 38.Bartkiewicz M, Houghton A, Baron R. Leucine zipper-mediated homodimerization of the adaptor protein c-Cbl. A role in c-Cbl’s tyrosine phosphorylation and its association with epidermal growth factor receptor. J Biol Chem. 1999;274:30887–30895. doi: 10.1074/jbc.274.43.30887. [DOI] [PubMed] [Google Scholar]
- 39.Kohler F, Storch B, Kulathu Y, Herzog S, Kuppig S, Reth M, Jumaa H. A leucine zipper in the N terminus confers membrane association to SLP-65. Nat Immunol. 2005;6:204–210. doi: 10.1038/ni1163. [DOI] [PubMed] [Google Scholar]