Abstract
Sirtuins are emerging as key regulators of many cellular functions including metabolism, cell growth, apoptosis, and genetic control of ageing. In mammals there are seven sirtuin analogues, SIRT1 to SIRT7. Among them SIRT3 is unique because this is the only analogue whose increased expression has been found to be associated with extended lifespan of humans. SIRT3 levels have been shown to be elevated by exercise and calorie restriction. Although the role of SIRT3 in cell biology is only beginning to be understood, initial studies have shown that SIRT3 plays a major role in free fatty acid oxidation and maintenance of cellular ATP levels. In the heart SIRT3 has been found to block development of cardiac hypertrophy and protect cardiomyocytes from oxidative stress-mediated cell death. Similarly, SIRT3 has been reported to have tumour-suppressive characteristics. In this article, we review the known effects of SIRT3 in different tissues and relate them to the protection of cardiomyocytes under stress.
Keywords: Sirtuins, SIRT3, Cardiac hypertrophy, Mitochondrial metabolism, Apoptosis
1. Introduction
The desire to live longer and probably forever has long fascinated mankind. Concoctions to prevent ageing and maintain youth have been described in medical books of ancient civilizations, including Charaka Samhita, the most ancient textbook of Ayurveda (an Indian system of traditional medicine), which is believed to have been written centuries before the birth of Christ. It seems our forefathers found a way to live longer and healthy by undergoing calorie restriction, a diet regimen that is considered to be impractical for modern society where food is surplus and time is scarce. Even though vaccination, antibiotics, better child care, and early disease-detection techniques in combination with modern drugs have helped us to increase our average lifespan, the quest to increase maximal lifespan still remains elusive. The major advances in ageing research that we have witnessed in the past two decades are the rediscovery of benefits of calorie restriction, and the delineation of the molecular mechanism involved in its protective effects. Many studies have proposed that the beneficial effect of calorie restriction is mediated through a set of genes collectively called sirtuins (SIRT1–7).1
Sirtuins belong to class III histone deacetylases that are dependent on nicotinamide adenine dinucleotide (NAD) for their activity. Seven different homologues of yeast Sir2 exist in mammals. Even though named as histone deacetylases, they have several distinct and overlapping non-histone protein targets and functions. SIRT1 is the most widely studied and the closest mammalian homologue of yeast Sir2. SIRT1 is present both in the nucleus and in the cytoplasm, and is shown to deacetylate many non-histone proteins including p53, Foxo, PGC1α, PPARγ LKB1, liver X receptor, hypoxia-inducible factor 2α, and TORC2 that are known to play a crucial role in cell survival, metabolism, and stress response.2–7 In the heart, low- to moderate overexpression of SIRT1 was found to be cardioprotective, whereas its high-level expression was associated with the development of cardiomyopathy.8 Similar to SIRT1, SIRT2 was also localized in the nucleus and in the cytoplasm and was shown to be important for chromosomal stability during mitosis.9,10 SIRT-3, -4, and -5 are localized predominantly in mitochondria and are referred to as mitochondrial stress sensors that can modulate the activity of several mitochondrial proteins involved in metabolism (reviewed in Huang et al.11). SIRT6 is a chromatin-associated sirtuin analogue involved in the regulation of diverse cellular functions like genomic stability, glucose homeostasis, and inflammation.12–14 SIRT6-deficient mice showed complex degenerative phenotype, enhanced rate of ageing, and shortened lifespan.15 The last member of this family, SIRT7 is found to be associated with nucleoli and condensed chromosomes.16,17 SIRT7-deficient mice have reduced lifespan, and develop cardiac hypertrophy and inflammatory cardiomyopathy.18 From these early studies, it seems that sirtuins have considerable redundancy in targets and regulate the fundamental aspects of longevity like cell survival, metabolism, and stress resistance.
Of the seven sirtuins, SIRT3 is the only sirtuin analogue whose increased expression was shown to be associated with longevity of humans.19,20 In a clinical study, it was reported that older individuals (60+ years) with a sedentary lifestyle had ∼40% reduced SIRT3 levels when compared with younger subjects, and after endurance exercise, the health benefits of older patients were accompanied by elevated levels of SIRT3.21 In another study, increased expression of SIRT3 due to polymorphism in the SIRT3 promoter was found to be associated with an extended lifespan of man, thus implicating a role of SIRT3 in the genetic control of human ageing.19,20 From these studies, the role of SIRT3 in cardioprotection is apparent as cardiovascular disease is the leading cause of mortality among adults above age 60; though very few studies have examined the role of SIRT3 in the heart, so far. In this review we discuss, our understanding of the role of SIRT3 in relation to the heart, and how SIRT3-mediated regulation of enzyme activity by protein-deacetylation reported in other tissues could correlate with dysfunction of the heart.
2. Subcellular localization of SIRT3
Understanding subcellular localization of a protein is important for delineating the physiological function of the protein, and also to identify its interacting partners to learn how they influence each others' activity under different physiological conditions. Subcellular localization of SIRT3 has been a subject of considerable debate. Several studies suggested a strictly mitochondrial localization of SIRT3, which was contradicted by others who found its presence in the nucleus and cytoplasm as well.22 Intertwined with disputes of localization are the reports regarding the size of SIRT3. The full-length human SIRT3 (hSIRT3) is a 44 kDa protein with an N-terminal mitochondrial localization sequence (MLS).23 Initially, it was considered that the nuclear form of SIRT3 is enzymatically inert. Following import to mitochondria, 142 amino acids from the N-terminal of full-length hSIRT3 are cleaved by matrix processing peptidase (MPP) present in the mitochondrial matrix to generate an active 28 kDa short form.23 This was confirmed by another study, which showed mitochondrial cristae as the seat of short form of hSIRT3, while overexpressed full-length hSIRT3 was localized in the cytoplasm.24 In contrast to this, Scher et al.25 reported that full-length hSIRT3 is a nuclear protein that translocates to the mitochondria upon stress. These studies were later challenged by Cooper and Spelbrink,26 who demonstrated that endogenous hSIRT3 was present exclusively in the mitochondria, whereas overexpression of hSIRT3 lacking MLS led to the expression of protein both in the cytoplasm and in the nucleus.
Contrary to hSIRT3, the mouse SIRT3 (mSIRT3) was initially reported to lack the mitochondrial localization signal.27–29 Yet, it was shown to be localized in the inner mitochondrial membrane, thus casting shadow over the previously identified MLS-mediated mechanism of mitochondrial localization and MPP-mediated protein cleavage. Amidst these controversies, we found that like hSIRT3, mSIRT3 is also expressed in two forms, an ∼44 kDa long form and a 28 kDa short form.30 In a later study, Cooper et al.31 identified three different splice variants for mSIRT3, designated as M1, M2, and M3. The translation of M1, M2, and M3 begins from Met1 (methionine1), Met15, and Met78, respectively. This study also showed that M3 lacks the mitochondrial leader peptide, and is equivalent to the human short form of SIRT3 (Met 143). M1 and M2 were found to be localized in the mitochondria, whereas, overexpression of M3 did not result in mitochondrial localization, but showed a uniform cytoplasmic distribution, and upon high level of overexpression it was found to be localized in the nucleus as well. Like hSIRT3, M1 and M2 were also found to be cleaved to generate identical shorter forms, similar in size to M3. We observed that in the heart, the endogenous long form of mSIRT3 was localized to the mitochondria, cytoplasm, and to a lesser extent in the nucleus; whereas, the short form of mSIRT3 was detected only in the mitochondria.30 There is no other report currently available regarding the localization of endogenous SIRT3 in the heart. With the deletion of mSIRT3, it was found that lysine acetylation increased only in the mitochondrial proteins, thus again providing evidence that mSIRT3 is the main deacetylase localized in the mitochondria.32 However, this study does not exclude the possibility of SIRT3 localization outside the mitochondria, as other sirtuins might have overtaken the effect of SIRT3 deficiency in other compartments. In a recent study, Bao et al.33 reported that after overexpression of long and short form of mSIRT3, both forms were detected in the nuclear and cytoplasmic fractions of mouse embryonic fibroblasts (MEFs) and H9C2 cells and both forms were enzymatically active. For clarity, subcellular localization of endogenous SIRT3 in human and mouse as reported is summarized in Table 1. Overall, these studies suggest the existence of similar mechanisms for subcellular localization, cleavage, and protein conservation between hSIRT3 and mSIRT3, except that no splice variants of hSIRT3 have been reported.
Table 1.
Subcellular localization of endogenous SIRT3 in human and mouse as reported
3. SIRT3, reactive oxygen species production, and cardiac hypertrophy
The heart produces and utilizes more energy than any other organ, and more than 90% of its energy is produced from mitochondrial respiration.34 Normal cardiac function could be maintained only if ATP is continuously re-synthesized.35 To meet this high energy demand, cardiomyocytes have the highest density of mitochondria among all mammalian organs and occupy ∼30% of the heart volume.34 Undesirable side effects of this high metabolic rate are the production of reactive oxygen species (ROS). These ROS, if unchecked can cause serious damage to cells through oxidation of proteins, lipids, and nucleic acids, by causing unwanted post-translational modification of proteins, altering the fluidity of the lipid membranes, and mutations in genomic and mitochondrial DNA.36 Cells are protected from the deleterious effect of ROS by antioxidant enzymes, such as superoxide dismutases (SODs), catalase, and peroxidases. Superoxides generated from mitochondrial respiration are converted to hydrogen peroxide by SOD, whereas catalase and glutathione peroxidase help to convert hydrogen peroxide to water. The expression of SOD, particularly MnSOD, and catalase has been shown to be regulated by the members of the family of forkhead transcription factors (Foxos). Foxo3a binds to the gene promoters of MnSOD and catalase, and transcriptionally upregulates their expression, thereby reducing the cellular ROS levels.37,38
Increased production of ROS has been strongly implicated in the development of cardiac hypertrophy.39,40 We have recently reported that SIRT3 blocks cardiac hypertrophic response through activation of Foxo3a-dependent antioxidant defence mechanism in mice.41 SIRT3-deficient mice though, looked normal in their routine activity, developed cardiac hypertrophy, and interstitial fibrosis at 8 weeks of age. These mice showed extreme hypertrophic response when challenged with hypertrophic agonists. On the other hand, transgenic mice overexpressing SIRT3 in the heart effectively blocked agonist-mediated cardiac hypertrophy. Cardiomyocytes cultured from SIRT3-deficient hearts showed increased ROS levels, when compared with their respective wild-type controls, suggesting that SIRT3 prevented cardiac hypertrophic response by scavenging cellular ROS. This observation was supported by findings showing reduced activation of the Ras oncoprotein and its downstream targets, namely MAPK/ERK and PI3K/Akt signalling pathways in SIRT3 transgenic mice, compared with wild-type controls, infused with hypertrophic agonists. In this study, SIRT3 used for the construction of transgenic mice lacks mitochondrial localization signal and hence was found to exert its function by deacetylating the transcription factor, Foxo3a, aiding its localization in the nucleus. Activation of Foxo3a resulted in the transcriptional upregulation of MnSOD and catalase. SIRT3-deficient mice showed reduced activity of MnSOD and catalase. On the other hand, SIRT3 transgenic mice had increased basal levels of both MnSOD and catalase, which remained unchanged when challenged with hypertrophic agonists, thus suggesting that SIRT3 elevates anti-oxidant defence mechanism of the heart.
These observations were supported by another study, where SIRT3 was shown to have the potential to suppress tumour progression. Kim et al.42 reported significantly increased ROS levels in SIRT3-deficient MEFs subjected to genotoxic and metabolic stress. The same study also showed that increased levels of ROS in SIRT3-deficient MEFs partly contributed to the tumour-permissive phenotype of cells. The tumour-suppressive characteristic of SIRT3 observed in this study also supports the anti-hypertrophic activity of this molecule reported by us before (Figure 1).41
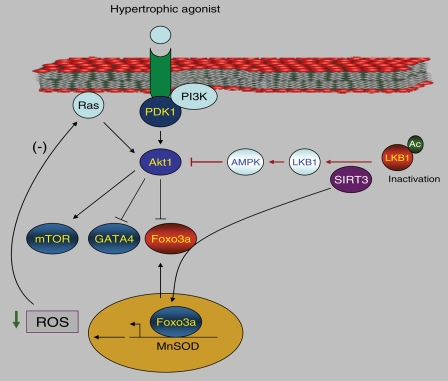
Figure 1.
Mechanisms of anti-hypertrophic effects of SIRT3. Akt activation is widely implicated in the development of cardiac hypertrophy. SIRT3 blocks cardiac hypertrophy by suppressing the Akt signalling in cardiomyocytes by, at least, two ways. First, it deacetylates and activates LKB1, thereby activating AMPK. AMPK activation is known to suppress Akt phosphorylation. Second, SIRT3-mediated deacetylation promotes nuclear localization of Foxo3a, leading to enhanced transcription of Foxo3a-dependent antioxidant genes, e.g. MnSOD, thereby reducing cellular ROS levels. Increased ROS levels are known to activate Ras, which in turn activates Akt. By blocking cellular ROS levels, SIRT3 shuts down the activity of Ras and thus the activity of Akt (for details see Sundaresan et al.41 and Pillai et al.56).
4. SIRT3 and metabolism
One of the hallmarks of cardiac remodelling associated with hypertrophy is the transcriptional reprogramming leading to foetal gene activation. This change in gene expression also causes considerable metabolic transformation in the heart. A normal healthy heart oxidizes both fatty acids and glucose simultaneously for its energy needs.43 But during cardiac hypertrophy, just like foetal heart, fatty acid oxidation becomes impaired, compromising the ability of the heart to oxidize long-chain fatty acids. This causes a switch in the myocardial energy substrate utilization from fatty acid β-oxidation to glycolysis.44 During the initial compensatory phase of hypertrophy, this switch in substrate preference increases the efficiency of the heart, but when hypertrophy prolongs, this metabolic adaptation becomes insufficient owing to decreased capacity to oxidize glucose, leading to heart dysfunction, and eventually to failure.44,45
Emerging evidence indicates that reduction in the activity of PGC-1α is associated with a shift from fatty acid oxidation to glycolytic metabolism in the heart.35,46 PGC1α has been shown to be downregulated in hypertrophied and failing hearts, as well as in cultured cardiomyocytes subjected to develop hypertrophy.47–51 PGC1α is an acetylated protein, and this post-translational modification inhibits its activity.52 Both SIRT1 and SIRT3 have been shown to activate PGC-1α in different cell types, though only SIRT1 was shown to deacetylate PGC1α.28,53,54 SIRT1 has been implicated in the regulation of systemic and hepatic glucose, lipid, and cholesterol homeostasis in mice. These effects of SIRT1 were found to be mediated through its ability to activate PGC-1α, and some of them were observed only under fasted conditions.5 Thus, it appears that SIRT1 is an important player in the regulation of freefatty acid oxidation and glucose metabolism, in response to mild stress (fasting).
Recent studies conducted with SIRT3-deficient mice complemented these findings and revealed the important role of SIRT3 in regulating freefatty acid metabolism under stress. Hirschey et al.55 observed that SIRT3 knockout mice had 50% reduction in freefatty acid oxidation, which resulted in the accumulation of long-chain fatty acids in the liver upon fasting. They also found that fasted mice had a 33% reduction in fatty acid oxidation in the heart. The defect in freefatty acid oxidation was correlated with increased acetylation of long-chain acyl-CoA dehydrogenase (LCAD) in SIRT3-deficient mice, which led to decreased enzymatic activity of LCAD by 40%. Since cardiac hypertrophy is also associated with reduced freefatty acid oxidation, and SIRT3 levels are reduced in hypertrophied/failing hearts,41 it seems to be of logic to assume that similar SIRT3-mediated mechanisms could operate in the heart to regulate free fatty acid oxidation.
One of the consequences of defective freefatty acid oxidation is reduced production of ATP. Several studies, including ours have shown a positive correlation between ATP depletion and cardiac hypertrophy.35,56 Basal ATP levels were found to be correlated with SIRT3 levels in several tissues. Many organs having lower ATP levels like pancreas, spleen, and skeletal muscle were shown to have reduced SIRT3 expression, while organs with higher ATP levels like heart, liver, and kidney showed higher levels of SIRT3 expression.57 SIRT3-deficient MEFs were also showed to have 30% reduced basal ATP levels. The hearts of SIRT3-deficient mice showed more than 50% reduction in ATP level, compared with wild-type mice.57 Furthermore, in stress conditions where ATP demand is high, increased expression levels of SIRT3 have been noticed. These studies demonstrated that SIRT3 is an important player in the regulation of cellular ATP levels.
Although the mechanism of SIRT3 in regulating the ATP biosynthesis is yet to be fully understood, several proteins of the mitochondrial complex 1 were found to be hyperacetylated in SIRT3-deficient mice. SIRT3 was also shown to directly interact with and deacetylate NDFU9 subunit of complex 1.57 In the liver, fasting ATP levels were found to be reduced by 43% in SIRT3-deficient mice, and that was associated with acetylation and decreased activity of LCAD.55 In a separate study, Kim et al.42 detected reduced activity of complex III in SIRT3-deficient Myc/Ras-transformed cells, compared with wild-type control cells. SIRT3 was also found to deacetylate and activate acetylcoenzyme A synthase 2 (AceCS2), which is highly expressed in the heart.58,59 AceCS2 plays an important role in the mitochondria by generating acetyl-CoA for the tricarboxylic acid (TCA) cycle. Also the intermediates of the TCA cycle have been shown to be deacetylated and activated by SIRT3. In vitro studies have shown that SIRT3 can deacetylate and activate glutamate dehydrogenase and isocitrate dehydrogenase 2.60 Furthermore, another TCA intermediate, succinate dehydrogenase was found to be acetylated in SIRT3 knockout mice, which led to decreased activity of the enzyme.61 There is also a report showing that SIRT3 enhanced the oxidative phosphorylation by modulating the activity of cyclophilin D. SIRT3-mediated deacetylation of cyclophilin D inhibits its enzymatic activity, thereby inducing its dissociation from ANT1 (adenine nucleotide translocator), which then promotes detachment of hexokinase-II from voltage-dependent anion channel. This process caused redistribution of hexokinase-II from mitochondria to cytosol, resulting in an increase in mitochondrial oxidative phosphorylation.62 These studies underscore a non-redundant and specific role of SIRT3 in maintaining the cellular energy balance, thus suggesting the immense possibility of finding other mitochondrial proteins whose activity could be modulated by SIRT3 in the heart.
Yet another mechanism through which SIRT3 regulates energy metabolism is by activating AMP-activated protein kinase (AMPK).54,56,63 AMPK is activated by its upstream kinase LKB1 in the α-subunit at threonine 172 (T172), which lies in the activation loop. Activated AMPK can upregulate the catabolic pathways such as cellular glucose uptake and fatty-acid oxidation resulting in the increased production of ATP, while switching off anabolic pathways that consume ATP. Since the heart produces and consumes more energy than any other organ, it is highly relevant that AMPK may act as a key regulator of energy metabolism in the heart.64 Consistent with this notion, activation of AMPK was shown to inhibit cardiac hypertrophy in both in vitro and in vivo models.65,66 Further, cardiac-specific deletion of LKB1 was found to induce cardiac hypertrophy and heart dysfunction.67 Recently, we have shown that SIRT3 can interact with and deacetylate LKB1 in the heart. This resulted in the activation of LKB1 which in turn activated AMPK. Activation of AMPK by exogenous supplementation of NAD rescued the heart from ATP depletion and blocked the activation of pro-hypertrophic Akt signalling (Figure 1).56 AMPK has also been shown to activate PGC1α, which has been implicated in blocking the cardiac hypertrophic response.66 These studies indicate that SIRT3-mediated AMPK activation could be another alternative mechanism through which SIRT3 can protect the heart from developing pathologic hypertrophy.
5. SIRT3 and apoptosis
Only very few studies have addressed the role of SIRT3 in apoptosis and the results are largely contradictory. SIRT3 has been shown to possess pro-apoptotic as well as anti-apoptotic activity depending upon cell type. Tumour cells (K562) stably made to deplete or to overexpress SIRT3 were not rescued from kaempferol (an oxidative stress-inducing agent) induced cell death.68 Allison and Milner reported that apoptosis of HCT116 cells induced by Bcl-2 silencing was prevented by depletion of SIRT3, thus suggesting pro-apoptotic effects of SIRT3 in cancer cells.69 Observations of these studies partially contradicted the role of SIRT3 in cell death as reported by other studies. Overexpression of Nampt, a stress- and nutrient-responsive gene, involved in the biosynthesis of NAD, has been shown to protect human fibrosarcoma cells against genotoxic agents. This protection was found to be mediated through activation of SIRT3 and SIRT4 in mitochondria.70 In cardiomyocytes, we observed that after oxidative stress SIRT3 levels were elevated in both the nucleus and mitochondria, and overexpression of SIRT3 protected cardiomyocytes from cell death.30 This protection was seen with the overexpression of wild-type SIRT3 and nuclear SIRT3 (lacking MLS), thus demonstrating that in addition to mitochondria, SIRT3 also plays a role in the nucleus to protect cells from death. Nuclear full-length SIRT3 has also been found to be enzymatically active and capable of deacetylating histones H3-K9ac and H4-K16ac both in vivo and in vitro,25 thus implicating that some of the anti-apopototoic effects of SIRT3 could be mediated through its ability to deacetylate nuclear targets.30 Like its homologue SIRT1, these observations raise the possibility of existence of several non-histone proteins as targets of SIRT3 regulating apoptosis. However, not much is known besides deacetylation of histones, which could explain the ability of nuclear SIRT3 to protect cells from genotoxic stress.
Mitochondria plays a prominent role in the induction of apoptosis after cellular stress. The initiating events of this process are translocation of a pro-apoptotic protein Bax from cytoplasm to mitochondria. Translocation of Bax to mitochondria causes release of cytochrome c, which activates caspase-9. Activated caspase-9 further cleaves pro-caspase-3 to caspase-3 resulting in the induction of apoptosis. Under survival conditions, Bax is in complex with Ku70, which prevents translocation of Bax to mitochondria and thereby blocking apoptosis. This Ku70–Bax interaction is lysine-acetylation-sensitive. Only deacetylated Ku70 forms complex with Bax, whereas acetylation of Ku70 releases Bax, thus enabling it to translocate to the mitochondria, where it initiates apoptotic cascade. Several studies have shown that SIRT1-mediated deacetylation of Ku70 sequesters Bax away from mitochondria.71,72 However, in cardiomyocytes SIRT1 was not found to be effective in deacetylating Ku70. Instead, we found that SIRT3 was capable of binding to and deacetylating Ku70. Accordingly, overexpression of SIRT3 blocked the translocation of Bax into mitochondria upon genotoxic stress.30 SIRT3 overexpression also rescued SIRT1-deficient HeLa cells from apoptosis induced by genotoxic stress, thus establishing a role of SIRT3 in deacetylating Ku70 and protecting cells from apoptosis.30 Increased activity of p53 after acetylation has also been implicated in apoptosis. Most recently, SIRT3 was found to directly interact with and deacetylate p53, thereby abrogating its activity to execute growth arrest and senescence in bladder carcinoma cells.73 From these studies, it appears that SIRT3 might have variable effects in many cancer cell lines but was found to be anti-apoptotic in cardiomyocytes.
6. Future perspectives
SIRT3 is a protein of tremendous potential, which can modulate a variety of cellular processes, including growth arrest, apoptosis, senescence, and metabolism. At this time a ‘pubmed’ search with keyword ‘SIRT3’ yielded only 75 results. Even though current research has identified several SIRT3 targets (Table 2) and broadened our understanding of this molecule, it seems like the tip of the iceberg. Moreover, very few studies have addressed the role of SIRT3 in the heart. Several other questions need to be resolved to fully understand its function in the heart. For example, the discovery that nuclear SIRT3 is enzymatically active, and that it can stimulate Foxo3a to transcriptionally activate the expression of antioxidant genes, suggests that SIRT3 has the potential to exert a complex influence on the transcription of genes, which has not yet been examined. It will be interesting to see if SIRT3 is capable of deacetylating and inactivating transcription factors like MEF2 or GATA4, whose overactivation by acetylation has been implicated in the progression of cardiac hypertrophy. In this context, it is important to recollect that the transgenic mice used in our study lack mitochondrial localization signal, and further studies utilizing full-length SIRT3 need to be conducted to understand the direct effect of SIRT3 on mitochondrial proteins in the heart. Even though SIRT3 has been shown to activate PGC1α, which is known to participate in mitochondrial biogenesis, a full understanding of the role of SIRT3 in mitochondrial regeneration and its association with the development of cardiac hypertrophy needs to be examined in detail. Because SIRT3 was found to regulate free fatty oxidation, there is a great possibility that during hypertrophy, change in SIRT3 levels dictates shift in cardiac metabolism from free fatty acids to glycolysis. The observation that SIRT3-deficient mice developed an early-age onset of hypertrophy associated with fibrosis warrants the need for studying SIRT3 gene mutations in familial hypertrophic cardiomyopathies, and also the role of SIRT3 in the differentiation of myoblasts and myofibroblasts. Regarding the post-translational modifications, there is a very high probability that SIRT3 could be post-translationally modified, which could influence its localization and/or function. By analysing the SIRT3 sequences, Onyango et al.24 have identified potential post-translational modification sites like glycosylation, phosphorylation, N-myristoylation, and amidation sites for SIRT3. However, so far no post-translational modifications of SIRT3 have been reported. Future research in this area will help to identify modifications that regulate SIRT3 activity, which might enable us to develop agents that can modulate its activity. Associated with this, is the need for identifying small molecular activators of SIRT3, which may lead to the development of new therapeutic agents for the treatment of heart failure and other diseases, where loss of SIRT3 seems to plays a leading role in pathogenesis.
Table 2.
Direct targets of SIRT3
| Substrates of SIRT3 | Effect of deacetylation | References |
|---|---|---|
| AceCS2 | Increased enzymatic activity | 23 |
| LCAD | Increased enzymatic activity | 55 |
| NDUFA9 | Increased enzymatic activity | 57 |
| Succinate dehydrogenase | Increased enzymatic activity | 61 |
| Isocitrate dehydrogenase | Increased enzymatic activity | 60 |
| Glutamate dehydrogenase | Increased enzymatic activity | 60 |
| LKB1 | Increased enzymatic activity | 56 |
| Foxo3a | Increased transcription activity | 41,74 |
| Ku70 | Increased binding with Bax | 30 |
| P53 | Increased protein degradation | 73 |
| MRPL10 | Decreased translational activity | 75 |
| Cyclophilin D | Decreased enzymatic activity | 62 |
| Histone 3,4 | Decreased transcription | 25 |
For additional substrates of SIRT3 identified by MALDI-TOF/TOF-MS/MS analysis see Law et al.76
Conflict of interest: None of the authors have conflict of interest.
Funding
These studies were supported by NIH grants RO1 HL-68083, HL-77788, and HL-83423.
References
- 1.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. doi:10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. doi:10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. doi:10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. doi:10.1016/S0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 5.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. ProcNatl Acad Sci. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. doi:10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. doi:10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 7.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. doi:10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 8.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. doi:10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 9.Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, et al. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2006;26:945–957. doi: 10.1038/sj.onc.1209857. doi:10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 10.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One. 2007;2:e784. doi: 10.1371/journal.pone.0000784. doi:10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbapap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. doi:10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombard DB. Sirtuins at the breaking point: SIRT6 in DNA repair. Aging (Albany NY) 2009;1:12–16. doi: 10.18632/aging.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. doi:10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. doi:10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 16.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. doi:10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. doi:10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. doi:10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 19.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. doi:10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, et al. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. doi:10.1016/S0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 21.Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, et al. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–2942. doi: 10.2337/db08-0349. doi:10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hallows WC, Albaugh BN, Denu JM. Where in the cell is SIRT3?–functional localization of an NAD+-dependent protein deacetylase. Biochem J. 2008;411:e11–e13. doi: 10.1042/BJ20080336. doi:10.1042/BJ20080336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. doi:10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. doi:10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. doi:10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper HM, Spelbrink JN. The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J. 2008;411:279–285. doi: 10.1042/BJ20071624. doi:10.1042/BJ20071624. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174–179. doi: 10.1016/j.bbrc.2007.11.122. doi:10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- 28.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. doi:10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 29.Yang YH, Chen YH, Zhang CY, Nimmakayalu MA, Ward DC, Weissman S. Cloning and characterization of two mouse genes with homology to the yeast Sir2 gene. Genomics. 2000;69:355–369. doi: 10.1006/geno.2000.6360. doi:10.1006/geno.2000.6360. [DOI] [PubMed] [Google Scholar]
- 30.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. doi:10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper HM, Huang JY, Verdin E, Spelbrink JN. A new splice variant of the mouse SIRT3 gene encodes the mitochondrial precursor protein. PLoS One. 2009;4:e4986. doi: 10.1371/journal.pone.0004986. doi:10.1371/journal.pone.0004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. doi:10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, et al. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J Cell Biochem. 2010;110:238–247. doi: 10.1002/jcb.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ventura-Clapier Re, Garnier A, Veksler V. Energy metabolism in heart failure. JPhysiol. 2004;555:1–13. doi: 10.1113/jphysiol.2003.055095. doi:10.1113/jphysiol.2003.055095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingwall JS. Energy metabolism in heart failure and remodelling. Cardiovasc Res. 2009;81:412–419. doi: 10.1093/cvr/cvn301. doi:10.1093/cvr/cvn301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanza IR, Nair KS. Mitochondrial function as a determinant of life span. Pflugers Arch. 2010;459:277–289. doi: 10.1007/s00424-009-0724-5. doi:10.1007/s00424-009-0724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Chiu J-F, Mossman BT, Fukagawa NK. Down-regulation of manganese-superoxide dismutase through phosphorylation of FOXO3a by Akt in explanted vascular smooth muscle cells from old rats. JBiol Chem. 2006;281:40429–40439. doi: 10.1074/jbc.M606596200. doi:10.1074/jbc.M606596200. [DOI] [PubMed] [Google Scholar]
- 38.Tan W-Q, Wang K, Lv D-Y, Li P-F. Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. JBiol Chem. 2008;283:29730–29739. doi: 10.1074/jbc.M805514200. doi:10.1074/jbc.M805514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang D. Cardiac hypertrophy and oxidative stress: a leap of faith or stark reality? Heart. 2002;87:316–317. doi: 10.1136/heart.87.4.316. doi:10.1136/heart.87.4.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zweier JL, Talukder MA. The role of oxidants and free radicals in reperfusion injury. Cardiovasc Res. 2006;70:181–190. doi: 10.1016/j.cardiores.2006.02.025. doi:10.1016/j.cardiores.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 41.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. doi:10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taegtmeyer H. Metabolism–the lost child of cardiology. J Am Coll Cardiol. 2000;36:1386–1388. doi: 10.1016/s0735-1097(00)00870-6. doi:10.1016/S0735-1097(00)00870-6. [DOI] [PubMed] [Google Scholar]
- 44.Leong HS, Brownsey RW, Kulpa JE, Allard MF. Glycolysis and pyruvate oxidation in cardiac hypertrophy–why so unbalanced? Comp Biochem Physiol A Mol Integr Physiol. 2003;135:499–513. doi: 10.1016/s1095-6433(03)00007-2. doi:10.1016/S1095-6433(03)00007-2. [DOI] [PubMed] [Google Scholar]
- 45.Taegtmeyer H. Switching metabolic genes to build a better heart. Circulation. 2002;106:2043–2045. doi: 10.1161/01.cir.0000036760.42319.3f. doi:10.1161/01.CIR.0000036760.42319.3F. [DOI] [PubMed] [Google Scholar]
- 46.Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: a question of balance. J Clin Invest. 2005;115:547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. doi:10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Barger PM, Brandt JM, Leone TC, Weinheimer CJ, Kelly DP. Deactivation of peroxisome proliferator-activated receptor-alpha during cardiac hypertrophic growth. J Clin Invest. 2000;105:1723–1730. doi: 10.1172/JCI9056. doi:10.1172/JCI9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanda H, Nohara R, Hasegawa K, Kishimoto C, Sasayama S. A nuclear complex containing PPARα/RXRα is markedly downregulated in the hypertrophied rat left ventricular myocardium with normal systolic function. Heart Vessels. 2000;15:191–196. doi: 10.1007/s003800070022. doi:10.1007/s003800070022. [DOI] [PubMed] [Google Scholar]
- 50.Lehman JJ, Boudina S, Banke NH, Sambandam N, Han X, Young DM, et al. The transcriptional coactivator PGC-1{alpha} is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol. 2008;295:H185–H196. doi: 10.1152/ajpheart.00081.2008. doi:10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehman JJ, Kelly DP. Transcriptional activation of energy metabolic switches in the developing and hypertrophied heart. Clin Exp Pharmacol Physiol. 2002;29:339–345. doi: 10.1046/j.1440-1681.2002.03655.x. doi:10.1046/j.1440-1681.2002.03655.x. [DOI] [PubMed] [Google Scholar]
- 52.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1[alpha] and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. doi:10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 53.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. doi:10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, III, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY) 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. doi:10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, et al. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285:3133–3144. doi: 10.1074/jbc.M109.077271. doi:10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, et al. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. doi:10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujino T, Kondo J, Ishikawa M, Morikawa K, Yamamoto TT. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. doi:10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- 59.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. doi:10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. doi:10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 61.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–311. doi: 10.1021/bi901627u. doi:10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. doi:10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Shi T, Fan GQ, Xiao SD. SIRT3 reduces lipid accumulation via AMPK activation in human hepatic cells. J Dig Dis. 2010;11:55–62. doi: 10.1111/j.1751-2980.2009.00416.x. doi:10.1111/j.1751-2980.2009.00416.x. [DOI] [PubMed] [Google Scholar]
- 64.Dyck JRB, Lopaschuk GD. AMPK alterations in cardiac physiology and pathology: enemy or ally? JPhysiol. 2006;574:95–112. doi: 10.1113/jphysiol.2006.109389. doi:10.1113/jphysiol.2006.109389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan AYM, Dolinsky VW, Soltys C-LM, Viollet B, Baksh S, Light PE, et al. Resveratrol inhibits cardiac hypertrophy via AMP-activated protein kinase and Akt. JBiol Chem. 2008;283:24194–24201. doi: 10.1074/jbc.M802869200. doi:10.1074/jbc.M802869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meng RS, Pei ZH, Yin R, Zhang CX, Chen BL, Zhang Y, et al. Adenosine monophosphate-activated protein kinase inhibits cardiac hypertrophy through reactivating peroxisome proliferator-activated receptor-alpha signaling pathway. Eur J Pharmacol. 2009;620:63–70. doi: 10.1016/j.ejphar.2009.08.024. doi:10.1016/j.ejphar.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 67.Ikeda Y, Sato K, Pimentel DR, Sam F, Shaw RJ, Dyck JRB, et al. Cardiac-specific deletion of LKB1 leads to hypertrophy and dysfunction. J Biol Chem. 2009;284:35839–35849. doi: 10.1074/jbc.M109.057273. doi:10.1074/jbc.M109.057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, et al. Kaempferol induces apoptosis in two different cell lines via Akt inactivation, Bax and SIRT3 activation, and mitochondrial dysfunction. J Cell Biochem. 2009;106:643–650. doi: 10.1002/jcb.22044. doi:10.1002/jcb.22044. [DOI] [PubMed] [Google Scholar]
- 69.Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- 70.Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. doi:10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. doi:10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 72.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 73.Li S, Banck M, Mujtaba S, Zhou M-M, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One. 2010;5:e10486. doi: 10.1371/journal.pone.0010486. doi:10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacobs KM, Pennington JD, Bisht KS, Aykin-Burns N, Kim HS, Mishra M, et al. SIRT3 interacts with the daf-16 homolog FOXO3a in the mitochondria, as well as increases FOXO3a dependent gene expression. Int J Biol Sci. 2008;4:291–299. doi: 10.7150/ijbs.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, et al. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 2010;285:7417–7429. doi: 10.1074/jbc.M109.053421. doi:10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM, Che CM, et al. Identification and characterization of proteins interacting with SIRT1 and SIRT3: implications in the anti-aging and metabolic effects of sirtuins. Proteomics. 2009;9:2444–2456. doi: 10.1002/pmic.200800738. doi:10.1002/pmic.200800738. [DOI] [PubMed] [Google Scholar]