Abstract
Ca2+-independent phospholipase A2β (iPLA2β) selectively hydrolyzes docosahexaenoic acid (DHA, 22:6n-3) in vitro from phospholipid. Mutations in the PLA2G6 gene encoding this enzyme occur in patients with idiopathic neurodegeneration plus brain iron accumulation and dystonia-parkinsonism without iron accumulation, whereas mice lacking PLA2G6 show neurological dysfunction and neuropathology after 13 months. We hypothesized that brain DHA metabolism and signaling would be reduced in 4-month-old iPLA2β-deficient mice without overt neuropathology. Saline or the cholinergic muscarinic M1,3,5 receptor agonist arecoline (30 mg/kg) was administered to unanesthetized iPLA2β−/−, iPLA2β+/−, and iPLA2β+/+ mice, and [1-14C]DHA was infused intravenously. DHA incorporation coefficients k* and rates Jin, representing DHA metabolism, were determined using quantitative autoradiography in 81 brain regions. iPLA2β−/− or iPLA2β+/− compared with iPLA2β+/+ mice showed widespread and significant baseline reductions in k* and Jin for DHA. Arecoline increased both parameters in brain regions of iPLA2β+/+ mice but quantitatively less so in iPLA2β−/− and iPLA2β+/− mice. Consistent with iPLA2β’s reported ability to selectively hydrolyze DHA from phospholipid in vitro, iPLA2β deficiency reduces brain DHA metabolism and signaling in vivo at baseline and following M1,3,5 receptor activation. Positron emission tomography might be used to image disturbed brain DHA metabolism in patients with PLA2G6 mutations.
Keywords: Ca2+-independent phospholipase A2, iPLA2 knockout mouse, brain imaging, muscarinic receptor, arecoline
Studies of isolated glial cells and in vitro enzymatic assays indicate that Ca2+-independent phospholipase A2 (iPLA2, EC 3.1.1.4) can selectively release docosahexaenoic acid (DHA, 22:6n-3) from the stereospecifically numbered (sn)-2 position of phospholipids and that iPLA2 can be activated via G-protein-coupled neuroreceptors in cells (1–4). In brain, iPLA2 has a postsynaptic location and is thought to participate in neurotransmission (5–10). In smooth muscle, iPLA2 also mediates arginine vasopressin-induced release of arachidonic acid (AA, 20:4n-6), another PUFA (11).
Two iPLA2 isoforms have been identified in mammalian brain: iPLA2β (also PARK14, PNPLA9, or iPLA2-VIA) and iPLA2γ (iPLA2-VIB also PNPLA8). iPLA2β is an 84−88 kDa enzyme localized in the cell cytosol and endoplasmic reticulum. It is not activated by extracellular-derived Ca2+ but may be activated when Ca2+ is released from intracellular stores to displace inhibitory calmodulin from it (6, 12–17). Mutations in the PLA2G6 gene encoding iPLA2β have been associated with infantile neuroaxonal dystrophy, idiopathic neurodegeneration with brain iron accumulation (18, 19), and adult-onset dystonia-parkinsonism without brain iron accumulation (20–22).
The contributions of the iPLA2 isoforms to in vivo brain DHA signaling and metabolism remain to be clarified. Thus, it would be of interest to examine these contributions with a validated in vivo imaging method and model (8, 23) in homozygous (iPLA2β−/−) and heterozygous (iPLA2β+/−) deficient compared with wild-type (iPLA2β+/+) mice (24). The method involves infusing the radiolabeled unesterified DHA intravenously in unanesthetized animals, determining radioactivity in different brain regions with quantitative autoradiography, then calculating regional brain DHA incorporation coefficients k* and rates Jin (product of k* and unesterified unlabeled plasma DHA).
Within minutes after [1-14C]DHA infusion, 80% of brain radioactivity is found as unchanged tracer in the sn-2 position of phospholipid and 10% is in triacylglycerol, with only about 10% consisting of aqueous radioactive metabolites (7, 10, 25, 26). Jin approximates the regional rate of brain DHA consumption, because unesterified but not esterified long-chain fatty acids enter the brain from plasma (27–29), and DHA, once lost by metabolism after being hydrolyzed from phospholipid, cannot be resynthesized de novo or significantly elongated in brain (<0.1%) from its precursor α-linolenic acid (α-LNA, 18:3n-3) (8, 30–32). Administration of the cholinergic muscarinic M1,3,5 receptor agonist, arecoline, increases [1-14C]DHA incorporation into synaptic membrane phospholipid (7, 10).
In the present study, we imaged k* and Jin for DHA in brains of unanesthetized iPLA2β−/−, iPLA2β+/−, and iPLA2β+/+ mice (24) at baseline and following administration of arecoline (7, 10, 33, 34). Based on the in vitro evidence cited above that iPLA2β selectively hydrolyzes DHA from phospholipid, we predicted that brain DHA signaling would be reduced at rest and following arecoline in the iPLA2β-deficient compared with wild-type mice. To minimize the effects of neuropathology that appear in older iPLA2β−/− mice, we studied 4-month-old mice free of significant histopathology or neurological abnormalities (35). An abstract of part of this work has been published (36).
MATERIALS AND METHODS
Animals
Procedures were performed under a protocol approved by the Animal Care and Use Committee of the Eunice Kennedy Shriver National Institute of Child Health and Human Development in accordance with National Institutes of Health guidelines (publication no. 86-23). Four-month-old male iPLA2β−/−, iPLA2β+/−, and littermate iPLA2β+/+ mice, derived from a C57BL/6 genetic background (24), were maintained in an animal facility with free access to water and food. The diet (PicoLab® Rodent Diet 20, 5053, LabDiet) contained soybean and fishmeal and 4.5% crude fat by weight. Gas-liquid chromatography showed that fatty acid concentrations (as percent of total fatty acid) were: 20.0% saturated, 22.2% monounsaturated, 47.8% linoleic, 5.1% α-LNA, 0.13% AA, 1.00% eicosapentaenoic, and 0.87% DHA (1.3 ± 0.0 μmol/g diet).
Surgical procedures and tracer infusion
A mouse was anesthetized with 2–3% halothane in O2, and PE 10 polyethylene catheters were inserted into the right femoral artery and vein. The wound site was closed with 454 Instant Adhesive (Loctite Corp., Hartford, CT), and the animal was wrapped loosely with the upper body remaining free in a fast-setting plaster cast taped to a wooden block and allowed to recover from anesthesia (3–4 h) in a warm environment.
Unanesthetized mice received intraperitoneally 0.9% NaCl (Abbott Laboratories, North Chicago, IL) or 30 mg/kg ip arecoline hydrobromide (Sigma, St. Louis, MO) in an injection volume of 0.01 ml/g body weight. The arecoline dose was chosen from a prior study in mice (34); lower doses gave less robust results (data not shown). Three minutes after injecting arecoline or saline, 45 μl [1-14C]DHA (300 μCi/kg; 56 mCi/mmol, >98% pure, Moravek Biochemicals, Brea, CA) in 5 mM HEPES buffer (pH 7.4) with 50 mg/ml fatty acid-free BSA was infused (3 min) via the femoral vein catheter (rate of 15 μl/min) with a Hamilton syringe and infusion pump (Harvard Apparatus, Model 22, Holliston, MA). Methylatropine bromide (Sigma), a competitive cholinergic muscarinic receptor antagonist that does not enter brain, was administered (4 mg/kg sc) 17 min before arecoline to block peripheral autonomic effects (7, 34). Ten arterial blood samples (15–20 μl) were collected (at 0, 0.25, 1.0, 1.5, 2.0, 2.8, 3.2, 5.0, 10, and 19 min) to determine the radioactivity of unesterified plasma DHA. At 20 min, the mouse was euthanized with Nembutal® (50 mg/kg, i.v.), and its brain was removed within 30 s, frozen in 2-methylbutane dry ice at −40°C, and stored at −80°C until sectioned.
Chemical analysis
Arterial blood samples collected before, during, and after [1-14C]DHA infusion were centrifuged immediately (30 s, 18,000 g). For each sample, total lipids were extracted (37) from plasma (5 µl) with chloroform-methanol (1 ml, 2:1, v/v) and 0.1 M KCl (0.5 ml). Radioactivity was determined in an organic phase aliquot (100 µl) by liquid scintillation spectrometry. After the 3 min [1-14C]DHA infusion, at least 98% of total plasma radioactivity was unmetabolized [1-14C]DHA, and 95% of the total brain radioactivity was in the form of esterified [1-14C]DHA in phospholipid (25). Concentrations of unlabeled unesterified DHA also were determined in arterial plasma (100 µl) to calculate Jin. Total lipids were extracted (37) and separated by thin layer chromatography on silica gel-60 plates by using the solvent system heptane:diethylether:glacial acetic acid (60:40:3, v/v/v). Unesterified fatty acids were scraped from the plate and converted to methyl ester derivatives (1% H2SO4 in methanol, 3 h, 70°C), which then were analyzed by gas chromatography with flame ionization detection and quantified relative to an internal standard, heptadecanoic acid (17:0).
Quantitative autoradiography
Frozen brains were cut in serial 20-μm-thick coronal sections on a cryostat at −20°C, then placed for 4 weeks with calibrated [14C]methylmethacrylate standards (Amersham, Arlington Heights, IL) on Ektascan C/RA film (Eastman Kodak Co., Rochester, NY). Radioactivity (nCi/g wet weight brain) in 81 identified regions (38) was measured bilaterally six times by quantitative densitometry by using the public domain NIH Image program 1.62. Regional DHA incorporation coefficients k* (ml/s/g wet weight brain) were calculated as (8, 23):
| (Eq. 1) |
where (nCi/g wet brain weight) is radioactivity of brain lipid at time 20 min (time of termination of experiment), (nCi/ml plasma) is the arterial plasma concentration of labeled unesterified DHA, and t (min) is time after beginning [1-14C]DHA infusion. Integrated plasma radioactivity due to unesterified [1-14C]DHA (input function) was determined by trapezoidal integration and used to calculate regional values of k*.
Regional incorporation rates of unesterified unlabeled DHA from plasma into brain, Jin (nmol/s/g), were calculated as:
| (Eq. 2) |
where cplasma equals unesterified unlabeled plasma DHA.
Statistical analyses
A one-way ANOVA with Tukey's post hoc test was used to compare mean body weights, plasma unesterified DHA concentrations, and baseline k* for DHA among the three genotypes by using GraphPad Prism (GraphPad Software, San Diego, CA). A two-way ANOVA (α = 0.01) was employed to examine effects of genotype (iPLA2β−/− or iPLA2β+/− vs. iPLA2β+/+) and drug (arecoline vs. saline) on the arterial input function and on k* by using SPSS 16.0 (SPSS Inc., Chicago, IL). In the absence of a significant interaction, for genotype main effects, Tukey's post hoc tests were performed to test differences in k* between the three genotype groups collapsed across drug. When an interaction was statistically significant, we performed Bonferroni's post hoc tests with correction for three comparisons (iPLA2β+/+ plus arecoline vs. iPLA2β+/+ saline, iPLA2β+/− plus arecoline vs. iPLA2β+/− saline, and iPLA2β−/− plus arecoline vs. iPLA2β−/− saline).
RESULTS
Body weight and plasma arterial input function
Mean body weight did not differ significantly among iPLA2β+/+ (25.6 ± 1.5 g; n = 13), iPLA2β+/− (27.2 ± 2.4 g; n = 13), and iPLA2β−/− (26.8 ± 1.8 g; n = 14) mice, as reported (39).
A two-way ANOVA did not reveal a significant main effect of arecoline (P = 0.07) or genotype (P = 0.21) or a significant genotype vs. arecoline interaction (P = 0.26) on integrated plasma arterial radioactivity (plasma input function in denominator of Eq. 1), thus on the DHA plasma half-life (40). Integrated plasma radioactivity [(nCi·s/ml) ± SD, n = 6–8] equaled: iPLA2β+/+ plus saline, 133,780 ± 88,104; iPLA2β+/+ plus arecoline, 88,369 ± 17,062; iPLA2β+/− plus saline, 95,936 ± 29,409; iPLA2β+/− plus arecoline, 68,053 ± 11,058; iPLA2β−/− plus saline, 102,899 ± 19,548; and iPLA2β−/− plus arecoline, 106,609 ± 28,012.
Plasma concentrations of unlabeled unesterified DHA
The mean unesterified DHA plasma concentration did not differ significantly (P > 0.05) among iPLA2β+/+ (20.85 ± 8.66 nmol/ml), iPLA2β+/− (23.92 ± 9.93 nmol/ml), and iPLA2β−/− (18.75 ± 12.34 nmol/ml) mice at baseline (in response to saline). An arecoline effect on plasma DHA was not determined, because the effect was statistically insignificant in a comparable prior study (34).
Regional DHA incorporation coefficients k*
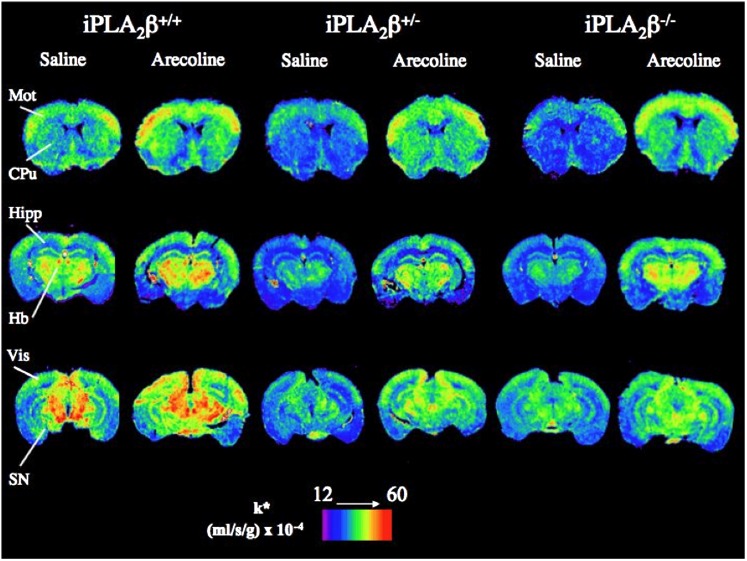
Figure 1 presents color-coded coronal autoradiographs representing k* for DHA from brains of iPLA2β-deficient and wild-type mice injected with saline or arecoline. iPLA2β+/− and iPLA2β−/− mice apparently had lower baseline (following saline) values of k* (Eq. 1) than did iPLA2β+/+ controls. Regional values of k* appear elevated in iPLA2β+/+ mice injected with arecoline compared with saline. The arecoline-induced elevations appear reduced or absent in the iPLA2β-deficient mice.
Fig. 1.
Autoradiographs of coronal brain sections showing effects of arecoline and iPLA2β genotype on regional DHA incorporation coefficients k* in mice. Values of k* [(ml/s/g wet weight brain) × 10−4] are given on a color scale. Abbreviations: CPu, caudate-putamen; Hb, habenular nuclei; Hipp, hippocampus; Mot, motor cortex; SN, substantia nigra; Vis, visual cortex.
Baseline.
In a one-way ANOVA with Tukey's post hoc test, we compared baseline values of k* for DHA among the three genotypes. Partial and total iPLA2β deletion significantly decreased baseline k* by 20–45% in 60 and 70 of 81 brain regions, respectively, compared with baseline k* in iPLA2β+/+ mice (data not shown). The baseline decreases were comparable in iPLA2β−/− and iPLA2β+/− mice.
Arecoline activation.
Mean DHA incorporation coefficients k* at baseline and following arecoline in each of 81 brain regions were compared among experimental groups and conditions using a two-way ANOVA. Thirty-five of the 81 regions did not have a statistically significant genotype × drug interaction, indicating that the iPLA2β genotype (heterozygous or homozygous) did not alter the arecoline response (data not shown). These regions included the median eminence, white matter, lateral and anterior arcuate nucleus, periventricular hypothalamus, mammillary nucleus, medial and lateral nuclei of the septum, nucleus accumbens, amygdala, CA1 to CA3 areas of the hippocampus, prefrontal cortex layers I and IV, primary olfactory cortex, globus pallidus, habenular nuclei, medial geniculate nucleus, substantia nigra, ventroposterior medial, and paraventricular and parafascicular thalamic nuclei. In the 35 regions, the main effect of arecoline was statistically significant, which means that increments in k* following arecoline were equally robust in the three genotypes. Increments (compared with saline) ranged from 32% in the median eminence to 170% in prefrontal cortex layer IV (mean = 78 ± 34%).
Data for the 46 remaining regions that had statistically significant genotype × drug interactions are summarized in Table 1. Arecoline compared with saline increased k* by 77% (auditory cortex layer IV) to 161% (olfactory tubercle) (mean = 108 ± 20%) in the 46 regions in the iPLA2β+/+ mice compared with 39% (somatosensory cortex layer IV) to 123% (visual cortex layer VI) (mean = 63 ± 18%) in the iPLA2β+/− mice and 46% (inferior colliculus) to 161% (frontal cortex 10 layer I) (mean = 78 ± 22%) in the iPLA2β−/− mice. The mean of the arecoline-induced increments in k* was significantly less (P < 0.001) in the iPLA2β+/− and iPLA2β−/− mice compared with wild-type mice (63 ± 18% vs. 108 ± 20%, and 78 ± 22% vs. 108 ± 20%). Furthermore, a Bonferroni's test corrected for three comparisons (α = 0.05/3) for each of the 46 regions showed that arecoline compared with saline significantly increased k* for DHA in each of the 46 regions in the iPLA2β+/+ mice and in 28 regions of the iPLA2β+/− mice and 36 regions of the iPLA2β−/− mice (Table 1).
TABLE 1.
DHA incorporation coefficients k* in iPLA2β+/+, iPLA2β+/−, and iPLA2β−/− mice in response to arecoline in regions with statistically significant genotype × drug interactions
| iPLA2β+/+ |
iPLA2β |
iPLA2β−1 |
||||
|---|---|---|---|---|---|---|
| Brain Region | Saline (n = 7) | Arecoline (n = 6) | Saline (n = 7) | Arecoline (n = 6) | Saline (n = 8) | Arecoline (n = 6) |
| Frontal cortex (10) | ||||||
| Layer I | 17.26 ± 2.43 | 43.59 ± 4.67*** | 13.89 ± 2.10 | 26.92 ± 7.05*** | 13.62 ± 2.77 | 35.49 ± 8.23*** |
| Layer IV | 23.11 ± 2.15 | 56.32 ± 10.80*** | 16.59 ± 3.05 | 26.02 ± 8.51* | 15.50 ± 4.49 | 30.47 ± 7.53*** |
| Frontal cortex (8) | ||||||
| Layer I | 20.24 ± 5.02 | 49.42 ± 10.44*** | 16.68 ± 3.44 | 26.47 ± 7.99* | 14.85 ± 4.03 | 26.37 ± 3.94** |
| Layer IV | 25.38 ± 4.42 | 56.43 ± 14.09*** | 19.52 ± 5.08 | 31.01 ± 7.47* | 16.18 ± 5.52 | 33.25 ± 6.98** |
| Pyriform cortex | 13.32 ± 2.87 | 29.88 ± 6.53*** | 14.83 ± 3.59 | 16.40 ± 2.63 | 11.22 ± 1.85 | 19.48 ± 4.48** |
| Anterior cingulate cortex | 24.53 ± 5.83 | 53.01 ± 7.23*** | 17.52 ± 3.42 | 25.33 ± 5.65* | 16.18 ± 2.59 | 27.50 ± 6.13** |
| Motor cortex | ||||||
| Layer I | 17.10 ± 3.87 | 38.85 ± 7.91*** | 13.85 ± 2.86 | 21.21 ± 5.13* | 12.19 ± 2.50 | 21.68 ± 2.33** |
| Layer II-III | 18.79 ± 3.66 | 40.45 ± 8.95*** | 15.38 ± 2.27 | 20.63 ± 3.58 | 12.35 ± 1.68 | 25.73 ± 4.12*** |
| Layer IV | 23.87 ± 4.46 | 48.30 ± 6.71*** | 17.38 ± 3.80 | 26.14 ± 7.79 | 15.15 ± 1.82 | 28.89 ± 5.91* |
| Layer V | 19.98 ± 3.76 | 41.87 ± 5.88*** | 13.64 ± 2.41 | 19.28 ± 4.11* | 12.40 ± 1.24 | 22.18 ± 4.26*** |
| Layer VI | 18.48 ± 4.08 | 38.19 ± 6.83*** | 13.77 ± 2.17 | 19.75 ± 4.73* | 11.29 ± 1.32 | 22.70 ± 4.04*** |
| Somatosensory cortex | ||||||
| Layer I | 20.69 ± 6.07 | 42.60 ± 4.62*** | 13.95 ± 2.80 | 22.09 ± 3.47* | 12.47 ± 1.69 | 24.15 ± 3.74*** |
| Layer II-III | 21.28 ± 5.16 | 45.63 ± 5.77*** | 16.33 ± 2.36 | 23.54 ± 4.83* | 15.55 ± 2.86 | 27.34 ± 3.41*** |
| Layer IV | 27.81 ± 5.45 | 56.28 ± 8.22*** | 19.63 ± 3.83 | 27.27 ± 6.29* | 18.66 ± 4.13 | 28.44 ± 3.95** |
| Layer V | 25.58 ± 4.51 | 51.77 ± 5.19*** | 17.76 ± 2.84 | 24.84 ± 6.01* | 16.84 ± 2.16 | 23.29 ± 4.47 |
| Layer VI | 23.20 ± 2.97 | 47.85 ± 6.62*** | 16.57 ± 2.47 | 24.21 ± 5.44* | 14.26 ± 2.10 | 25.23 ± 6.72** |
| Auditory cortex | ||||||
| Layer I | 21.55 ± 3.99 | 41.05 ± 6.87*** | 14.26 ± 4.04 | 22.85 ± 5.16** | 14.32 ± 2.41 | 20.06 ± 4.79 |
| Layer IV | 25.83 ± 4.16 | 45.68 ± 7.94*** | 14.29 ± 3.47 | 23.23 ± 4.72** | 16.79 ± 2.48 | 21.50 ± 4.22 |
| Visual cortex | ||||||
| Layer I | 21.52 ± 2.44 | 42.42 ± 7.26*** | 13.69 ± 4.21 | 24.62 ± 4.97*** | 14.21 ± 2.48 | 21.55 ± 3.83* |
| Layer IV | 21.52 ± 2.70 | 49.95 ± 7.92*** | 15.58 ± 5.00 | 29.33 ± 3.56*** | 15.13 ± 3.15 | 23.94 ± 4.19 |
| Layer VI | 23.25 ± 2.11 | 42.19 ± 5.80*** | 12.36 ± 2.60 | 27.56 ± 3.62*** | 13.58 ± 2.62 | 21.37 ± 3.48*** |
| Preoptic area (LPO/MPO) | 16.01 ± 4.19 | 29.25 ± 3.46*** | 13.36 ± 2.59 | 15.58 ± 2.32 | 12.32 ± 3.90 | 16.93 ± 3.50 |
| Olfactory tubercle | 17.35 ± 2.68 | 45.28 ± 8.80*** | 15.70 ± 2.79 | 21.55 ± 5.96 | 15.37 ± 5.04 | 25.84 ± 6.64** |
| Diagonal band ventral | 22.42 ± 2.66 | 45.49 ± 9.71*** | 15.49 ± 5.01 | 23.52 ± 8.01 | 15.23 ± 3.38 | 18.99 ± 7.65 |
| Hippocampus | ||||||
| Dentate gyrus | 21.94 ± 2.09 | 39.16 ± 5.61*** | 16.01 ± 5.91 | 20.62 ± 4.15 | 12.65 ± 2.21 | 21.55 ± 3.46** |
| SLM | 28.44 ± 2.14 | 55.89 ± 10.30*** | 17.13 ± 4.29 | 29.18 ± 5.07** | 16.11 ± 2.39 | 28.24 ± 1.87** |
| Caudate putamen | ||||||
| Ventral | 19.88 ± 3.80 | 41.69 ± 7.92*** | 13.59 ± 1.54 | 23.51 ± 7.00** | 13.40 ± 2.33 | 23.65 ± 5.18*** |
| Lateral | 19.45 ± 3.57 | 43.43 ± 8.07*** | 13.94 ± 2.24 | 23.92 ± 6.57** | 13.96 ± 2.59 | 22.35 ± 4.05** |
| Medial | 18.90 ± 3.90 | 40.91 ± 9.06*** | 14.49 ± 2.16 | 22.96 ± 7.14* | 15.46 ± 2.46 | 24.04 ± 5.38* |
| Lateral geniculate nu | 32.73 ± 3.36 | 62.51 ± 10.24*** | 21.58 ± 4.13 | 37.16 ± 7.37** | 19.52 ± 3.51 | 35.21 ± 9.86** |
| Thalamus | ||||||
| Ventroposterior lateral nu | 28.88 ± 4.15 | 57.47 ± 9.48*** | 23.45 ± 5.28 | 29.75 ± 6.26 | 17.64 ± 4.40 | 32.36 ± 8.89*** |
| Paratenial nu | 22.68 ± 5.37 | 42.71 ± 7.63*** | 19.26 ± 2.55 | 24.25 ± 9.00 | 15.68 ± 3.29 | 27.59 ± 4.61** |
| Anteroventral nu | 38.27 ± 9.31 | 74.67 ± 11.29*** | 26.24 ± 5.36 | 36.71 ± 10.30 | 24.70 ± 3.76 | 37.03 ± 7.08* |
| Anteromedial nu | 25.86 ± 5.52 | 51.43 ± 8.39*** | 20.49 ± 3.97 | 26.37 ± 7.68 | 17.43 ± 3.46 | 35.35 ± 8.16*** |
| Reticular nu | 28.78 ± 5.54 | 53.21 ± 8.81*** | 19.68 ± 2.61 | 28.50 ± 6.11 | 17.56 ± 2.85 | 33.54 ± 8.17*** |
| Subthalamic nu | 30.42 ± 3.72 | 70.68 ± 11.19*** | 22.24 ± 3.45 | 28.39 ± 8.70 | 21.99 ± 5.11 | 26.72 ± 8.93 |
| Hypothalamus | ||||||
| Supraoptic nu | 16.65 ± 3.05 | 34.50 ± 4.42*** | 13.26 ± 1.38 | 17.27 ± 4.17 | 13.52 ± 4.62 | 17.82 ± 1.91 |
| Posterior | 25.27 ± 4.77 | 46.65 ± 8.50*** | 15.76 ± 4.08 | 22.96 ± 5.91 | 15.25 ± 3.47 | 24.84 ± 3.57** |
| Interpeduncular nu | 39.44 ± 5.13 | 77.35 ± 9.91*** | 27.20 ± 6.92 | 36.91 ± 8.63 | 25.77 ± 3.54 | 37.41 ± 10.28 |
| Pretectal area | 27.51 ± 4.22 | 59.79 ± 6.65*** | 16.05 ± 3.86 | 26.97 ± 5.79** | 16.30 ± 3.35 | 30.45 ± 4.30 ** |
| Gray layer sup colliculus | 24.72 ± 3.33 | 49.41 ± 6.45*** | 15.68 ± 2.87 | 26.67 ± 7.10** | 15.90 ± 2.04 | 26.81 ± 3.23*** |
| Superior colliculus | 24.80 ± 2.47 | 51.92 ± 11.22*** | 21.09 ± 6.30 | 28.69 ± 6.71 | 15.82 ± 2.80 | 30.00 ± 5.34*** |
| Inferior colliculus | 43.06 ± 9.39 | 82.49 ± 13.77*** | 26.60 ± 4.82 | 44.20 ± 8.36** | 26.15 ± 6.91 | 38.30 ± 5.82** |
| Flocculus | 30.51 ± 5.40 | 57.19 ± 7.23*** | 20.05 ± 4.49 | 33.69 ± 4.76** | 19.33 ± 4.60 | 29.28 ± 9.72* |
| Molecular layer cerebellar gray matter | ||||||
| 33.28 ± 9.21 | 75.97 ± 5.55*** | 25.69 ± 2.86 | 38.64 ± 6.09** | 24.31 ± 4.84 | 37.70 ± 8.05* | |
| Non-blood-brain barrier regions | ||||||
| Subfornical organ | 20.66 ± 5.47 | 51.42 ± 12.90*** | 14.22 ± 2.92 | 21.35 ± 6.14 | 15.42 ± 3.12 | 23.18 ± 5.19 |
Data are mean ± SD. k* = (ml/s/g) × 10−4. Mice were given methylatropine (4 mg/kg, subcutaneously) or saline 17 min before administration of saline or arecoline (30 mg/kg i.p.). [14C]DHA infusion was started 3 min after arecoline administration. In cases of statistically significant genotype × arecoline interactions, main effects are not reported, and Bonferroni's post tests were performed. *P < 0.05; **P < 0.01; ***P < 0.001; iPLA2β+/+ plus arecoline vs. iPLA2β+/+ saline; iPLA2β+/− plus arecoline vs. iPLA2β+/− saline, iPLA2β−/− plus arecoline vs. iPLA2β−/− saline. nu, nucleus; SLM, stratum lacunosum-moleculae of hippocampus.
Regional DHA incorporation rates
Because the mean total (labeled and unlabeled) unesterified plasma DHA concentration did not differ significantly among genotypes (see above), the statistical significance of group differences in incorporation rates Jin corresponded generally to the differences in regional values of k*, because Jin is the product of k* and the unesterified unlabeled plasma DHA concentration (Eq. 2). In iPLA2β+/+ mice, baseline Jin ranged from 311 ± 49 × 10−4 nmol/s/g in the piriform cortex to 898 ± 196 × 10−4 nmol/s/g in the inferior colliculus. In iPLA2β−/− mice, the range was 267 ± 38 × 10−4 nmol/s/g in the internal capsule to 581 ± 55 × 10−4 nmol /s/g in the inferior colliculus. In iPLA2β+/− mice, baseline Jin ranged from 166 ± 28 × 10−4 nmol/s/g in the periventricular of the hypothalamus to 487 ± 128 × 10−4 nmol/s/g in the inferior colliculus. Similarly, in response to arecoline, means for Jin decreased significantly in iPLA2β−/− and iPLA2β+/− compared with iPLA2β+/+ mice.
DISCUSSION
Regional brain incorporation coefficients k* and rates Jin for DHA at baseline (following saline) were reduced significantly in 4-month-old, unanesthetized, male iPLA2β−/− and iPLA2β+/− mice compared with iPLA2β+/+ mice. Muscarinic M1.3,5 receptor activation by arecoline significantly increased k* and Jin for DHA in multiple brain regions in the wild-type mice, as previously reported in rodents (7, 10, 34), but the increments were significantly less on average or statistically insignificant in many brain regions with a significant genotype × drug interaction (e.g., Table 1) of the iPLA2β−/− and iPLA2β+/− mice.
Brain regions in which genotype × drug interactions were statistically significant, shown in Table 1, roughly overlap with sites having high densities of postsynaptic M1,3,5 receptors (41, 42). These regions include neocortical projection regions, parts of the hippocampus, and the caudate-putamen. Low M1,3,5 receptor densities are reported in the thalamus, brainstem, and hypothalamus, where genotype × drug interactions often were statistically insignificant. The arecoline-induced increments in DHA incorporation in these latter regions may have represented downstream effects of direct activation elsewhere (43).
Because DHA cannot be synthesized de novo in vertebrates (30) and only a negligible amount (<0.1%) is elongated in brain from precursor α-LNA (8, 28, 31, 32), the lower values of k* and Jin at baseline and following arecoline in the iPLA2β−/− and iPLA2β+/− compared with iPLA2β+/+ mice represent reduced brain DHA consumption under resting (steady-state) and agonist stimulation conditions, respectively. In iPLA2β−/− mice, these reductions are associated with a reduced DHA concentration in brain ethanolamine glycerophospholipid (Y. Cheon, A. Taha, H. Y. Kim, and S. I. Rapoport, unpublished observations). A role for iPLA2β in regulating brain DHA metabolism is consistent with evidence that brain DHA turnover is reduced, as are the brain DHA concentration and iPLA2β mRNA, protein, and activity levels, in rats fed a low n-3 PUFA diet lacking DHA (31, 44).
Increased incorporation of labeled unesterified DHA from plasma into the sn-2 position of synaptic membrane phospholipids of brain has been demonstrated directly by chemical analysis in unanesthetized rats given arecoline (7, 10). Our new data suggest that a congenital absence of iPLA2β reduces this incorporation, as well as baseline signaling. The effects did not seem to depend on whether the mice had a partial or full iPLA2β deletion, possibly because of additional compensatory neuroplastic adaptive responses associated with the full deletion (45, 46).
Because the total iPLA2β deletion did not abolish the regional DHA responses to arecoline seen in iPLA2β+/+ mice, other enzymes likely contributed to the arecoline signal in the deficient mice. One candidate is iPLA2γ, which provides residual iPLA2 activity in the iPLA2β−/− mouse (Y. Cheon, A. Taha, H. Y. Kim, and S. I. Rapoport, unpublished observations) (24). Other candidates include plasmalogen-selective PLA2 and cytosolic PLA2 (cPLA2)γ, Ca2+-dependent secretory sPLA2, and phospholipase C (1, 3, 15, 47).
In addition to releasing DHA following arecoline activation of muscarinic M1,3,5 receptors in vivo (7, 10), iPLA2β can do so following agonist activation of serotonergic 5-HT2A/2C, bradykinin B2 or purigenic P2Y receptors in glial cells in vitro (2, 4). These latter receptors, as well as M1,3,5 receptors, activate effector enzymes by G-protein-coupled mechanisms (48–52). Muscarinic agonists also can release Ca2+ from intracellular stores through the inositol-1,4,5-phosphate receptor (53, 54) to indirectly activate iPLA2β (16, 55). On the other hand, activation of ionotropic glutamatergic N-methyl-d-aspartate receptors by N-methyl-d-aspartate, which allows extracellular Ca2+ into the cell, did not produce a measurable brain DHA signal in unanesthetized rats while producing a robust AA signal (17, 56). These data are consistent with iPLA2 being DHA selective and independent of extracellular Ca2+, but with cPLA2-IVA being AA selective and Ca2+-dependent (4, 6, 47, 57, 58).
Although we considered iPLA2β deletion effects on brain DHA signaling in this paper, based on in vitro evidence of the enzyme's selectivity for DHA (1–4, 6), AA signaling may be disturbed as well, because brain activities of sPLA2 and cPLA2-IV, which release AA from phospholipids (59), are elevated in iPLA2β−/− mice, whereas the brain AA concentration in phospholipid is reduced (Y. Cheon, A. Taha, H. Y. Kim, and S. I. Rapoport, unpublished observations). These enzyme changes would represent additional indirect compensatory responses in the full knockout condition (45, 46).
iPLA2β is reported to modulate apoptosis, cell proliferation, membrane fusion, behavior, memory, and motor function (14, 60–62). Some of these effects may be due to its influence on brain DHA metabolism and signaling. iPLA2β mRNA is reduced in the hippocampus of aged rats (63), but its brain mRNA and protein levels are increased in multiple sclerosis patients and in mice with experimental autoimmune encephalomyelitis (for which pretreatment with a selective iPLA2 inhibitor was helpful) (64).
Because DHA is a precursor of antiinflammatory neuroprotectins and resolvins, the reduced brain DHA metabolism in the iPLA2β-deficient mice may increase their vulnerability to neuroinflammation (9, 65, 66). Our observations also suggest that brain DHA signaling and metabolism would be altered in patients with a PLA2G6 mutation (18–22). This could be tested directly by quantitatively imaging regional brain DHA incorporation in these patients with positron emission tomography (67).
In summary, a congenital partial or complete absence of iPLA2β in 4-month-old mice reduced brain DHA signaling and metabolism at baseline and following M1,3,5 receptor activation. Studies in 13-month-old iPLA2β-deficient mice with neurological and behavioral impairments that correlate with brain accumulation of ubiquitin-containing tubulovesicular membranes (35, 60) may identify additional brain lipid metabolic disturbances. A detailed analysis of brain enzyme activity, lipid composition (68), and PUFA metabolism of mice at both ages could be informative and clinically relevant.
Supplementary Material
Acknowledgments
The authors thank Dr. Eugene Streicher for proofreading the manuscript.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- α-LNA
- α-linolenic
- DHA
- docosahexaenoic acid
- PLA2
- phospholipase A2
- cPLA2
- Ca2+-dependent cytosolic PLA2
- iPLA2
- Ca2+-independent PLA2
- sPLA2
- secretory PLA2
- sn
- stereospecifically numbered
This research was supported by the Intramural Research Program of the National Institute on Aging, the National Institutes of Health, and by United States Public Health Service Grants R37-DK34388, P41-RR00954, P60-DK20579, and P30-DK56341 to J.T. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or USPHS.
REFERENCES
- 1.Strokin M., Sergeeva M., Reiser G. 2007. Prostaglandin synthesis in rat brain astrocytes is under the control of the n-3 docosahexaenoic acid, released by group VIB calcium-independent phospholipase A2. J. Neurochem. 102: 1771–1782. [DOI] [PubMed] [Google Scholar]
- 2.Garcia M. C., Kim H. Y. 1997. Mobilization of arachidonate and docosahexaenoate by stimulation of the 5–HT2A receptor in rat C6 glioma cells. Brain Res. 768: 43–48. [DOI] [PubMed] [Google Scholar]
- 3.Lucas K. K., Dennis E. A. 2005. Distinguishing phospholipase A2 types in biological samples by employing group-specific assays in the presence of inhibitors. Prostaglandins Other Lipid Mediat. 77: 235–248. [DOI] [PubMed] [Google Scholar]
- 4.Strokin M., Sergeeva M., Reiser G. 2003. Docosahexaenoic acid and arachidonic acid release in rat brain astrocytes is mediated by two separate isoforms of phospholipase A2 and is differently regulated by cyclic AMP and Ca2+. Br. J. Pharmacol. 139: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ong W. Y., Yeo J. F., Ling S. F., Farooqui A. A. 2005. Distribution of calcium-independent phospholipase A2 (iPLA2) in monkey brain. J. Neurocytol. 34: 447–458. [DOI] [PubMed] [Google Scholar]
- 6.Rosa A. O., Rapoport S. I. 2009. Intracellular- and extracellular-derived Ca2+ influence phospholipase A2-mediated fatty acid release from brain phospholipids. Biochim. Biophys. Acta. 1791: 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeGeorge J. J., Nariai T., Yamazaki S., Williams W. M., Rapoport S. I. 1991. Arecoline-stimulated brain incorporation of intravenously administered fatty acids in unanesthetized rats. J. Neurochem. 56: 352–355. [DOI] [PubMed] [Google Scholar]
- 8.Rapoport S. I. 2001. In vivo fatty acid incorporation into brain phospholipids in relation to plasma availability, signal transduction and membrane remodeling. J. Mol. Neurosci. 16: 243–261. [DOI] [PubMed] [Google Scholar]
- 9.Horrocks L. A., Farooqui A. A. 2004. Docosahexaenoic acid in the diet: its importance in maintenance and restoration of neural membrane function. Prostaglandins Leukot. Essent. Fatty Acids. 70: 361–372. [DOI] [PubMed] [Google Scholar]
- 10.Jones C. R., Arai T., Rapoport S. I. 1997. Evidence for the involvement of docosahexaenoic acid in cholinergic stimulated signal transduction at the synapse. Neurochem. Res. 22: 663–670. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins C. M., Han X., Mancuso D. J., Gross R. W. 2002. Identification of calcium-independent phospholipase A2 (iPLA2) beta, and not iPLA2gamma, as the mediator of arginine vasopressin-induced arachidonic acid release in A-10 smooth muscle cells. Enantioselective mechanism-based discrimination of mammalian iPLA2s. J. Biol. Chem. 277: 32807–32814. [DOI] [PubMed] [Google Scholar]
- 12.Wolf M. J., Gross R. W. 1996. The calcium-dependent association and functional coupling of calmodulin with myocardial phospholipase A2. Implications for cardiac cycle-dependent alterations in phospholipolysis. J. Biol. Chem. 271: 20989–20992. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins C. M., Wolf M. J., Mancuso D. J., Gross R. W. 2001. Identification of the calmodulin-binding domain of recombinant calcium-independent phospholipase A2β. implications for structure and function. J. Biol. Chem. 276: 7129–7135. [DOI] [PubMed] [Google Scholar]
- 14.Akiba S., Sato T. 2004. Cellular function of calcium-independent phospholipase A2. Biol. Pharm. Bull. 27: 1174–1178. [DOI] [PubMed] [Google Scholar]
- 15.Ma Z., Turk J. 2001. The molecular biology of the group VIA Ca2+-independent phospholipase A2. Prog. Nucleic Acid Res. Mol. Biol. 67: 1–33. [DOI] [PubMed] [Google Scholar]
- 16.Park K. M., Trucillo M., Serban N., Cohen R. A., Bolotina V. M. 2008. Role of iPLA2 and store-operated channels in agonist-induced Ca2+ influx and constriction in cerebral, mesenteric, and carotid arteries. Am. J. Physiol. Heart Circ. Physiol. 294: H1183–H1187. [DOI] [PubMed] [Google Scholar]
- 17.Ramadan E., Rosa A. O., Chang L., Chen M., Rapoport S. I., Basselin M. 2010. Extracellular-derived calcium does not initiate in vivo neurotransmission involving docosahexaenoic acid. J. Lipid Res. 51: 2334–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan N. V., Westaway S. K., Morton J. E., Gregory A., Gissen P., Sonek S., Cangul H., Coryell J., Canham N., Nardocci N., et al. 2006. PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat. Genet. 38: 752–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory A., Westaway S. K., Holm I. E., Kotzbauer P. T., Hogarth P., Sonek S., Coryell J. C., Nguyen T. M., Nardocci N., Zorzi G., et al. 2008. Neurodegeneration associated with genetic defects in phospholipase A2. Neurology. 71: 1402–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayflick S. J. 2009. Dystonia-parkinsonism disease gene discovery: expect surprises. Ann. Neurol. 65: 2–3. [DOI] [PubMed] [Google Scholar]
- 21.Paisan-Ruiz C., Bhatia K. P., Li A., Hernandez D., Davis M., Wood N. W., Hardy J., Houlden H., Singleton A., Schneider S. A. 2009. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann. Neurol. 65: 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider S. A., Hardy J., Bhatia K. P. 2009. Iron accumulation in syndromes of neurodegeneration with brain iron accumulation 1 and 2: causative or consequential? J. Neurol. Neurosurg. Psychiatry. 80: 589–590. [DOI] [PubMed] [Google Scholar]
- 23.Robinson P. J., Noronha J., DeGeorge J. J., Freed L. M., Nariai T., Rapoport S. I. 1992. A quantitative method for measuring regional in vivo fatty-acid incorporation into and turnover within brain phospholipids: review and critical analysis. Brain Res. Brain Res. Rev. 17: 187–214. [DOI] [PubMed] [Google Scholar]
- 24.Bao S., Miller D. J., Ma Z., Wohltmann M., Eng G., Ramanadham S., Moley K., Turk J. 2004. Male mice that do not express group VIA phospholipase A2 produce spermatozoa with impaired motility and have greatly reduced fertility. J. Biol. Chem. 279: 38194–38200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bazinet R. P., Rao J. S., Chang L., Rapoport S. I., Lee H. J. 2005. Chronic valproate does not alter the kinetics of docosahexaenoic acid within brain phospholipids of the unanesthetized rat. Psychopharmacology (Berl.). 182: 180–185. [DOI] [PubMed] [Google Scholar]
- 26.Nariai T., Greig N. H., DeGeorge J. J., Genka S., Rapoport S. I. 1993. Intravenously injected radiolabeled fatty acids image brain tumour phospholipids in vivo: differential uptakes of palmitate, arachidonate and docosahexaenoate. Clin. Exp. Metastasis. 11: 141–149. [DOI] [PubMed] [Google Scholar]
- 27.Smith Q. R., Nagura H. 2001. Fatty acid uptake and incorporation in brain: studies with the perfusion model. J Mol Neurosci. 16: 167–172; discussion 215–21. [DOI] [PubMed] [Google Scholar]
- 28.Purdon D., Arai T., Rapoport S. 1997. No evidence for direct incorporation of esterified palmitic acid from plasma into brain lipids of awake adult rat. J. Lipid Res. 38: 526–530. [PubMed] [Google Scholar]
- 29.Chen C. T., Ma D. W., Kim J. H., Mount H. T., Bazinet R. P. 2008. The low density lipoprotein receptor is not necessary for maintaining mouse brain polyunsaturated fatty acid concentrations. J. Lipid Res. 49: 147–152. [DOI] [PubMed] [Google Scholar]
- 30.Holman R. T. 1986. Control of polyunsaturated acids in tissue lipids. J. Am. Coll. Nutr. 5: 183–211. [DOI] [PubMed] [Google Scholar]
- 31.DeMar J. C., Jr, Ma K., Bell J. M., Rapoport S. I. 2004. Half-lives of docosahexaenoic acid in rat brain phospholipids are prolonged by 15 weeks of nutritional deprivation of n-3 polyunsaturated fatty acids. J. Neurochem. 91: 1125–1137. [DOI] [PubMed] [Google Scholar]
- 32.Demar J. C., Jr, Ma K., Chang L., Bell J. M., Rapoport S. I. 2005. α-Linolenic acid does not contribute appreciably to docosahexaenoic acid within brain phospholipids of adult rats fed a diet enriched in docosahexaenoic acid. J. Neurochem. 94: 1063–1076. [DOI] [PubMed] [Google Scholar]
- 33.Jones C. R., Arai T., Bell J. M., Rapoport S. I. 1996. Preferential in vivo incorporation of [3H]arachidonic acid from blood into rat brain synaptosomal fractions before and after cholinergic stimulation. J. Neurochem. 67: 822–829. [DOI] [PubMed] [Google Scholar]
- 34.Basselin M., Villacreses N. E., Langenbach R., Ma K., Bell J. M., Rapoport S. I. 2006. Resting and arecoline-stimulated brain metabolism and signaling involving arachidonic acid are altered in the cyclooxygenase-2 knockout mouse. J. Neurochem. 96: 669–679. [DOI] [PubMed] [Google Scholar]
- 35.Malik I., Turk J., Mancuso D. J., Montier L., Wohltmann M., Wozniak D. F., Schmidt R. E., Gross R. W., Kotzbauer P. T. 2008. Disrupted membrane homeostasis and accumulation of ubiquitinated proteins in a mouse model of infantile neuroaxonal dystrophy caused by PLA2G6 mutations. Am. J. Pathol. 172: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapoport S. I., Ramadan E., Kim H. W., Turk J., Basselin M. 2010. Decreased Brain Docosahexaenoic Acid Signaling in iPLA2-VIA(β)-Deficient Mice. 41st Annual Meeting of the American Society of Neurochemistry, Santa Fe, NM: p. 86 (PSM04-13). [Google Scholar]
- 37.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 38.Franklin K. B. J., Paxinos G. 1997. The Mouse Brain in Stereotaxic Coodinates. Academic Press, Inc., San Diego. [Google Scholar]
- 39.Ramanadham S., Yarasheski K. E., Silva M. J., Wohltmann M., Novack D. V., Christiansen B., Tu X., Zhang S., Lei X., Turk J. 2008. Age-related changes in bone morphology are accelerated in group VIA phospholipase A2 (iPLA2β)-null mice. Am. J. Pathol. 172: 868–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rapoport S. I., Fitzhugh R., Pettigrew K. D., Sundaram U., Ohno K. 1982. Drug entry into and distribution within brain and cerebrospinal fluid: 14C-urea pharmacokinetics. Am. J. Physiol. 242: R339–R348. [DOI] [PubMed] [Google Scholar]
- 41.Levey A. I. 1993. Immunological localization of m1-m5 muscarinic acetylcholine receptors in peripheral tissues and brain. Life Sci. 52: 441–448. [DOI] [PubMed] [Google Scholar]
- 42.Porter A. C., Bymaster F. P., DeLapp N. W., Yamada M., Wess J., Hamilton S. E., Nathanson N. M., Felder C. C. 2002. M1 muscarinic receptor signaling in mouse hippocampus and cortex. Brain Res. 944: 82–89. [DOI] [PubMed] [Google Scholar]
- 43.van Koppen C. J., Kaiser B. 2003. Regulation of muscarinic acetylcholine receptor signaling. Pharmacol. Ther. 98: 197–220. [DOI] [PubMed] [Google Scholar]
- 44.Rao J. S., Ertley R. N., DeMar J. C., Jr, Rapoport S. I., Bazinet R. P., Lee H. J. 2007. Dietary n-3 PUFA deprivation alters expression of enzymes of the arachidonic and docosahexaenoic acid cascades in rat frontal cortex. Mol. Psychiatry. 12: 151–157. [DOI] [PubMed] [Google Scholar]
- 45.Gingrich J. A., Hen R. 2000. The broken mouse: the role of development, plasticity and environment in the interpretation of phenotypic changes in knockout mice. Curr. Opin. Neurobiol. 10: 146–152. [DOI] [PubMed] [Google Scholar]
- 46.Routtenberg A. 2002. Targeting the “species gene ensemble”. Hippocampus. 12: 105–108. [DOI] [PubMed] [Google Scholar]
- 47.Six D. A., Dennis E. A. 2000. The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim. Biophys. Acta. 1488: 1–19. [DOI] [PubMed] [Google Scholar]
- 48.Cooper J. R., Bloom F. E., Roth R. H. 2003. The Biochemical Basis of Neuropharmacology. 8th ed Oxford University Press, Oxford. [Google Scholar]
- 49.Murray-Whelan R., Reid J. D., Piuz I., Hezareh M., Schlegel W. 1995. The guanine-nucleotide-binding protein subunit G alpha i2 is involved in calcium activation of phospholipase A2. Effects of the dominant negative G alpha i2 mutant, [G203T]G alpha i2, on activation of phospholipase A2 in Chinese hamster ovary cells. Eur. J. Biochem. 230: 164–169. [DOI] [PubMed] [Google Scholar]
- 50.Bayon Y., Hernandez M., Alonso A., Nunez L., Garcia-Sancho J., Leslie C., Sanchez Crespo M., Nieto M. L. 1997. Cytosolic phospholipase A2 is coupled to muscarinic receptors in the human astrocytoma cell line 1321N1: characterization of the transducing mechanism. Biochem. J. 323: 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun G. Y., Xu J., Jensen M. D., Yu S., Wood W. G., Gonzalez F. A., Simonyi A., Sun A. Y., Weisman G. A. 2005. Phospholipase A2 in astrocytes: responses to oxidative stress, inflammation, and G protein-coupled receptor agonists. Mol. Neurobiol. 31: 27–41. [DOI] [PubMed] [Google Scholar]
- 52.Felder C. C. 1995. Muscarinic acetylcholine receptors: signal transduction through multiple effectors. FASEB J. 9: 619–625. [PubMed] [Google Scholar]
- 53.Fisher S. K., Domask L. M., Roland R. M. 1989. Muscarinic receptor regulation of cytoplasmic Ca2+ concentrations in human SK-N-SH neuroblastoma cells: Ca2+ requirements for phospholipase C activation. Mol. Pharmacol. 35: 195–204. [PubMed] [Google Scholar]
- 54.Yamamoto K., Hashimoto K., Isomura Y., Shimohama S., Kato N. 2000. An IP3-assisted form of Ca2+-induced Ca2+ release in neocortical neurons. Neuroreport. 11: 535–539. [DOI] [PubMed] [Google Scholar]
- 55.Singaravelu K., Lohr C., Deitmer J. W. 2006. Regulation of store-operated calcium entry by calcium-independent phospholipase A2 in rat cerebellar astrocytes. J. Neurosci. 26: 9579–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basselin M., Chang L., Bell J. M., Rapoport S. I. 2006. Chronic lithium chloride administration attenuates brain NMDA receptor-initiated signaling via arachidonic acid in unanesthetized rats. Neuropsychopharmacology. 31: 1659–1674. [DOI] [PubMed] [Google Scholar]
- 57.Weichel O., Hilgert M., Chatterjee S. S., Lehr M., Klein J. 1999. Bilobalide, a constituent of Ginkgo biloba, inhibits NMDA-induced phospholipase A2 activation and phospholipid breakdown in rat hippocampus. Naunyn Schmiedebergs Arch. Pharmacol. 360: 609–615. [DOI] [PubMed] [Google Scholar]
- 58.Clark J. D., Schievella A. R., Nalefski E. A., Lin L. L. 1995. Cytosolic phospholipase A2. J. Lipid Mediat. Cell Signal. 12: 83–117. [DOI] [PubMed] [Google Scholar]
- 59.Dennis E. A. 1994. Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 269: 13057–13060. [PubMed] [Google Scholar]
- 60.Shinzawa K., Sumi H., Ikawa M., Matsuoka Y., Okabe M., Sakoda S., Tsujimoto Y. 2008. Neuroaxonal dystrophy caused by group VIA phospholipase A2 deficiency in mice: a model of human neurodegenerative disease. J. Neurosci. 28: 2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaeffer E. L., Gattaz W. F. 2005. Inhibition of calcium-independent phospholipase A2 activity in rat hippocampus impairs acquisition of short- and long-term memory. Psychopharmacology (Berl.). 181: 392–400. [DOI] [PubMed] [Google Scholar]
- 62.Fujita S., Ikegaya Y., Nishiyama N., Matsuki N. 2000. Ca2+-independent phospholipase A2 inhibitor impairs spatial memory of mice. Jpn. J. Pharmacol. 83: 277–278. [DOI] [PubMed] [Google Scholar]
- 63.Aid S., Bosetti F. 2007. Gene expression of cyclooxygenase-1 and Ca2+-independent phospholipase A2 is altered in rat hippocampus during normal aging. Brain Res. Bull. 73: 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalyvas A., Baskakis C., Magrioti V., Constantinou-Kokotou V., Stephens D., Lopez-Vales R., Lu J. Q., Yong V. W., Dennis E. A., Kokotos G., et al. 2009. Differing roles for members of the phospholipase A2 superfamily in experimental autoimmune encephalomyelitis. Brain. 132: 1221–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Serhan C. N. 2007. Resolution phase of inflammation: novel endogenous anti-inflammatory and proresolving lipid mediators and pathways. Annu. Rev. Immunol. 25: 101–137. [DOI] [PubMed] [Google Scholar]
- 66.Bazan N. G. 2005. Neuroprotectin D1 (NPD1): a DHA-derived mediator that protects brain and retina against cell injury-induced oxidative stress. Brain Pathol. 15: 159–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umhau J. C., Zhou W., Carson R. E., Rapoport S. I., Polozova A., Demar J., Hussein N., Bhattacharjee A. K., Ma K., Esposito G., et al. 2009. Imaging incorporation of circulating docosahexaenoic acid into the human brain using positron emission tomography. J. Lipid Res. 50: 1259–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bao S., Li Y., Lei X., Wohltmann M., Jin W., Bohrer A., Semenkovich C. F., Ramanadham S., Tabas I., Turk J. 2007. Attenuated free cholesterol loading-induced apoptosis but preserved phospholipid composition of peritoneal macrophages from mice that do not express group VIA phospholipase A2. J. Biol. Chem. 282: 27100–27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.