Abstract
The focus of the present study was to define the human plasma lipidome and to establish novel analytical methodologies to quantify the large spectrum of plasma lipids. Partial lipid analysis is now a regular part of every patient's blood test and physicians readily and regularly prescribe drugs that alter the levels of major plasma lipids such as cholesterol and triglycerides. Plasma contains many thousands of distinct lipid molecular species that fall into six main categories including fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterols, and prenols. The physiological contributions of these diverse lipids and how their levels change in response to therapy remain largely unknown. As a first step toward answering these questions, we provide herein an in-depth lipidomics analysis of a pooled human plasma obtained from healthy individuals after overnight fasting and with a gender balance and an ethnic distribution that is representative of the US population. In total, we quantitatively assessed the levels of over 500 distinct molecular species distributed among the main lipid categories. As more information is obtained regarding the roles of individual lipids in health and disease, it seems likely that future blood tests will include an ever increasing number of these lipid molecules.
Keywords: human lipidome, metabolomics, reference material, mass spectrometry
In this era of genomics, transcriptomics, and proteomics, metabolomics is emerging as an important component of the omics evolution (1). Of the four kinds of biological molecules that comprise the human body, i.e., nucleic acids, amino acids (proteins), carbohydrates (sugars), and lipids (fats), lipids stand out among the various cellular metabolites in the sheer number of distinct molecular species. Using state-of-the-art lipidomics approaches made possible by newly developed instrumentation, protocols, and bioinformatics tools (2), the LIPID MAPS Consortium is carrying out comprehensive analyses of the mammalian lipidome (3). As an emerging ‘omics’ field, lipidomics provides a powerful approach to understanding lipid biology (4).
The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) in collaboration with the National Institute of Standards (NIST) recently produced a human plasma standard reference material (SRM 1950) for metabolite analysis. The SRM was prepared by obtaining plasma samples from 100 individuals between 40 and 50 years of age, whose ethnicity was representative of the US population and that included an equal number of men and women. The intent of the NIDDK/NIST project was to provide a reference material that would be publically available to researchers and that could be used by the clinical chemistry community to identify plasma metabolites for diagnostic purposes.3 Signature metabolites could then be further probed for their usefulness as disease biomarkers.
The LIPID MAPS Consortium has developed innovative lipidomics techniques based on liquid chromatography coupled to mass spectroscopy to probe biological systems (3) and has undertaken the task of analyzing the NIDDK/NIST SRM by systematically identifying and quantifying the lipid molecular species of the mammalian lipidome (5, 6). We report here for the first time an in-depth lipid profile of human plasma that reveals the enormous structural diversity of lipids comprising the six major lipid categories as defined by LIPID MAPS.4 Lipidomics analysis of the sample was challenging due to the extremely large number of lipid species in human plasma and the analytical complexity of the molecular species present. Nevertheless, we report the quantitative levels of over 500 different lipid molecular species present in this human reference plasma sample.
EXPERIMENTAL PROCEDURES
Experimental details are provided online as supplemental material.
RESULTS AND DISCUSSION
Plasma lipid distribution by category
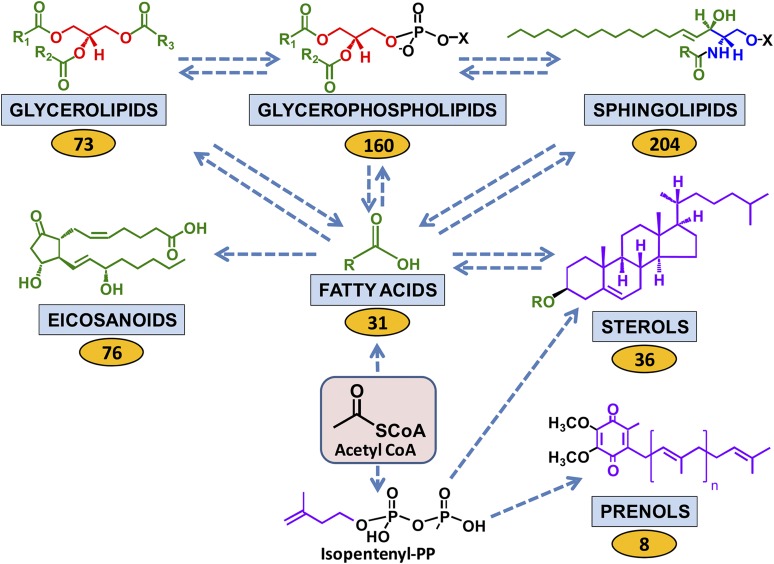
The results are presented and discussed below for each main lipid category and the diversity of lipid molecular species is summarized in Fig. 1. The most number of lipid molecular species analyzed was for the sphingolipid category but the sterols, including cholesterol and its esters, were the most abundant of the lipids on a molar basis (nmol/ml) followed by triglycerides, glycerophospholipids, free fatty acyls, sphingolipids, diacylglycerols, and prenols, though on a weight basis (mg/dl) the glycerophospholipids were the most abundant, as shown in Table 1. Complete data sets are also available on the LIPID MAPS/Nature Lipidomics Gateway, www.lipidmaps.org.
Fig. 1.
Human plasma lipid diversity. Relationships among the major mammalian lipid categories are shown starting with the 2-carbon precursor acetyl CoA, which is the building block in the biosynthesis of fatty acids. Fatty acyl subsituents in turn may be transferred to be part of the complex lipids, namely sphingolipids, glycerolipids, glycerophospholipids, and sterols (as steryl esters). Some fatty acids are converted to eicosanoids. A second major biosynthetic route from acetyl CoA generates the 5-carbon isoprene precursor isopentenyl pyrophosphate, which provides the building blocks for the prenol and sterol lipids. Fatty acyl-derived subsituents are colored green; isoprene-derived atoms are colored purple; glycerol and serine-derived groups are colored red and blue, respectively. Arrows denote multistep transformations among the major lipid categories starting with acetyl CoA. Values in yellow ovals represent the number of analytes within each lipid category that were quantified by mass spectrometry in the human plasma sample (see text).
TABLE 1.
Lipid categories and species quantified in the human plasma SRM
| Lipid Category | Number of Species | Sum (nmol/ml) | Sum (mg/dl) |
|---|---|---|---|
| Fatty Acyls | |||
| Fatty Acids | 31 | 214 | 5.82 |
| Eicosanoids | 76 | 0.071 | 0.002 |
| Total | 107 | 214 | 5.82 |
| Glycerolipids | |||
| Triacylglycerols | 18 | 1058 | 90.6 |
| 1,2-Diacylglycerols | 28 | 39 | 2.36 |
| 1,3-Diacylglycerols | 27 | 13 | 0.805 |
| Total | 73 | 1110 | 93.7 |
| Glycerophospholipids | |||
| PE | 38 | 435 | 32.7 |
| LPE | 7 | 36.6 | 1.78 |
| PC | 31 | 1974 | 157 |
| LPC | 12 | 103 | 5.25 |
| PS | 20 | 7.00 | 0.559 |
| PG | 16 | 6.12 | 0.480 |
| PA | 15 | 2.50 | 0.173 |
| PI | 19 | 31.5 | 2.74 |
| N-acyl-PS | 2 | 0.013 | 0.001 |
| Total | 160 | 2596 | 201 |
| Sphingolipids | |||
| Sphingomyelins | 101 | 303.468 | 22.817 |
| Monohexosylceramides | 56 | 2.3135 | 0.180 |
| Ceramides | 41 | 11.586 | 0.732 |
| Sphingoid Bases | 6 | 0.5678 | 0.02029 |
| Total | 204 | 318 | 23.7 |
| Sterol Lipids* | |||
| Free Sterols | 14 | 826 | 31.8 |
| Esterified Sterols | 22 | 2954 | 114 |
| Total | 36 | 3780 | 146 |
| Prenol Lipids | |||
| Dolichols | 6 | 0.025 | 0.003 |
| Coenzyme-Q | 2 | 4.59 | 0.394 |
| Total |
8 |
4.62 |
0.397 |
| Total | 588 | 8023 | 471 |
Sterols in plasma (nmol/ml) were determined as described in the supplementary experimental procedures. Weight calculations (mg/dl) were based on the sterol backbone for sterol esters. Additionally, total sterols were also measured by clinical laboratories following procedures certified by the Center for Disease Control and they reported similar plasma levels, though they are lower than the national average (National Health and Nutrition Examination Survey 2003–2006). The SRM consists of plasma pooled from individuals within a narrow age range. Thus, cholesterol values may deviate from other averages that include individuals from a wider age spectrum.
Several simplifying assumptions were necessary to generate the data in the molecular detail presented throughout this study. First, only a small number of isotope-labeled internal and reference standards are available for mass spectrometry-based quantitative analyses for the most abundant lipids in plasma, i.e., cholesteryl esters, triglycerides, and glycerophospholipids. Therefore, we assumed that the behavior of these standards with respect to extraction and mass spectrometry was representative for each class of abundant lipid. Second, for the triglycerides and glycerophospholipids, isobaric (same molecular weight) molecular species exist, which further confounds precise quantitation. We realize that these simplifying assumptions may have introduced errors into the accuracy with respect to absolute quantitation for each lipid species. Nonetheless, the total values obtained for individual lipid classes are similar to those measured by techniques that disregard molecular species identity. The information provided in the supplementary tables thus provides an accurate picture of the relative amounts of unique lipid molecular species in the NIST plasma sample. When using the data as a reference, one should keep in mind that the plasma samples were kept frozen for some time during the collection effort. Although every precaution was taken to minimize the effects of storage and thawing on the integrity of the sample, some oxidation or hydrolysis of lipids cannot be entirely ruled out. Physical strain imposed during the process of blood drawing may lead to the activation of leukocytes and induce the formation of metabolites of the arachidonic acid cascade. This may potentially contribute to the amount of eicosanoids that were measured in the plasma samples. Leukocyte activation is of particular concern when hemolysis occurs; therefore, all individual plasma samples that displayed visible indications of hemolysis were discarded and not included to generate the pool of the plasma SRM.
Fatty acyls
Although free fatty acids represent only a small fraction of total fatty acids in plasma, they represent a highly metabolically active lipid class. Adipose tissue is the main source of plasma free fatty acids, which have a distribution that is closely related to the fatty acid composition of the diet (7). Under normal conditions the release of fatty acids from adipose tissue is tightly regulated to meet the energy demands of tissues; however, in several metabolic disorders this homeostasis is compromised and shifts toward increased lipolysis leading to the release of excess free fatty acids relative to tissue needs. As shown in Table 1 and supplementary Table IA, human plasma of healthy individuals after overnight fasting contains an average of 214 nmol/ml of free fatty acids. Oleic acid (18:1) is the major constituent followed by palmitic acid (16:0) and stearic acid (18:0) (supplementary Table IA). Together, these three species comprise about 78% of all free fatty acids in the circulation. Linoleic acid (18:2) and arachidonic acid (20:4) are the main PUFAs (about 8% of the total), but the nutritionally essential α-linolenic acid (18:3ω-3), eicosapentaenoic acid (20:5, EPA), and docosahexaenoic acid (22:6, DHA) are also present at significant levels, together making up about 1% of all free fatty acids.
Eicosanoids are a class of bioactive lipid mediators that are derived from the metabolism of arachidonic acid or related PUFAs (8). Three classes of enzymes including cyclooxygenases (COX-1 and COX-2), lipoxygenases (LOXs), and cytochrome P450 epoxygenases (CYPs) synthesize eicosanoids (9). In addition, PUFAs can undergo autoxidation to produce bioactive lipids through a nonenzymatic pathway. PUFA metabolites derived from all three enzymatic pathways and through autoxidation and lipid peroxidation were detected in the SRM (Table 1 and supplementary Table IB). Typically, eicosanoids have a short half-life and undergo further enzymatic or nonenzymatic modifications that are often accompanied by a change of pharmacological potency. Notably, 15deoxy Δ12,14 PGD2 (15d PGD2), a degradation product of prostaglandin D2 (PGD2), was one of the major COX metabolites found in the human plasma sample. The formation of 15d PGD2 is of particular interest because, like 15deoxy Δ12,14 PGJ2 (15d PGJ2), this eicosanoid robustly stimulates peroxisome proliferator-activated receptor-γ activity, which has been implicated in the regulation of inflammatory responses. The hydroxylated products of arachidonic acid, 5-hydroxy-eicosatetraenoic acid (5-HETE) and 12-HETE, and of linoleic acid, 9-hydroxy-octadecadienoic acid (9-HODE) and 13-HODE, were the major metabolites of the LOX pathway detected in the sample. In fact, the 5-LOX product 5-HETE represented the single most prominent eicosanoid found in the SRM. We also identified some prominent metabolites derived from the CYP pathway including 12,13-dihydroxy-octadec-9-enoic acid (12,13 DiHOME), which is produced by the action of epoxide hydrolase from the corresponding 12,13 epoxide of linoleic acid, the epoxide 14,15-epoxy-5,8,11-eicosatrienoic acid (14,15 EpETrE) formed from arachidonic acid and the corresponding vicinal diol 14,15-dihydroxy-5,8,11-eicosatrienoic acid (14,15 DiHETrE). Some autoxidation products were also detected. Interestingly, the majority of these lipid peroxidation products are derived from DHA, even though this 22:6 fatty acid represents a minor fraction of all free PUFAs in the plasma.
Glycerolipids
Glycerolipids encompassed a high proportion of total lipids present in plasma. Of these, the most abundant members were triacylglycerols (TAGs), which were present at a concentration of 1.1 µmol/ml (90.6 mg/dl) in the plasma SRM (Table 1). The absolute concentration of TAGs is dependent on food intake because the synthesis and packaging of TAG into lipoprotein particles (chiefly chylomicrons and very low density lipoprotein) is the major mechanism by which these lipids are distributed to tissues in the body. We found that about 50% of all TAGs had a total of 52 fatty acyl carbon atoms and that most fatty acyl groups had multiple double bonds. Although the analysis of TAGs did not allow definition of all molecular species, previous studies have revealed that each molecular subset (e.g., 52:3 TAG) is a complex mixture of multiple isobaric entities (10). For example, the three abundant species that make up 52:3 TAG are 16:1/18:1/18:1, 16:1/18:0/18:2, and 16:0/18:1/18:2, not considering isomers that differ at the position on the glycerol carbon atoms where acylation occurs. Including minor fatty acyl species such as 18:3, 20:1, 20:2, and 20:3 fatty acids in the calculation expands the number of possible nonisomeric and distinct molecular species to over seven. Applying chromatographic retention times and considering all isobaric species, it was possible to detect over 200 individual molecular species of TAGs in the plasma sample (supplementary Table IIA). In addition, several ether-linked glycerolipids were detected as alkylether triacylglycerols (ether 50:3, ether 50:2, ether 52:4, ether 52:3, ether 54:6, ether 54:5, and ether 54:4). Diacylglycerols (DAGs) were also present in the plasma sample but at substantially lower levels. Both 1,2-DAGs (supplementary Table IIB) and 1,3-DAGs (supplementary Table IIC) were quantified and found to contain between 30 and 40 total acyl carbon atoms esterified to the glycerol backbone. 1,2-DAGs were about three times as abundant as 1,3-DAGs in this sample.
Glycerophospholipids
Over 200 species of glycerophospholipids were detected and identified in the plasma SRM. Due to the presence of isobaric species with the same m/z, out of all species identified, 158 glycerophosphate (PA), glycerophosphocholine (PC), glycerophosphoethanolamine (PE), glycerophosphoglycerol (PG), glycerophosphoinositol (PI), and glycerophosphoserine (PS) species were quantified and are summarized in Table 1. The composition by subclass and quantities of individual glycerophospholipid species are reported in supplementary Table IIIA. By mass, the overwhelming majority of glycerophospholipids in human plasma are PCs and PEs. These two classes were also found to contain substantial amounts of ether-linked lipids (PEe/ p = 43% of PE by mass; PCe/p = 5.4% of PC by mass; e designates the plasmanyl and p designates the plasmenyl analogues of glycerophospholipids). The amount of total glycerophospholipid is similar to that previously reported using different analytical approaches (11, 12).
In eukaryotes, the polyglycerophospholipid cardiolipin is synthesized and localized in the mitochondrion (13, 14) particularly the inner mitochondrial membrane (15). As plasma is free of cells and subcellular organelles like mitochondria, cardiolipin was not detected. The limit of detection of plasma cardiolipin was found to be 4 nM using synthetic cardiolipin standards.
Evidence for the existence of N-acylphosphatidylserine (N-acyl-PS) was first reported by Nelson in 1970 in sheep erythrocytes (16). As part of the LIPID MAPS consortium's search for novel lipids, Guan et al. (17) in 2007 reported the identification of a family of N-acyl-PS molecules present in mouse and pig brain, yeast, and mouse RAW264.7 macrophage tumor cells. The role of N-acyl-PS in animal cells was not investigated, but it was proposed that N-acyl-PS may function as a precursor of the bioactive signaling lipid N-acyl-L-serine (17). Building on this work, we probed the human plasma SRM for the presence of the two most abundant ions of N-acyl-PS previously detected, i.e., 58:1 and 60:2, named for the number of carbons and double bonds each contains. Both ions containing a complex mixture of isobaric species were detected, with 58:1 making up 83% of the total N-acyl-PS detected (supplementary Table IIIB).
Sphingolipids
We identified over 200 individual sphingolipids in the human plasma SRM. These results are summarized in Table 1 and levels of subspecies are shown in supplementary Table IV. Sphingomyelins (SM) accounted for the largest fraction of sphingolipids in plasma, and ∼100 subspecies were sufficiently abundant for their amounts to be estimated (supplementary Table IV). This number is about twice that of previous estimates of plasma SM subspecies (18–21). Sphingosine was the most common sphingoid base, accounting for ∼61% of the total, followed by sphingadiene (18%) and sphinganine (9.5%); the remainder had other even- and odd-chain lengths, with the 16-carbon-chain length sphingosine accounting for ∼10%. The fatty acids of SM ranged in chain length from 13 to 28 carbons, of which over half were saturated (with 16:0 comprising about one-third of all SM subspecies), about 20% were monounsaturated (the most prevalent being 24:1), and there were small amounts of fatty acids with two (about 3%) and three (<1%) double bonds. Our approaches would not have detected 3-O-acyl-SM, which has been reported in trace amounts (22), because the acyl-group would be hydrolyzed during extraction (23). These proportions are likely to vary among individuals because diet can affect the types and amounts of SM in plasma (24–26).
The majority of the ceramide monohexoses (CMHs) also had sphingosine as the most prevalent sphingoid base and fatty acid chain lengths ranging from 14 to 26 carbons, but fewer subspecies were quantified because the amounts were much lower than SM (∼1%). The major backbone subspecies were similar to SM except that the proportion of very-long-chain fatty acids is higher for CMH (supplementary Table IV). The CMH of human plasma was comprised of both glucosyl- (GlcCer) and galactosyl-ceramides (GalCer), as has been reported before (27–29), with higher proportions of GlcCer. More complex glycosphingolipids such as lactosylceramide were also detected but these were not quantified because the internal standards are not yet available.
Plasma contained small amounts of free ceramide (4% of SM) with approximately the same spectrum of subspecies as were seen in SM and CMH, although many of the minor subspecies were present in amounts too low for quantitation (supplementary Table IV). A noteworthy feature of the free ceramides was that the fatty acid distribution differed substantially from that seen in SM or CMH with very little palmitic acid (∼5%) compared with the very-long-chain fatty acids (i.e., 24:0, ∼33%, and 24:1, ∼12%). A similar distinction has been reported in other analyses of human plasma (30, 31). The N-acyl-derivatives of two novel categories of ceramides with 1-deoxy- and 1-(desoxymethyl)-sphingoid base backbones were also detected but not quantified (32).
Free sphingoid bases were found in the human plasma SRM, with sphingosine 1-phosphate as the major species, followed by lower amounts of sphingosine, sphinganine, and sphinganine 1-phosphate. In addition to d18:1 sphingosine 1-phosphate (d indicates the presence of two hydroxyl groups, followed by the chain length and number of double bonds), there were smaller amounts of other chain lengths (d16:1 to d19:1). A similar profile has been seen in other studies (33–36), although the amounts vary by several fold, which might be due to differences in diet, timing of collection, or storage of the sample.
Sterol lipids
Sterols in plasma exist in both free and fatty-acyl esterified forms. For quantification of total (esterified and free) sterols, plasma was subjected to base hydrolysis, extracted with organic solvent, and sterols were partially purified by solid-phase extraction (37–39). Fourteen different sterols of endogenous and exogenous sources (diet) were detected at concentrations spanning five orders of magnitude (supplementary Table VA). Cholesterol was the most abundant sterol at 3.76 µmol/ml (145 mg/dl) followed by several sterol intermediates arising from the cholesterol biosynthetic pathway, including lathosterol, desmosterol, and 7-dehydrocholesterol, which were present at concentrations ranging from ∼2 to 6 nmol/ml. Two plant sterols derived from the diet, campesterol and sitosterol, were also within this range. Oxysterols, which are derivatives of cholesterol with an extra hydroxyl group on the side chain or ring structures, and the biosynthetic intermediate lanosterol, were the least abundant sterols at 0.02–0.5 nmol/ml.
Free sterols were extracted and isolated using similar methods (37–39) but without the base hydrolysis step (supplementary Table VB). Cholesterol again was the most abundant free sterol at 0.82 µmol/ml. Lathosterol and desmosterol, both intermediates in cholesterol biosynthesis, were present at ∼1–2.5 nmol/ml. Another biosynthetic intermediate, 7-dehydrocholesterol, was not detected as a free sterol. The diet-derived sterols, campesterol and sitosterol, were detected at ∼1nmol/ml. Free oxysterol levels ranged between ∼0.001 and 0.01nmol/ml. Depending on the sterol, between 2 and 45% were present in free form; 22% of cholesterol was found in the free form. An aliquot of the NIDDK/NIST reference plasma was also submitted for total (esterified and free) cholesterol measurement using a colorimetric assay-based clinical instrument, which returned an average value of 3.86 µmol/ml. This concentration is consistent with the mass spectrometry-based measurement of 3.76 µmol/ml.
We also quantified cholesteryl esters (CEs) without prior hydrolysis and found that at 3.6 µmol/l (230 mg/dl), the neutral CEs represented the single most abundant lipid class in this human plasma sample. The complexity of molecular species was far less than that observed in other neutral lipids such as TAGs and DAGs in that a single molecular species, 18:2 CE, comprised over 50% of this class (supplementary Table VC). Also, 20:4 CE was a major component (6.9% mol fraction of total CE). Considering the absolute concentration of these polyunsaturated CE species in plasma, it is likely that a majority of arachidonate found in cells is delivered directly as 20:4 CE or is made from 18:2 CE. Subtracting the value of free cholesterol measured in the plasma SRM from the total cholesterol value (supplementary Tables VA, B) yields a predicted value of 2.94 µmol/ml for the CE content of the sample. This value is smaller than the CE content determined directly (3.6 µmol/ml). We believe that the smaller value of 2.94 µmol/ml is more correct and that the difference arises from the use of a single internal standard (13C18:1 CE) to estimate the quantities of all CE species as opposed to using multiple sterol standards that more accurately reflect recovery and chromatographic behavior of individual sterols. Should it become important to measure individual CE species with greater accuracy in the future, appropriate standards will be developed.
Prenol lipids
Prenol lipids are synthesized from five carbon isoprene units. Two classes of prenol lipids were included in this analysis, dolichols and ubiquinones (Table 1). Dolichols are a group of α-saturated polyprenols, with the number of isoprene repeats typically varying from 14 to 24 (40). Changes in serum and plasma dolichol levels may have clinical significance (40, 41) and have been shown to increase in a number of human diseases (40, 42). Free dolichols consisting of 16–20 isoprenoid units were detected in the human plasma SRM (supplementary Table VIA). Consistent with previous reports (40, 42), dolichol-19 was the most abundant species, making up 53% of the total free dolichol detected. A total of 25.1 pmol/ml of free dolichol was detected in the human plasma SRM, comparable to previously published plasma levels (43).
Ubiquinones are a group of 1,4-benzoquinones modified with repeating isoprene units. The number of isoprene repeats varies among organisms, with yeast and bacteria possessing 6, 7, and 8 repeats (CoQ6, CoQ7, and CoQ8, respectively) and rats, mice, and humans possessing 9 and 10 repeats (CoQ9 and CoQ10, respectively) (44). Our analysis of human plasma SRM focused on CoQ9 and CoQ10, which are essential components of the mitochondrial electron transfer chain and thus required for ATP synthesis. Clinical evidence suggests that CoQ9 and CoQ10 also possess cardioprotective properties, preventing ischemic damage to heart tissue and reducing ventricular dysfunction and cardiac arrhythmias associated with heart failure and acute myocardial infarction (44–46). Because the biosynthesis of CoQ is dependent upon the availability of farnesyl pyrophosphate produced by the mevalonate pathway, inhibition of the rate-limiting enzyme in this pathway, HMG-CoA reductase, by the cholesterol lowering statin drugs may lead to reductions in serum CoQ levels (46, 47). Both CoQ9 and CoQ10 were detected in the human plasma SRM samples (supplementary Table VIB). Consistent with previous reports (44, 46, 47), CoQ10 was the more abundant of the two ubiquinones, making up 94% of the total plasma CoQ. A total of 4.6 µmol/l of plasma ubiquinone was detected, slightly higher than previously published values (43, 47, 48).
CONCLUSION
The analysis of just two main plasma lipids (triglycerides and cholesterol, the latter in both the LDL and HDL subfractions) and sporadically vitamin D has been considered an integral component of medical diagnosis and treatment for some time. Indeed, it is now well established that intracellular lipid metabolism is dramatically perturbed in numerous metabolic diseases that have genetic and/or dietary and nutritional as well as life style origins and may have implications for the treatment of metabolic diseases including Type 2 diabetes and cardiovascular disease as well as autoimmune disorders including rheumatoid arthritis, neurodegenerative diseases, kidney and liver disorders, and numerous other lipodystrophys. The causal link between lipids and these various disorders has brought about an increasing comprehension of the diverse functions of lipids in many cellular metabolic pathways. Aside from their role as building blocks of membranes of cells and organelles and as energy storage entities, lipids perform important functions in signaling and metabolic regulation. Their diversity in function is reflected by an enormous variation in structure; however, restraints imposed by previously available analytical procedures have limited the number of lipid species that could be reliably assessed in clinical laboratories. Typical blood tests include total triglycerides and cholesterol, but as we report here, it is now technically feasible to quantify human plasma lipids with a much greater depth and accuracy. The amazing complexity of the human plasma lipidome establishes it as a rich source of molecules that can be evaluated for the clues that they provide about human physiology, nutrition, and disease. Analysis of a patient's plasma lipidome will in the future of clinical medicine provide a valuable approach for detecting and monitoring human diseases and their treatment efficacy.
Supplementary Material
Acknowledgments
We extend our appreciation to Dr. Arthur Castle, Program Director, Metabolomics and Informatics, NIDDK, for logistic support in reference material development and making it available to LIPID MAPS as part of the National Institutes of Health Roadmap initiative.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- CMH
- ceremide monohexose
- DAG
- diacylglycerol
- NIDDK
- National Institute of Diabetes and Digestive and Kidney Diseases
- NIST
- National Institute of Standards
- SRM
- standard reference material
- TAG
- triacylglycerol
Further information is available at www.nist.gov/cstl/analytical/organic/metabolitesinserum.cfm
See the LIPID MAPS/Nature Lipidomics Gateway, www.lipidmaps.org
This study was supported by the LIPID MAPS Large Scale Collaborative Grant GM069338 from the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of experimental procedures, references, and 15 tables.
REFERENCES
- 1.Dennis E. A. 2009. Lipidomics joins the omics evolution. Proc. Natl. Acad. Sci. USA. 106: 2089–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipidomics and Bioactive Lipids: Mass-Spectrometry-Based Lipid Analysis. 2007. Methods Enzymol. 43: (special issue). 1–387. [Google Scholar]
- 3.Dennis E. A., Brown H. A., Deems R. A., Glass C. K., Merrill A. H., Murphy R. C., Raetz R. H., Shaw W., Subramaniam S., Russel D. W., et al. 2005. The LIPID MAPS approach to lipidomics. Functional Lipidomics. Feng L., Prestwich G., CRC Press/Taylor & Francis Group, Boca Raton, FL. [Google Scholar]
- 4.Brown H. A., Murphy R. C. 2009. Working towards an exegesis for lipids in biology. Nat. Chem. Biol. 5: 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H., Jr, Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46: 839–861. [DOI] [PubMed] [Google Scholar]
- 6.Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., Shimizu T., Spener F., van Meer G., Wakelam M. J., Dennis E. A. 2009. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50(Suppl): S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertzel A. V., Thompson B. R., Wiczer B. M., Bernlohr D. A. 2008. Lipid metabolism in adipose tissue. Biochemistry of Lipids, Lipoproteins and Membranes. Vance D. E., Vance J. E., Elsevier, Amsterdam: 277–304. [Google Scholar]
- 8.Funk C. D. 2001. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 294: 1871–1875. [DOI] [PubMed] [Google Scholar]
- 9.Buczynski M. W., Dumlao D. S., Dennis E. A. 2009. Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology. J. Lipid Res. 50: 1015–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAnoy A. M., Wu C. C., Murphy R. C. 2005. Direct qualitative analysis of triacylglycerols by electrospray mass spectrometry using a linear ion trap. J. Am. Soc. Mass Spectrom. 16: 1498–1509. [DOI] [PubMed] [Google Scholar]
- 11.Dougherty R. M., Galli C., Ferro-Luzzi A., Iacono J. M. 1987. Lipid and phospholipid fatty acid composition of plasma, red blood cells, and platelets and how they are affected by dietary lipids: a study of normal subjects from Italy, Finland, and the USA. Am. J. Clin. Nutr. 45: 443–455. [DOI] [PubMed] [Google Scholar]
- 12.Bradamante S., Barchiesi E., Barenghi L., Zoppi F. 1990. An alternative expeditious analysis of phospholipid composition in human blood plasma by 31P NMR spectroscopy. Anal. Biochem. 185: 299–303. [DOI] [PubMed] [Google Scholar]
- 13.Schlame M., Ren M., Xu Y., Greenberg M. L., Haller I. 2005. Molecular symmetry in mitochondrial cardiolipins. Chem. Phys. Lipids. 138: 38–49. [DOI] [PubMed] [Google Scholar]
- 14.Schlame M. 2008. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 49: 1607–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houtkooper R. H., Vaz F. M. 2008. Cardiolipin, the heart of mitochondrial metabolism. Cell. Mol. Life Sci. 65: 2493–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson G. J. 1970. Studies on the lipids of sheep red blood cells. IV. The identification of a new phospholipid. N-acyl phosphatidyl serine. Biochem. Biophys. Res. Commun. 38: 261–265. [DOI] [PubMed] [Google Scholar]
- 17.Guan Z., Li S., Smith D. C., Shaw W. A., Raetz C. R. 2007. Identification of N-acylphosphatidylserine molecules in eukaryotic cells. Biochemistry. 46: 14500–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuelsson B., Samuelsson L. 1969. Separation and identification of ceramides derived from human plasma sphingomyelins. J. Lipid Res. 10: 47–55. [PubMed] [Google Scholar]
- 19.Hirvisalo E. L., Renkonen O. 1970. Composition of human serum sphingomyelins. J. Lipid Res. 11: 54–59. [PubMed] [Google Scholar]
- 20.Kuksis A., Stachnyk O., Holub B. J. 1969. Improved quantitation of plasma lipids by direct gas-liquid chromatography. J. Lipid Res. 10: 660–667. [PubMed] [Google Scholar]
- 21.Vieu C., Terce F., Chevy F., Rolland C., Barbaras R., Chap H., Wolf C., Perret B., Collet X. 2002. Coupled assay of sphingomyelin and ceramide molecular species by gas liquid chromatography. J. Lipid Res. 43: 510–522. [PubMed] [Google Scholar]
- 22.Kramer J. K., Blackwell B. A., Dugan M. E., Sauer F. D. 1996. Identification of a new sphingolipid 3-O-acyl-D-erythro-sphingomyelin in newborn pig and infant plasma. Biochim. Biophys. Acta. 1303: 47–55. [DOI] [PubMed] [Google Scholar]
- 23.Shaner R. L., Allegood J. C., Park H., Wang E., Kelly S., Haynes C. A., Sullards M. C., Merrill A. H., Jr 2009. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 50: 1692–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nye W. H. 1969. Variability of plasma phospholipids in normal adults. Am. J. Clin. Nutr. 22: 5–7. [DOI] [PubMed] [Google Scholar]
- 25.Li Z., Basterr M. J., Hailemariam T. K., Hojjati M. R., Lu S., Liu J., Liu R., Zhou H., Jiang X. C. 2005. The effect of dietary sphingolipids on plasma sphingomyelin metabolism and atherosclerosis. Biochim. Biophys. Acta. 1735: 130–134. [DOI] [PubMed] [Google Scholar]
- 26.Katsikas H., Wolf C. 1995. Blood sphingomyelins from two European countries. Biochim. Biophys. Acta. 1258: 95–100. [DOI] [PubMed] [Google Scholar]
- 27.Wells H. W., Jones M. 1973. Galactosylceramides in human plasma. Am. J. Clin. Pathol. 60: 890–896. [DOI] [PubMed] [Google Scholar]
- 28.Vance D. E., Sweeley C. C. 1967. Quantitative determination of the neutral glycosyl ceramides in human blood. J. Lipid Res. 8: 621–630. [PubMed] [Google Scholar]
- 29.Koscielak J., Maslinski W., Zielenski J., Zdebska E., Brudzynski T., Miller-Podraza H., Cedergren B. 1978. Structures and fatty acid compositions of neutral glycosphingolipids of human plasma. Biochim. Biophys. Acta. 530: 385–393. [DOI] [PubMed] [Google Scholar]
- 30.Gorska M., Dobrzyn A., Zendzian-Piotrowska M., Namiot Z. 2002. Concentration and composition of free ceramides in human plasma. Horm. Metab. Res. 34: 466–468. [DOI] [PubMed] [Google Scholar]
- 31.Ichi I., Nakahara K., Miyashita Y., Hidaka A., Kutsukake S., Inoue K., Maruyama T., Miwa Y., Harada-Shiba M., Tsushima M., et al. 2006. Association of ceramides in human plasma with risk factors of atherosclerosis. Lipids. 41: 859–863. [DOI] [PubMed] [Google Scholar]
- 32.Zitomer N. C., Mitchell T., Voss K. A., Bondy G. S., Pruett S. T., Garnier-Amblard E. C., Liebeskind L. S., Park H., Wang E., Sullards M. C., et al. 2009. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: a novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J. Biol. Chem. 284: 4786–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caligan T. B., Peters K., Ou J., Wang E., Saba J., Merrill A. H., Jr 2000. A high-performance liquid chromatographic method to measure sphingosine 1-phosphate and related compounds from sphingosine kinase assays and other biological samples. Anal. Biochem. 281: 36–44. [DOI] [PubMed] [Google Scholar]
- 34.Berdyshev E. V., Gorshkova I. A., Garcia J. G., Natarajan V., Hubbard W. C. 2005. Quantitative analysis of sphingoid base-1-phosphates as bisacetylated derivatives by liquid chromatography-tandem mass spectrometry. Anal. Biochem. 339: 129–136. [DOI] [PubMed] [Google Scholar]
- 35.Jiang X., Han X. 2006. Characterization and direct quantitation of sphingoid base-1-phosphates from lipid extracts: a shotgun lipidomics approach. J. Lipid Res. 47: 1865–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scherer M., Schmitz G., Liebisch G. 2009. High-throughput analysis of sphingosine 1-phosphate, sphinganine 1-phosphate, and lysophosphatidic acid in plasma samples by liquid chromatography-tandem mass spectrometry. Clin. Chem. 55: 1218–1222. [DOI] [PubMed] [Google Scholar]
- 37.McDonald J. G., Thompson B. M., McCrum E. C., Russell D. W. 2007. Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzymol. 432: 145–170. [DOI] [PubMed] [Google Scholar]
- 38.Dzeletovic S., Breuer O., Lund E., Diczfalusy U. 1995. Determination of cholesterol oxidation products in human plasma by isotope dilution-mass spectrometry. Anal. Biochem. 225: 73–80. [DOI] [PubMed] [Google Scholar]
- 39.Xu F., Rychnovsky S. D., Belani J. D., Hobbs H. H., Cohen J. C., Rawson R. B. 2005. Dual roles for cholesterol in mammalian cells. Proc. Natl. Acad. Sci. USA. 102: 14551–14556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiota Y., Kiyota K., Kobayashi T., Kano S., Kawamura M., Matsushima T., Miyazaki S., Uchino K., Hashimoto F., Hayashi H. 2008. Distribution of dolichol in the serum and relationships between serum dolichol levels and various laboratory test values. Biol. Pharm. Bull. 31: 340–347. [DOI] [PubMed] [Google Scholar]
- 41.Sakakihara Y., Hoa T. J., Kamoshita S. 1989. Age-associated decrease of total dolichol in human serum. Clin. Chim. Acta. 184: 333–334. [DOI] [PubMed] [Google Scholar]
- 42.Swiezewska E., Danikiewicz W. 2005. Polyisoprenoids: structure, biosynthesis and function. Prog. Lipid Res. 44: 235–258. [DOI] [PubMed] [Google Scholar]
- 43.Elmberger P. G., Engfeldt P., Dallner G. 1988. Presence of dolichol and its derivatives in human blood. J. Lipid Res. 29: 1651–1662. [PubMed] [Google Scholar]
- 44.Lekli I., Das S., Mukherjee S., Bak I., Juhasz B., Bagchi D., Trimurtulu G., Krishnaraju A. V., Sengupta K., Tosaki A., et al. 2008. Coenzyme Q9 provides cardioprotection after converting into coenzyme Q10. J. Agric. Food Chem. 56: 5331–5337. [DOI] [PubMed] [Google Scholar]
- 45.Singh R. B., Wander G. S., Rastogi A., Shukla P. K., Mittal A., Sharma J. P., Mehrotra S. K., Kapoor R., Chopra R. K. 1998. Randomized, double-blind placebo-controlled trial of coenzyme Q10 in patients with acute myocardial infarction. Cardiovasc. Drugs Ther. 12: 347–353. [DOI] [PubMed] [Google Scholar]
- 46.Overvad K., Diamant B., Holm L., Holmer G., Mortensen S. A., Stender S. 1999. Coenzyme Q10 in health and disease. Eur. J. Clin. Nutr. 53: 764–770. [DOI] [PubMed] [Google Scholar]
- 47.Ghirlanda G., Oradei A., Manto A., Lippa S., Uccioli L., Caputo S., Greco A. V., Littarru G. P. 1993. Evidence of plasma CoQ10-lowering effect by HMG-CoA reductase inhibitors: a double-blind, placebo-controlled study. J. Clin. Pharmacol. 33: 226–229. [DOI] [PubMed] [Google Scholar]
- 48.Mabuchi H., Nohara A., Kobayashi J., Kawashiri M. A., Katsuda S., Inazu A., Koizumi J. 2007. Effects of CoQ10 supplementation on plasma lipoprotein lipid, CoQ10 and liver and muscle enzyme levels in hypercholesterolemic patients treated with atorvastatin: a randomized double-blind study. Atherosclerosis. 195: e182–e189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.