Abstract
The R46L variant in the proprotein-convertase subtilisin-kexin type 9 (PCSK9) gene was associated with reduced levels of LDL and total cholesterol and with a lower risk of coronary artery disease. We investigated the association of R46L with myocardial infarction (MI) in 1,880 Italian patients with premature MI and 1,880 controls. A trend toward a protective effect of the L46 allele was observed [odds ratio (OR) = 0.75, 95% confidence interval (CI) = 0.49–1.13; P = 0.17], although the association with MI was not significant. This is probably due to the combined effect of the low frequency of R46L among Italians and of the young age of the analyzed cohort for whom the impact of coronary atherosclerosis is less important. This hypothesis was indirectly confirmed by the significant association found after including 1,056 additional older controls (OR = 0.67, 95% CI = 0.46-0.97; P = 0.036). LDL cholesterol was significantly lower in L46 carriers (116.2 ± 34.7 mg/dl) than in noncarriers (137.4 ± 47.3 mg/dl; P = 0.00022); a similar reduction was observed for total cholesterol (191.7 ± 37.7 vs. 211.7 ± 49 mg/dl; P = 0.00019). Analysis of 23 additional polymorphisms in the PCSK9 region identified another single nucleotide polymorphism (SNP) (rs11206510) associated with cholesterol levels. We confirmed that the L46 allele not only decreases LDL cholesterol but also protects against MI. Moreover, we replicated the association of total and LDL cholesterol with the SNP rs11206510.
Keywords: proprotein-convertase subtilisin-kexin type 9 genetic variants, low density lipoprotein, association study
Experimental, genetic, and epidemiologic data have demonstrated the association between plasma levels of LDL and HDL cholesterol and coronary artery disease (CAD) incidence worldwide (1–3). High serum concentrations of cholesterol lead to accelerated atherosclerosis, resulting from cholesterol deposition in arterial walls, thereby increasing the risk of CAD. A meta-analysis on 150,000 individuals, including 3,000 with CAD-related deaths, showed that both LDL and HDL cholesterol levels are independently associated with CAD risk (4).
Although smoking, diet, and physical activity all have a role in determining individual lipid profiles, it has been estimated that roughly 50% of the inter-individual variability in lipid and lipoprotein levels is genetically determined (5, 6), and it is clear that LDL cholesterol, HDL cholesterol, and triglyceride concentrations are strongly influenced by the genetic constitution of each individual. Furthermore, genetic variants that increase LDL cholesterol concentrations have also been associated with increased susceptibility to CAD (7). Thus, the available evidence demonstrates not only that genetic variants account for a substantial fraction of individual variation in lipid concentrations but also that lipid concentrations are associated with the risk of CAD.
Several genetic defects responsible for rare Mendelian forms of severe hypercholesterolemia or hypocholesterolemia have been identified; among them, in 2003, pro-protein convertase subtilisin-kexin type 9 (PCSK9) emerged as the third gene involved in autosomal dominant hypercholesterolemia (8). PCSK9, also called neural apoptosis-regulated convertase 1, is the ninth member of the mammalian proprotein convertase family (9), which is involved in activation, inactivation, and regulation of cellular localization of secretory proteins (10, 11). In particular, PCSK9 plays a critical role in cholesterol metabolism by controlling LDL receptor (LDLR) protein levels and, hence, the levels of LDL particles that circulate in blood. PCSK9 mutations causing autosomal dominant hypercholesterolemia are very rare and account for a much smaller percentage of dominant hypercholesterolemia than do mutations in LDLR and apolipoprotein B (8, 12).
After the first report of gain-of-function mutations leading to autosomal dominant hypercholesterolemia, loss-of-function mutations associated with lower cholesterol levels were described (13), and a spectrum of sequence variations in PCSK9 with variable frequencies and effect sizes were shown to contribute to interindividual differences in LDL cholesterol levels in the general population (14). Concerning PCSK9 loss-of-function mutations, a large 15-year prospective study showed that two nonsense mutations (Y142X and C679X), present in 2.6% of African-Americans, are associated with a 28% reduction in mean LDL cholesterol levels and an 88% reduction in the risk of CAD. In the same study, Caucasians with the R46L variation (found with a frequency of 3.2%) had a 47% reduction in CAD despite having a mean reduction in LDL cholesterol levels of only 15% (15). Up to now, the reduction in CAD associated with the PCSK9-R46L mutation was replicated only in two case-control studies: McPherson and Kavaslar (16) reported a lower frequency of the L46 allele in patients with premature CAD (2.4%) compared with healthy elderly individuals (4.5%); more recently, Kathiresan et al. (17) performed a large-scale study providing strong replication evidence that heterozygosity for the R46L variation protects against myocardial infarction (MI). Moreover, during the final drafting of this paper, a very large study performed by Benn et al. (18) on 45,699 individuals from 3 independent Danish studies found a significant association of the L46 allele with a 58% reduction in risk of MI only in the largest cohort.
In this work, we aimed to determine the relative frequency of the PCSK9-R46L allele and its association with premature MI and plasma lipid levels in the Italian population. Moreover, taking advantage of the genome-wide association study (GWAS) performed on our cohort of MI patients and controls (19), we investigated the effects of common variants in PCSK9 on cholesterol levels.
METHODS
Study populations
The Institutional Review Boards of the participating hospitals approved the study; all participants gave their written informed consent and every attempt was made to ensure anonymity.
This study has been conducted as an extension of the previously selected Atherosclerosis, Thrombosis, and Vascular Biology Italian Study Group (ATVB)-cohort (20) collected between January 1997 and January 2007 by a network of 125 Italian coronary units. The 1,880 Caucasian patients were individuals with early-onset MI (first event before the age of 45) who underwent coronary arteriography at the time of hospitalization. Acute MI was defined as resting chest pain lasting more than 30 min, associated with persistent electrocardiographic changes, and confirmed by an increased total creatine kinase or its myocardial-type fraction by more than twice the upper normal limits. The 1,880 controls were healthy subjects unrelated to the cases but individually matched with them in terms of age, sex, and geographical origin, who were enrolled from the staff of the participating hospitals; they declared to have no personal or family history of thromboembolic disease.
All participants filled out a standardized questionnaire concerning traditional cardiovascular risk factors. The collected data included: i) age (mean age 39.6 ± 4.9); ii) sex (88.8% men, 11.2% women); iii) smoking status defined as current, former, or never smoker on the basis of self-reports (44.9, 42.6, and 12.5%, respectively, for patients; 30.4, 18, and 51.6% for controls); iv) hypercholesterolemia defined either as fasting serum cholesterol level >5.2 mmol/L or as the taking of anti-hypercholesterolemic medications (61.5% of patients and 45.5% of controls showed hypercholesterolemia); v) diabetes defined by a fasting serum glucose level of 125 mg/dl or higher, a nonfasting glucose level of 200 mg/dl, or by the use of hypoglycemic agents (7.6% of patients and 0.7% of controls were diabetic); vi) hypertension defined by a systolic blood pressure of 140 mm Hg or higher, a diastolic blood pressure of 90 mm Hg or higher, or by taking antihypertensive medications (27.6% cases and 9.1% of controls suffered from hypertension); and vii) body mass index (BMI) (mean BMI 26.77 ± 4.2 and 25.01 ± 3.3 for patients and controls, respectively).
A supplementary control Caucasian population, composed of 1,056 unrelated individuals enrolled from the staff of the participating hospitals and required to have no history of atherothrombosis, was also collected. This control population had a mean age 15 years higher than the ATVB cohort (53.9 ± 11.5) and a similar gender distribution (males: 80.7%).
Genotyping of rs11591147 (the PCSK9 variant R46L) by real-time PCR
The R46L variant was genotyped using an in-house developed Taqman assay. Oligonucleotide primers and probes were designed with the Beacon Designer software (Premier Biosoft International, Palo Alto, CA) and purchased from Sigma-Genosys (Haverhill, Suffolk, UK). Their sequences can be provided on request.
PCRs were performed on 10 ng of genomic DNA in a 12.5 µL final volume containing 6.25 µL 2× Premix Ex Taq (Takara Biomedicals, Shiga, Japan), 480 nM primers, 240 nM wild-type probe (6-Fam-labeled), 240 nM mutant probe (Hex-labeled). After DNA denaturation at 95°C for 10 s, 45 cycles of a two-step PCR protocol were carried out in an iQ5 thermal cycler (Bio-Rad, Hercules, CA) as follows: denaturation at 95°C for 5 s and annealing/extension at 57°C for 30 s. Fluorescence was measured during the PCR annealing/extension step. Allele discrimination was performed using the iQ5 Cycler software (Bio-Rad).
Genotyping SNPs covering the PCSK9 gene by the GeneChip Mapping 6.0 Array
A GWAS was performed on the ATVB cohort in the frame of the MIGen project (19) using the GeneChip Mapping 6.0 Array from Affymetrix (Santa Clara, CA). For methodological details, see Kathiresan et al. (19). For the present study, 23 single nucleotide polymorphisms (SNPs) distributed along a genomic region comprising the whole PCSK9 gene and its flanking regions (19 kb upstream and 11 kb downstream) were analyzed for association with MI, LDL, and total cholesterol levels. A call rate >97%, a minor allele frequency >1%, and values of P > 0.001 for test of deviation from Hardy-Weinberg equilibrium were used as quality-control criteria.
Statistical analysis
All procedures were performed using either the R program or the software package PLINK v. 1.04 (21).
The association of traditional risk factors with MI was assessed by testing for difference between cases and controls by means of a chi-square test for categorical data (diabetes, hypertension, hypercholesterolemia, smoking) or by using the Student's t-test for continuous data (i.e., BMI, which was analyzed as a quantitative variable after having controlled that its departure from linearity was not statistically significant).
For each SNP, a standard case-control analysis using allelic chi-square test (1 d.f.) was used to provide asymptotic P values, odds ratios (ORs), and 95% confidence intervals (CIs) for minor alleles. The association test was performed (when possible) conditional on the matching by using the Cochran-Mantel-Haenszel statistic. Adjustment for classical cardiovascular risk factors was performed by adding those covariates in a multiple logistic-regression model. Power estimates indicated that if each analyzed polymorphism (allele frequency of 2%) were to directly confer a 1.5-fold increase in the relative risk of MI, the case and control groups used in this research would be of sufficient size to have 82% power to detect a significant association at the 0.05 level in the “initial” paired case-control population sample.
Furthermore, total and LDL cholesterol values, available for 3,453 (1,710 cases + 1,743 controls) and 2,495 (1,353 cases + 1,142 controls) subjects, respectively, were adjusted for gender, age, age2, and MI status. The multivariable-adjusted residual lipid concentrations served as phenotype values for all subsequent genotype-phenotype analyses.
Values of P < 0.05 were considered to indicate statistical significance.
The linkage disequilibrium (LD) structure of the PCSK9 locus was assessed with the software package Haploview version 4.1 (22). LD blocks were defined according to the criteria of Gabriel et al. (23). Haplotype-based association analysis was hence performed considering only those SNPs mapping within the same LD block using the PLINK software.
The study was conducted according to the STrengthening the REporting of Genetic Association Studies (STREGA) guidelines (24).
RESULTS
Association analysis of the R46L variant with MI and lipid levels
The study sample consisted of 1,670 men and 210 women with early-onset MI and an equal number of age- and sex-matched controls; all traditional risk factors (see Methods for details on their distributions) were strongly associated with MI. In particular, diabetes and hypertension were the risk factors with the strongest effect [OR = 11.4 (95% CI = 5.9–21.8) and 3.8 (95% CI = 3.1–4.6), respectively]; smoking (grouping together former and never smokers vs. current smokers), BMI, and hypercholesterolemia were also significantly associated (P < 0.001), but with a lower effect size.
The functional missense variant R46L within the PCSK9 gene was genotyped in the entire ATVB cohort to investigate its association with premature MI in the Italian population. The genotyping success rate exceeded 98% and the accuracy was >99% according to random duplicated genotyping of 5% of samples. No significant deviation from Hardy-Weinberg equilibrium was observed either in cases or in controls (or in both).
Frequency of the L46 allele was higher in controls than in cases (1.42% vs. 1.04%) and a clear trend toward a protective effect of this allele was observed, even though the association with lower MI risk did not reach statistical significance (OR = 0.75; 95% CI = 0.49–1.13; P = 0.17) (Table 1). After adjustment for covariates, the association remained not significant (OR = 0.78; 95% CI = 0.49–1.25; P = 0.31) (Table 1).
TABLE 1.
Association of the PCSK9 missense variant R46L with early-onset MI
| Minor Allele Frequency (%) |
||||||
|---|---|---|---|---|---|---|
| Study | N. Cases | N. Controls | Cases | Controls | OR (95% CI) | P |
| ATVB cases vs. ATVB ctrl | 1,872 | 1,865 | 1.04 | 1.42 | 0.75 (0.49-1.13) | 0.170 |
| Adjusted Values: | 0.78 (0.49-1.25) | 0.310 | ||||
| ATVB cases vs. Suppl-ctrl | 1,872 | 996 | 1.04 | 1.81 | 0.57 (0.36-0.90) | 0.019 |
| ATVB cases vs. ATVB ctrl+Suppl-ctrl | 1,872 | 2,861 | 1.04 | 1.55 | 0.67 (0.46-0.97) | 0.036 |
ATVB ctrl = controls collected in the frame of the ATVB study (20) on premature MI (mean age 39.6 ± 4.9); Suppl-ctrl = supplementary control group (mean age 53.9 ± 11.5). Significant results are in bold; adjustment took into account hypertension, smoking, diabetes, hypercholesterolemia, and BMI.
The observed lack of association might be due to the combined effect of the young age of the analyzed cohort (in whom the impact of coronary atherosclerosis is known to be less important) and of the lower frequency of the L46 allele observed in the Italian population compared with that reported in other Caucasian populations (i.e., 2–4.5%) (15–17). To confirm this hypothesis, a supplementary control population was analyzed; it included 1,056 controls with a mean age 15 years higher than the ATVB cohort and with the same male/female ratio. The R46L variant was genotyped in this additional cohort, which was used as such or together with the ATVB control group in association analyses with MI; in both cases, the minor allele was significantly associated with a reduced risk of MI (OR = 0.57, 95% CI = 0.36–0.90, P = 0.019, using as control group only the supplementary control cohort; OR = 0.67, 95% CI = 0.46–0.97, P = 0.036, using as control groups ATVB + supplementary cohort) (Table 1).
We also compared the distribution of plasma LDL and total cholesterol levels between homozygous wild-type individuals and subjects carrying the L46 allele at the heterozygous state (no homozygotes were detected). Age, sex distribution, BMI, and prevalence of diabetes, hypertension, and smoking habits were not significantly different between individuals carrying the L46 allele and those without it (Table 2).
TABLE 2.
Cardiovascular risk factors among carriers and noncarriers of the L46 allele
| Characteristics | Noncarriers (n = 3,645) | Carriers (n = 92) | P |
|---|---|---|---|
| Age (years)a | 36.64 ± 4.9 | 38.89 ± 5.2 | 0.15b |
| Male sex (%) | 88.8 | 87.0 | 0.83b |
| Diabetes (%) | 3.1 | 2.2 | 1.00c |
| Hypertension (%) | 18.2 | 17.6 | 1.00c |
| BMI (kg/m2) | 25.88 ± 3.9 | 26.06 ± 3.6 | 0.65b |
| Smoking (%)d | 38.1 | 38.5 | 1.00c |
Data are shown either as mean ± standard deviation or as %.
Age at onset for cases.
Continuous data were tested using two-tailed Student's t-test.
Categorical data were tested using a chi-square test.
Non and former smokers were aggregated in the single category of non smokers.
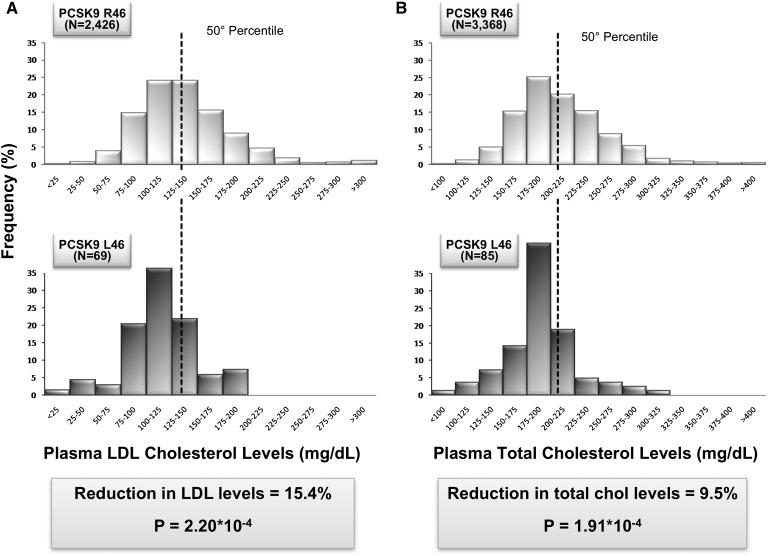
LDL cholesterol levels were significantly lower in L46 carriers (mean: 116.2 ± 34.7 mg/dl) than in noncarriers (mean: 137.4 ± 47.3 mg/dl) (difference = 21.2; P = 2.2×10−4; 95% CI = 9.9–32.4); a similar shift toward lower levels in total cholesterol was also observed (191.7 ± 37.7 in carriers vs. 211.7 ± 49.0 mg/dl in noncarriers; difference = 20; P = 1.91×10−4; 95% CI = 9.5-30.5) (Fig. 1).
Fig. 1.
Distribution of plasma LDL and total cholesterol levels in the ATVB population according to the presence or absence of the L46 allele. A: Distribution of plasma LDL cholesterol levels among 2,426 subjects who did not carry the L46 allele (top) is compared with the distribution of levels among the 69 individuals heterozygous for the L46 allele (bottom). B: Distribution of plasma total cholesterol levels among 3,368 subjects who did not carry the L46 allele (top) is compared with the distribution of levels among the 85 individuals heterozygous for the L46 allele (bottom).
Association analysis of SNP variants in the PCSK9 locus with MI and lipid levels
In order to search for additional variants/haplotypes in the PCSK9 locus that might be associated with premature MI and/or with LDL and total cholesterol levels, we carried out a fine mapping of the region. To this purpose, genotyping data on 23 SNPs covering the PCSK9 gene were extracted from the GWAS performed on the ATVB cohort in the frame of the MIGen project (19).
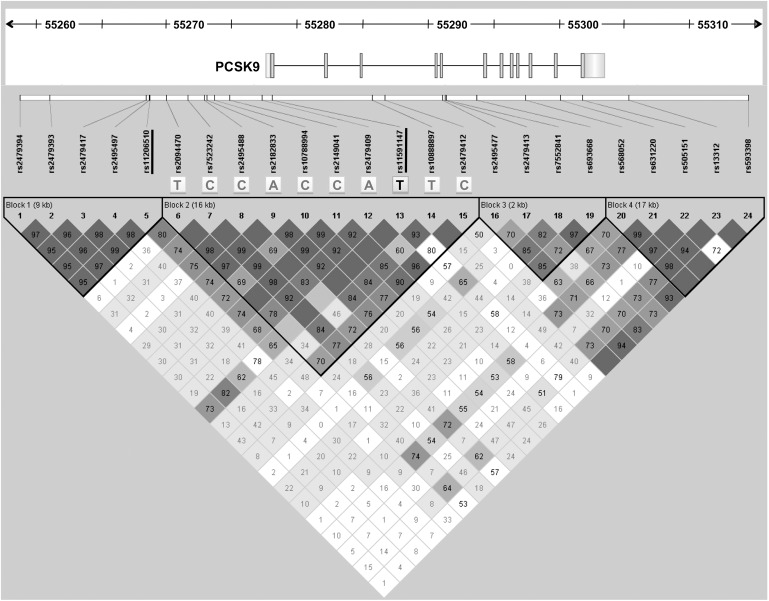
None of the 23 analyzed SNPs was nominally associated with premature MI (Table 3). Nonetheless, haplotype analysis revealed that the R46L variation is located on a unique haplotype (block 2) spanning about 16 kb (Fig. 2),which was slightly associated with premature MI (P = 0.055, ATVB cases vs. ATVB controls).
TABLE 3.
Association of SNPs located in the PCSK9 locus with early-onset MI, and LDL and total cholesterol levels
| Position on chr 1 (bp) | Minor Allele | Minor Allele Frequency (%) |
MI |
LDL Cholesterol |
Total Cholesterol |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Cases | Controls | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | ||
| rs2479394 | 55,258,652 | G | 31.1 | 30.4 | 0.518 | 1.04 (0.93–1.16) | 0.222 | 1.07 (0.96–1.19) | 0.862 | 1.01 (0.92–1.11) |
| rs2479393 | 55,260,957 | A | 30.0 | 28.9 | 0.362 | 1.05 (0.94–1.18) | 0.154 | 1.08 (0.97–1.2) | 0.721 | 1.02 (0.92–1.12) |
| rs2479417 | 55,268,332 | T | 38.0 | 36.7 | 0.227 | 1.07 (0.96–1.19) | 0.345 | 1.05 (0.95–1.16) | 0.488 | 1.03 (0.94–1.13) |
| rs2495497 | 55,268,583 | T | 13.6 | 12.8 | 0.646 | 1.04 (0.89–1.20) | 0.332 | 1.07 (0.93–1.24) | 0.325 | 1.07 (0.94–1.21) |
| rs11206510 | 55,268,627 | C | 18.9 | 20.6 | 0.144 | 0.91 (0.80–1.03) | 1.89E-03 | 0.82 (0.73–0.93) | 8.12E-05 | 0.80 (0.72–0.89) |
| rs2094470 | 55,269,890 | C | 6.5 | 5.9 | 0.182 | 1.15 (0.93–1.42) | 0.680 | 0.96 (0.79–1.17) | 0.603 | 1.05 (0.87–1.26) |
| rs7523242 | 55,271,537 | T | 22.4 | 21.0 | 0.384 | 1.06 (0.93–1.19) | 0.305 | 1.06 (0.94–1.20) | 0.256 | 1.06 (0.96–1.19) |
| rs2495488 | 55,272,794 | T | 5.6 | 5.5 | 0.653 | 1.05 (0.84–1.31) | 0.908 | 0.99 (0.80–1.22) | 0.694 | 1.04 (0.86–1.26) |
| rs2182833 | 55,273,017 | G | 22.8 | 21.7 | 0.457 | 1.05 (0.93–1.18) | 0.465 | 1.04 (0.93–1.18) | 0.310 | 1.06 (0.95–1.18) |
| rs10788994 | 55,273,564 | T | 22.8 | 21.5 | 0.401 | 1.05 (0.93–1.19) | 0.334 | 1.06 (0.94–1.19) | 0.225 | 1.07 (0.96–1.19) |
| rs2149041 | 55,274,725 | G | 22.3 | 21.0 | 0.406 | 1.06 (0.93–1.20) | 0.205 | 1.08 (0.96–1.22) | 0.192 | 1.08 (0.96–1.20) |
| rs2479409 | 55,277,238 | G | 34.2 | 33.3 | 0.442 | 1.04 (0.94–1.16) | 0.133 | 1.08 (0.98–1.20) | 0.025 | 1.11 (1.01–1.22) |
| rs10888897 | 55,277,985 | T | 42.0 | 43.5 | 0.256 | 0.94 (0.85–1.04) | 0.974 | 1.00 (0.91–1.11) | 0.954 | 1.00 (0.92–1.10) |
| rs2479412 | 55,285,649 | G | 4.8 | 4.7 | 0.903 | 0.99 (0.78–1.25) | 0.404 | 1.10 (0.88–1.37) | 0.155 | 1.16 (0.94–1.43) |
| rs2495477 | 55,286,661 | G | 39.4 | 42.2 | 0.105 | 0.92 (0.83–1.02) | 0.446 | 0.96 (0.87–1.07) | 0.121 | 0.93 (0.85–1.02) |
| rs2479413 | 55,291,055 | T | 31.8 | 33.8 | 0.180 | 0.93 (0.83–1.03) | 0.759 | 0.98 (0.88–1.09) | 0.366 | 0.96 (0.87–1.05) |
| rs7552841 | 55,291,270 | T | 40.2 | 39.9 | 0.792 | 0.99 (0.89–1.09) | 0.971 | 1.00 (0.90–1.11) | 0.456 | 1.04 (0.94–1.13) |
| rs693668 | 55,291,340 | G | 35.6 | 37.8 | 0.273 | 0.94 (0.85–1.05) | 0.404 | 0.96 (0.86–1.06) | 0.203 | 0.94 (0.86–1.03) |
| rs568052 | 55,293,697 | G | 29.2 | 30.0 | 0.569 | 0.97 (0.86–1.08) | 0.377 | 0.95 (0.85–1.06) | 0.174 | 0.93 (0.84–1.03) |
| rs631220 | 55,297,430 | A | 15.8 | 16.9 | 0.397 | 0.94 (0.82–1.08) | 0.399 | 0.94 (0.82–1.08) | 0.158 | 0.91 (0.81–1.04) |
| rs505151 | 55,300,067 | G | 3.8 | 3.2 | 0.390 | 1.13 (0.85–1.50) | 0.166 | 1.20 (0.93–1.55) | 0.098 | 1.22 (0.96–1.54) |
| rs13312 | 55,301,775 | G | 17.7 | 19.1 | 0.403 | 0.95 (0.83–1.08) | 0.721 | 0.98 (0.86–1.11) | 0.350 | 0.95 (0.84–1.06) |
| rs593398 | 55,305,330 | C | 3.6 | 3.4 | 0.676 | 1.06 (0.81–1.39) | 0.182 | 1.20 (0.92–1.58) | 0.474 | 1.09 (0.86–1.40) |
P-values are presented as noncorrected for multiple testing. Significant results are indicated in bold.
Fig. 2.
Linkage disequilibrium structure of the PCSK9 locus. The gene structure of PCSK9 is drawn to scale; the gene size is indicated by the ruler at the top of the figure. Pairwise LD values [D'/logarithm of odds (LOD)], estimated for the genotyped SNPs, are represented by boxes: dark gray indicates strong LD, light gray intermediate, whereas white denotes no LD. D' values are shown within the boxes; empty cells are in complete LD. The R46L variant (rs11591147) and the rs11206510 polymorphism located in the promoter region are underlined. The haplotype in block 2 slightly associated with MI is also indicated.
The association between each of the 23 SNPs with residuals of either LDL or total cholesterol showed that the minor allele of the rs11206510 SNP is significantly associated with lower concentration of both LDL (OR = 0.82, 95% CI = 0.73-0.93, P = 1.89 × 10−3) and total cholesterol (OR = 0.80, 95% CI = 0.72–0.89, P = 8.12 × 10−5) (Table 3). Each copy of the minor C allele (frequency: 19.77% in the entire cohort) decreased LDL cholesterol concentration by ∼3.5 mg/dl (mean plasma levels: TT 138.4 ± 45.8 mg/dl, TC 133.2 ± 47.6 mg/dl, CC 128.7 ± 38.3 mg/dl), a similar reduction was also observed for total cholesterol (mean plasma levels: TT 213.7 ± 48.3 mg/dl, TC 207.0 ± 49.1 mg/dl, CC 199.8 ± 40.3 mg/dl).
To assess whether this result represented an independent association signal or simply reflected LD with the R46L variant, we calculated LD between the two associated SNPs. Pairwise LD analysis showed only a low degree of LD between the two SNPs (r2 = 0.02, D′=0.66) (Fig. 2). Because the two SNPs were not completely independent, we also tested the association between rs11206510 and LDL and total cholesterol, adding the allelic dosage of R46L as a covariate. In both cases, the association remained statistically significant (OR = 0.85, 95% CI = 0.75–0.96, P = 0.01, and OR = 0.82, 95% CI = 0.73–0.92, P = 4.9 × 10−4 for LDL and total cholesterol, respectively).
DISCUSSION
Several PCSK9 loss-of-function mutations associated with low LDL cholesterol plasma levels have been described, but only few studies have investigated the association between variations in this gene and the risk of CAD. Besides the seminal paper by Cohen et al. (15), to date only three studies have positively replicated the association between PCSK9 and cardiovascular risk (16, 17, 25). Moreover, a very recent work by Benn et al. (18) showed a significant association between the L46 allele and a reduction in risk of both ischemic heart disease and MI.
The primary aim of this work was to replicate the association between the R46L variation and the risk of premature MI in the Italian population. The distribution of plasma LDL and total cholesterol levels between wild-type and R46L heterozygous subjects showed a significant association of the L46 allele with lower concentration of both LDL and total cholesterol. Strikingly, the reduction in LDL and total cholesterol was identical to that reported in the literature (15): 15.4% and 9.5%, respectively, confirming the robustness of this association.
Despite the confirmed importance of R46L in determining cholesterol levels, in this study, the association between this variation and premature MI did not reach statistical significance. Possible explanations could be: insufficient sample size, given the low frequency of the R46L variation in the Italian population, and the mean age of the analyzed cohort. In fact, the effect of plasma LDL cholesterol levels might be more difficult to detect in young people, who have been exposed to low/high LDL cholesterol levels for a shorter period of time (26). This hypothesis was corroborated by the significant association (P = 0.036) observed by comparing the same patient cohort with a supplementary control sample extended to reach a total of 2,861 individuals (newly added individuals having a mean age of 54 years, 15 years higher than the initially analyzed control population). Our results are consistent with previous findings suggesting that the protective effect of LDL-lowering variants in PCSK9 may increase with aging (27, 28). The availability of the genotyping data obtained from the GWAS performed on our cohort in the frame of the MIGen project gave us the opportunity to explore the PCSK9 locus in greater details. The 23 selected SNPs, covering a ∼56 kb region spanning from ∼19 kb upstream to ∼11 kb downstream of the PCSK9 gene (Fig. 2), were tested for association with LDL and total cholesterol and with premature MI. The rs11206510 SNP, located ∼9 kb upstream of the PCSK9 gene, was strongly related to LDL and total cholesterol (reduction of ∼3.5 mg/dl per allele for both traits). This variation was previously found to be genome-wide significantly associated with LDL cholesterol in two large meta-analyses (25, 29). Moreover, a meta-analysis performed on the six MIGen populations (including the ATVB cohort) showed a highly significant association of rs11206510 with MI (19). This SNP shows only a low degree of LD with the R46L functional variation, and its association with LDL and total cholesterol remains statistically significant even after correction for the allelic dosage of R46L. It is therefore conceivable that an additional functional variant in the promoter region (either rs11206510 itself or in LD with it) might influence the expression level of the gene and hence the LDL cholesterol levels. In support of a possible direct effect of the T>C transition located 9,472 nucleotides upstream of the PCSK9 ATG start codon, an in-silico analysis with the Patch 1.0 software querying the TRANSFAC 6.0 database, showed that the C allele causes the disappearance of a putative glucocorticoid responsive element (GAAAGT), which might be involved in the regulation of PCSK9 expression.
Besides rs11206510, the rs505151 SNP is the only PCSK9 variation previously associated with LDL cholesterol and CAD included in the Affymetrix 6.0 array. The minor allele of this nonsynonymous coding SNP, causing the E670G amino acid change, was associated with increased LDL cholesterol levels and with coronary atherosclerosis in the Lipoprotein Coronary Atherosclerosis Study population from Houston (30). This association was not confirmed by subsequent investigations (14, 31); however, a very recent work by Norata et al. (32) reported a positive association between the E670G polymorphism and an increased progression of intima media thickness of the common carotid artery in the Italian population. In our cohort, we did not observe any association of rs505151 either with MI or with LDL or total cholesterol levels.
The 23 analyzed SNPs were also used to construct the LD structure of the PCSK9 region, which showed high concordance with HapMap data. The L46 allele lies within a single haplotype, spanning ∼16 kb and comprising 10 SNPs. Only two of the 85 subjects carrying the R46L variation had a single difference in this haplotype. These data, together with the uneven distribution of the R46L variation in different ethnic groups, suggest that the mutation probably arose in a single common founder. The haplotype encompassing the R46L variation was slightly associated with premature MI (P = 0.055), whereas the association with the single R46L mutation did not reach statistical significance even though a clear trend toward a protective effect of the L46 allele was observed. This difference is mainly due to the greater power provided by haplotype-based association analysis (33).
In conclusion, our study confirms the association of the L46 allele of PCSK9 with decreased LDL and total cholesterol levels and highlights the importance of the length of exposure to higher cholesterol concentrations to reveal the protective effect of the PCSK9 L46 variant on MI.
Footnotes
Abbreviations:
- BMI
- body mass index
- CAD
- coronary artery disease
- CI
- confidence interval
- GWAS
- genome-wide association study
- LD
- linkage disequilibrium
- LDLR
- LDL receptor
- MI
- myocardial infarction
- OR
- odds ratio
- PCSK9
- proprotein-convertase subtilisin-kexin type 9
- SNP
- single nucleotide polymorphism
REFERENCES
- 1.Kuulasmaa K., Tunstall-Pedoe H., Dobson A., Fortmann S., Sans S., Tolonen H., Evans A., Ferrario M., Tuomilehto J. 2000. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA Project populations. Lancet. 355: 675–687. [DOI] [PubMed] [Google Scholar]
- 2.Law M. R., Wald N. J., Rudnicka A. R. 2003. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: systematic review and meta-analysis. BMJ. 326: 1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke R., Emberson J. R., Parish S., Palmer A., Shipley M., Linksted P., Sherliker P., Clark S., Armitage J., Fletcher A., et al. 2007. Cholesterol fractions and apolipoproteins as risk factors for heart disease mortality in older men. Arch. Intern. Med. 167: 1373–1378. [DOI] [PubMed] [Google Scholar]
- 4.Lewington S., Whitlock G., Clarke R., Sherliker P., Emberson J., Halsey J., Qizilbash N., Peto R., Collins R. 2007. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet. 370: 1829–1839. [DOI] [PubMed] [Google Scholar]
- 5.Pollin T. I., Hsueh W. C., Steinle N. I., Snitker S., Shuldiner A. R., Mitchell B. D. 2004. A genome-wide scan of serum lipid levels in the Old Order Amish. Atherosclerosis. 173: 89–96. [DOI] [PubMed] [Google Scholar]
- 6.Pilia G., Chen W. M., Scuteri A., Orru M., Albai G., Dei M., Lai S., Usala G., Lai M., Loi P., et al. 2006. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breslow J. L. 2000. Genetics of lipoprotein abnormalities associated with coronary artery disease susceptibility. Annu. Rev. Genet. 34: 233–254. [DOI] [PubMed] [Google Scholar]
- 8.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., et al. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. [DOI] [PubMed] [Google Scholar]
- 9.Seidah N. G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S. B., Stifani S., Basak A., Prat A., Chretien M. 2003. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 100: 928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seidah N. G., Prat A. 2002. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 38: 79–94. [DOI] [PubMed] [Google Scholar]
- 11.Taylor N. A., van de Ven W. J. M., Creemers J. W. M. 2003. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 17: 1215–1227. [DOI] [PubMed] [Google Scholar]
- 12.Horton J. D., Cohen J. C., Hobbs H. H. 2007. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem. Sci. 32: 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37: 161–165. [DOI] [PubMed] [Google Scholar]
- 14.Kotowski I. K., Pertsemlidis A., Luke A., Cooper R. S., Vega G. L., Cohen J. C., Hobbs H. H. 2006. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 78: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen J. C., Boerwinkle E., Mosley T. H., Jr, Hobbs H. H. 2006. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354: 1264–1272. [DOI] [PubMed] [Google Scholar]
- 16.McPherson R., Kavaslar N. 2007. Statins for primary prevention of coronary artery disease. Lancet. 369: 1078. [DOI] [PubMed] [Google Scholar]
- 17.Kathiresan S; the Myocardial Infarction Genetics Consortium. 2008. A PCSK9 missense variant associated with a reduced risk of early-onset myocardial infarction. N. Engl. J. Med. 358: 2299–2300. [DOI] [PubMed] [Google Scholar]
- 18.Benn M., Nordestgaard B. G., Grande P., Schnohr P., Tybjaerg-Hansen A. 2010. PCSK9 R46L, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses. J. Am. Coll. Cardiol. 55: 2833–2842. [DOI] [PubMed] [Google Scholar]
- 19.Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., et al. 2009. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41: 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atherosclerosis, Thrombosis, and Vascular Biology Italian Study Group. 2003. No evidence of association between prothrombotic gene polymorphisms and the development of acute myocardial infarction at a young age. Circulation. 107: 1117–1122. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett J. C., Fry B., Maller J., Daly M. J. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 21: 263–265. [DOI] [PubMed] [Google Scholar]
- 23.Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. 2002. The structure of haplotype blocks in the human genome. Science. 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- 24.Little J., Higgins J. P., Ioannidis J. P., Moher D., Gagnon F., von Elm E., Khoury M. J., Cohen B., Davey-Smith G., Grimshaw J., et al. 2009. STrengthening the REporting of Genetic Association studies (STREGA) - an extension of the STROBE statement. Eur. J. Clin. Invest. 39: 247–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lloyd-Jones D. M., Wilson P. W., Larson M. G., Leip E., Beiser A., D'Agostino R. B., Cleeman J. L., Levy D. 2003. Lifetime risk of coronary heart disease by cholesterol levels at selected ages. Arch. Intern. Med. 163: 1966–1972. [DOI] [PubMed] [Google Scholar]
- 27.Polisecki E., Peter I., Robertson M., McMahon A. D., Ford I., Packard C., Shepherd J., Jukema J. W., Blauw G. J., Westendorp R. G., et al. PROSPER Study Group 2008. Genetic variation at the PCSK9 locus moderately lowers low-density lipoprotein cholesterol levels, but does not significantly lower vascular disease risk in an elderly population. Atherosclerosis. 200: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C. C., Fornage M., Lloyd-Jones D. M., Wei G. S., Boerwinkle E., Liu K. 2009. Longitudinal association of PCSK9 sequence variations with low-density lipoprotein cholesterol levels: the Coronary Artery Risk Development in Young Adults Study. Circ. Cardiovasc. Genet. 2: 354–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S. N., Ballantyne C. M., Gotto A. M., Jr, Tan Y., Willerson J. T., Marian A. J. 2005. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J. Am. Coll. Cardiol. 45: 1611–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scartezini M., Hubbart C., Whittall R. A., Cooper J. A., Neil A. H., Humphries S. E. 2007. The PCSK9 gene R46L variant is associated with lower plasma lipid levels and cardiovascular risk in healthy UK men. Clin. Sci. (Lond.). 113: 435–441. [DOI] [PubMed] [Google Scholar]
- 32.Norata G. D., Garlaschelli K., Grigore L., Raselli S., Tramontana S., Meneghetti F., Artali R., Noto D., Cefalù A. B., Buccianti G., et al. 2010. Effects of PCSK9 variants on common carotid artery intima media thickness and relation to ApoE alleles. Atherosclerosis. 208: 177–182. [DOI] [PubMed] [Google Scholar]
- 33.Morris R. W., Kaplan N. L. 2002. On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet. Epidemiol. 23: 221–233. [DOI] [PubMed] [Google Scholar]