Abstract
A precise and sensitive method for measuring cellular free and esterified cholesterol is required in order to perform studies of macrophage cholesterol loading, metabolism, storage, and efflux. Until now, the use of an enzymatic cholesterol assay, commonly used for aqueous phase plasma cholesterol assays, has not been optimized for use with solid phase samples such as cells, due to inefficient solubilization of total cholesterol in enzyme compatible solvents. We present an efficient solubilization protocol compatible with an enzymatic cholesterol assay that does not require chemical saponification or chromatographic separation. Another issue with enzyme compatible solvents is the presence of endogenous peroxides that interfere with the enzymatic cholesterol assay. We overcame this obstacle by pretreatment of the reaction solution with the enzyme catalase, which consumed endogenous peroxides resulting in reduced background and increased sensitivity in our method. Finally, we demonstrated that this method for cholesterol quantification in macrophages yields results that are comparable to those measured by stable isotope dilution gas chromatography with mass spectrometry detection. In conclusion, we describe a sensitive, simple, and high-throughput enzymatic method to quantify cholesterol in complex matrices such as cells.
Keywords: cell cholesterol assay, cholesterol solubilization, catalase
Cholesterol accumulation in macrophage foam cells is one of the earliest histological features of atherosclerosis in the arterial intima (1). In order to perform studies of macrophage cholesterol loading, metabolism, storage, and efflux, one requires a precise, accurate, and sensitive method for measuring cellular free and esterified cholesterol levels. The most commonly used assays to quantify cholesterol levels can be separated into two groups: 1) analytical methods such as gas-liquid chromatography or liquid chromatography coupled with flame ionization or mass spectrometry detection and quantification (2–4); and 2) enzymatic assays, which can be colorimetric (5, 6) or fluorometric (7). However, the chromatographic and mass spectrometry methods require specialized equipment and chemical saponification of cholesterol esters; furthermore, these methods are time consuming and require processing samples one at a time. In contrast, the colorimetric and fluorometric enzymatic methods are simple, sensitive, and relatively fast assays that can process many samples at once. The principal of a sensitive fluorometric assay for the measurement of total cholesterol is shown below, with free cholesterol measured by omitting the first esterase step, and cholesterol mass in cholesterol esters calculated as the difference between the total and free cholesterol contents.
The enzymatic method is the principal method used for measuring cholesterol levels in plasma, where cholesterol is solubilized in an aqueous solution through its incorporation in lipoproteins. However, in order to accurately measure the cholesterol content of cells or tissue by this method, one must identify a suitable solvent that allows good recovery of free and esterified cholesterol in solution and that is compatible with the enzymes and substrates. Previously, the use of the enzymatic cholesterol assay for cells and tissues has been limited due to the inadequate quantitative recovery of free cholesterol and cholesterol esters in enzyme compatible solvents; in fact, a methods paper has argued that the enzymatic method cannot be used for this purpose due to poor extraction and solubilization, which led to an underestimation of the cholesterol mass compared with an analytical method (8). Here, we have identified a suitable and simple solvent system that yields excellent cholesterol recovery. Although this solvent initially led to high background in the enzymatic cholesterol assay, we discovered that pretreatment of this solvent with the enzyme catalase consumed endogenous peroxides, thus resulting in our ability to measure cellular cholesterol content by the simple fluorometric enzymatic assay. We demonstrate that this method for cholesterol quantification in macrophages yields results that are comparable to those measured by gas chromatography-mass spectrometry (GC-MS).
MATERIALS AND METHODS
Reagents
[14C]Cholesterol, [3H]cholesterol, and [3H]cholesteryl oleate were purchase from Perkin Elmer. Bovine liver catalase, cholesterol oxidase from Streptomyces sp., cholesterol esterase from Pseudomonas sp., horseradish peroxidase (HRP), deuterated cholesterol [2H 2,2,3,4,4,6], and SylonTM HTP (HMDS+TMCS+Pyridine) were purchased from Sigma-Aldrich. 10-acetyl-3,7-dihydroxyphenoxazine (ADHP) was purchased from AnaSpec; ADHP is also commonly known as Amplex® Red (Invitrogen). Black polystyrene 96-well plates were purchased from Whatman. Human LDL (1.019 < d < 1.063 g/ml, adjusted with KBr) was prepared by ultracentrifugation and acetylated as described previously (9). Cholesterol and cholesteryl oleate standards were prepared by dissolving 4 mg of desiccated products (Sigma-Aldrich) in 1 ml chloroform. The bicinchoninic acid (BCA) protein assay was performed using the BCA assay kit from Pierce.
Verification of cholesterol extraction and recovery
Acetylated LDL (AcLDL) was incubated for 10 min at 37°C in the presence of [3H]cholesterol. The radiolabeled AcLDL at 100 µg/ml was added for 2 days to RAW264.7 mouse macrophages cultured in 12-well dishes. To determine if solvent extraction of the cellular total cholesterol was quantitative, we compared the recovery of [3H] dpm after solvent extraction with the recovery from a whole cell lysate. After two washes with PBS, the cells were extracted by addition of 1 ml hexane:isopropanol (3:2, v:v) directly to the wells. The extracts were transferred to scintillation vials and dried overnight. Cells in other wells were dissolved in 1 ml of 0.2N sodium hydroxide, incubated at 37°C for 3 h, and 200 µL was transferred to a vial and the pH neutralized by the addition of acetic acid. Four milliliters of scintillation fluid were added to the dried cholesterol extract or cell lysate and radioactivity assessed using a LS6500 scintillation counter (Beckman).
Following the extraction of cellular total cholesterol in hexane:isopropanol, the total cholesterol must be resolubilized in a solvent system that is compatible with the assay enzymes and substrates. After testing several solvent systems, we selected isopropanol:Nonidet P-40 (NP40; 9:1, v:v). In order to assess the recovery of total cholesterol in this solvent, known amounts of [14C]cholesterol and [3H]cholesteryl oleate were added to the hexane:isopropanol extract of cholesterol loaded but unlabeled RAW264.7 cells and these were dried down together in microfuge tubes. One milliliter of isopropanol:NP40 was added to the microfuge tubes and vortexed. The free cholesterol and cholesteryl oleate radioactivity recovered in an aliquot of this solvent was determined by scintillation counting and compared with the added radioactivity.
Fluorometric cholesterol assay
Cholesterol loading and extraction.
RAW 264.7 mouse macrophages were plated in 6-well dishes unless otherwise specified and cholesterol loaded by incubation with the specified concentration of AcLDL for 24 h. Unloaded cells were used as a control. Total cell cholesterol was extracted from the cells using 1 ml hexane:isopropanol (3:2, v:v). The extracts were transferred to microfuge tubes and dried. After solvent extraction, any residual solvent remaining on the cells was evaporated at room temperature. Then the protein from the same wells was dissolved by addition of 1.4 ml of 0.2N NaOH. The plate was incubated at 37°C for 3 h then rocked for 5 min at room temperature. The protein lysates were transferred to microfuge tubes and protein concentration was determined using the BCA assay.
Free cholesterol quantification.
Cholesterol standards, samples, and blanks were dissolved in isopropanol:NP40 (9:1, v:v) and treated similarly, using 1 ml to redissolve each well of cells from a 6-well dish. In a black 96-well plate, 10 µL of a 100 U/ml catalase solution was distributed in each well and 40 µL of each sample was mixed followed by 15 min incubation at 37°C in order to eliminate any peroxides present in reagents or samples. Then 150 µL of reagent A (0.1 M potassium phosphate buffer, pH 7.4, 0.25 M NaCl, 5 mM cholic acid, 0.1% Triton X-100, 0.3 U/ml cholesterol oxidase, 1.3 U/ml HRP, and 0.4 mM ADHP) was added and mixed in each well. The plate was incubated at 37°C for an additional 15 min and fluorescence was read at an excitation wavelength of 530 nm and an emission wavelength of 580 nm. The cholesterol mass of the 40 μl aliquot of the unknown samples was determined by linear regression using the fluorescence emission of the blanks and the 40 μl cholesterol standards (20 to 800 ng range). The final cholesterol mass in the 1 ml samples was calculated by multiplying by 25.
Total cholesterol quantification.
The same procedure was followed for total cholesterol except that reagent A was supplemented with 0.67 U/ml cholesterol esterase, yielding a final concentration of 0.5 U/ml.
Cholesterol esters quantification.
The cholesterol mass in cholesterol esters was determined by subtracting the free cholesterol values from the total cholesterol values.
GC-MS
Total cholesterol extraction and cholesterol esters saponification.
Macrophages were scraped from each well of a 6-well plate and were resuspended in 400 µL water and 100 µL of 1 µg/ml deuterated internal cholesterol standard ([2H 2,2,3,4,4]cholesterol) in isopropanol. Two milliliters of hexane:isopropanol (3:2) and 20 μl of acetic acid were then added to the cell suspension. After vortexing and centrifugation, cholesterol and its derivatives present in the organic phase were collected. The aqueous phase was reextracted by the addition of 1ml hexane followed by vortexing and centrifugation. The hexane layer was collected and combined with the previous organic phase. The remaining aqueous phase containing the extracted cells was used to determine protein concentration by the BCA assay. The combined organic extract was divided into two parts. One part was dried under nitrogen for free cholesterol quantification. The other part was dried under nitrogen and cholesterol esters were saponified in 100 µL 0.5 M potassium hydroxide in methanol for 1 h at 37°C followed by adding 100 µL of 1 M hydrochloric acid, 300 µL water, and 1 ml isopropanol:hexane:acetic acid (40:10:1, v:v:v). The total cholesterol was extracted by two successive additions of 1 ml hexane and dried under nitrogen.
Derivatization of cholesterol to TMS derivatives.
50 µL Sylon™ HTP was added to the dried cholesterol preparations and trimethylsilyl (TMS) derivatives were formed in a 1 h incubation at 90°C. Calibration curves were prepared after TMS derivatization of varying cholesterol amounts plus 100 ng of stable deuterium-labeled cholesterol internal standard.
GC-MS.
One microliter of the TMS-derivatized sample or calibrator was injected onto 6890/5973 GC-MS equipped with an automatic liquid sampler (Agilent Technologies) using the positive ion chemical ionization mode with methane as the reagent gas. The source temperature was set at 230°C. The electron energy was 240 eV and the emission current was 300 µA. The cholesterol TMS ethers were separated on a J and W Scientific (Folsom, CA) DB-1 column (20 m, 0.18 mm inner diameter, 0.18 µm film thickness). The injector and the transfer line temperatures were maintained at 250°C. The initial GC oven temperature was set at 230°C and increased 20°C/min to 270°C followed by increases of 4°C/min to 300°C. The total ion mass spectra of TMS derivatives were recorded in the mass range m/z 200 to 500. The GC chromatograms were extracted at m/z = 329 and 335 for cholesterol and [2H 2,2,3,4,4]cholesterol, respectively, and the peak areas were integrated. Then cholesterol content in each sample was calculated by stable isotope dilution analysis.
Combined enzymatic and GC-MS assay
In order to further validate the enzymatic assay, we performed both analyses on the same cell extracts using two internal standards. RAW 264.7 cells were untreated or cholesterol loaded by 24 h incubation with varying amounts of AcLDL as described above. Cells were washed in DMEM twice. One milliliter of hexane:isopropanol (3:2, v:v), 2 nCi of [3H]cholesterol, and 100 ng of deuterated internal cholesterol standard ([2H 2,2,3,4,4]cholesterol) were added to each well. After mixing for 5 min, half of the extract was transferred to a microfuge tube for processing using the enzymatic assay as described above. The remaining half of the extract was transferred to a glass tube for processing using the GC-MS assay as described above. The [3H]cholesterol standard was used to calculate the fraction of the total cellular extract that was input into the enzymatic assay. The deuterated cholesterol standard was used for the GC-MS cholesterol calculations as described above.
RESULTS
Cholesterol extraction and recovery
We first validated that hexane:isopropanol (3:2) yielded quantitative recovery of total [3H]cholesterol from cholesterol loaded RAW264.7 cells. The extracted dpm represented 98.6 ± 2% (N = 4 ± SD) of the total cells lysate control. For the enzymatic cholesterol assay, the dried hexane:isopropanol extract must then be redissolved in a solvent compatible with the enzymes and substrates. Cullen et al. (8) reported that isopropanol, ethanol, or 10% triton X-100, all suitable for enzyme activity, allowed at best 50% free cholesterol recovery and 20% for cholesterol esters. We tested the suitability of isopropanol:NP40 (9:1) for sterol extraction by adding it to a dried cell hexane:isopropanol extract containing known amounts of [14C]cholesterol and [3H]cholesteryl oleate. We observed that the isopropanol:NP40 mixture yielded 102 ± 3% and 96 ± 7% recovery of free cholesterol and cholesterol esters, respectively (N = 4, ± SD).
Enzymatic assay optimization
Catalase.
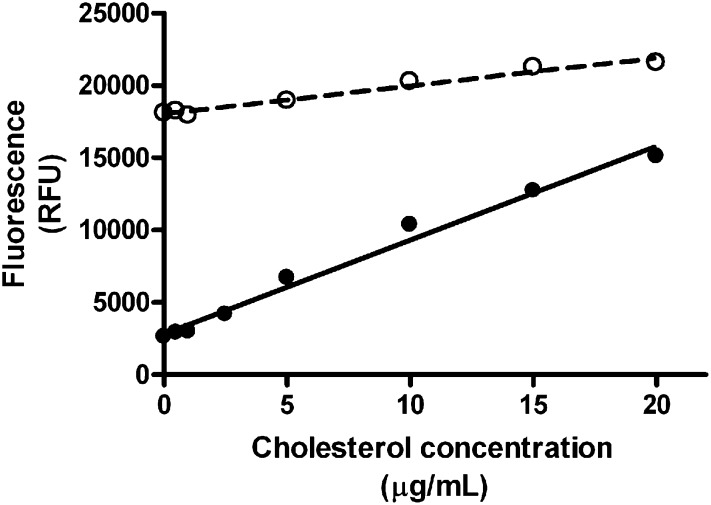
One of the limitations for the use of our enzymatic method resides on the apparent contamination of the isopropanol:NP40 mixture with endogenous peroxides, a common problem with some solvents, resulting in high fluorescence background of sample blanks. In order to counteract this problem, we pretreated the blanks, standards, and samples with a final concentration of 20 U/ml of catalase for 15 min at 37°C before cholesterol quantification. This greatly reduced the background fluorescence by at least 75%. Standards curves were prepared in presence or absence of catalase and the results are shown in Fig. 1. Doubling the amount of catalase to 40 U/ml did not alter the results (data not shown); thus, 20 U/ml was retained as the standard dose. The data showed that catalase treatment not only reduced background but also improved the sensitivity of the assay as shown by the higher slope obtained with this treatment.
Fig. 1.
Standard curves for the determination of cholesterol. Cholesterol standard curves were prepared with (closed circles) or without (open circles) preincubation with catalase 20U/ml.
Sensitivity and reproducibility.
We wanted to determine the lowest number of unloaded cells that our technique was sensitive enough to detect. We measured total cholesterol in different numbers of cells recovered from a T75 flask (104 to 105 cells) and found we could detect both protein (4.88 ± 0.02 µg) and total cholesterol (88 ± 10 ng) in 8 × 104 cells, yielding 18.1 ± 2.0 µg total cholesterol/mg cell protein. To determine the reproducibility of this assay, one unloaded and one AcLDL loaded (100 µg/ml) cell extract were subjected to total and free cholesterol assays using six technical replicates each. The coefficient of variation for the technical replicates of these four assays ranged from 2.2% to 4.4%, thus showing excellent technical reproducibility. In this experiment, the unloaded and AcLDL loaded cells yielded 22.4 and 128.6 µg total cholesterol/mg cell protein, respectively.
Esterase efficiency.
In order to quantify total cholesterol, cholesterol esters need to be hydrolyzed into free cholesterol by cholesterol esterase. Thus, we needed to make sure that the concentration of cholesterol esterase as well as the incubation time were sufficient to achieve the hydrolysis of all cholesterol esters into free cholesterol. Using the cholesteryl oleate standard, we assayed 400 ng cholesterol (corresponding to 672 ng cholesteryl oleate) in the presence of 0.5 U/ml esterase. We obtained 393.5 ± 9.9 ng corresponding to 98.3 ± 2.5% of the theoretical amount; thus, the esterase dose used could efficiently convert up to 672 ng of cholesteryl oleate into free cholesterol in our assay.
Comparison of the enzymatic and the GC-MS assays
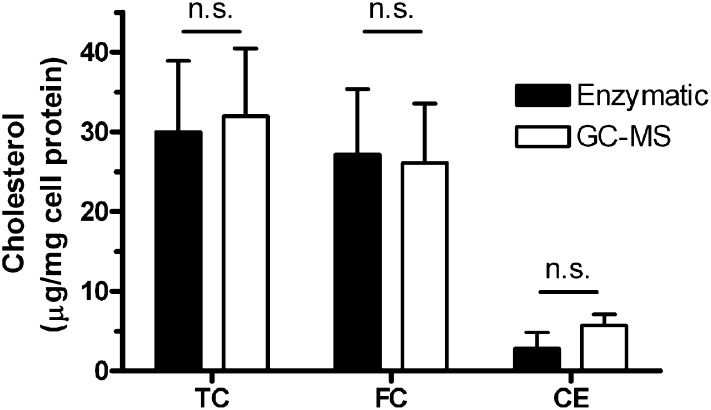
To compare the efficiency and accuracy of the enzymatic assay with the GC-MS assay, we performed three separate experiments with unloaded cells in which duplicate or triplicate wells were assayed for cholesterol by each method. The total cholesterol levels were 30.0 ± 8.9 and 32.0 ± 8.5 µg/mg cell protein for the enzymatic and GC-MS assays, respectively (Fig. 2, not significant). In these unloaded cells, the bulk of the cholesterol was detected as free cholesterol with similar values in both assays (Fig. 2). Although the overall cholesterol esters levels were low in these unloaded cells, they were somewhat higher in the GC-MS assay; however, this difference was not significant (Fig. 2).
Fig. 2.
Cholesterol quantification in cholesterol unloaded RAW 264.7 mouse macrophages. Total cholesterol (TC), free cholesterol (FC), and cholesterol esters (CE) were quantified in unloaded cells using the enzymatic (filled bars) or the GC-MS (open bars) method. Each bar represents the mean ± SD of three separate experiments, each done with duplicate or triplicate wells. No significant differences were observed between the two methods.
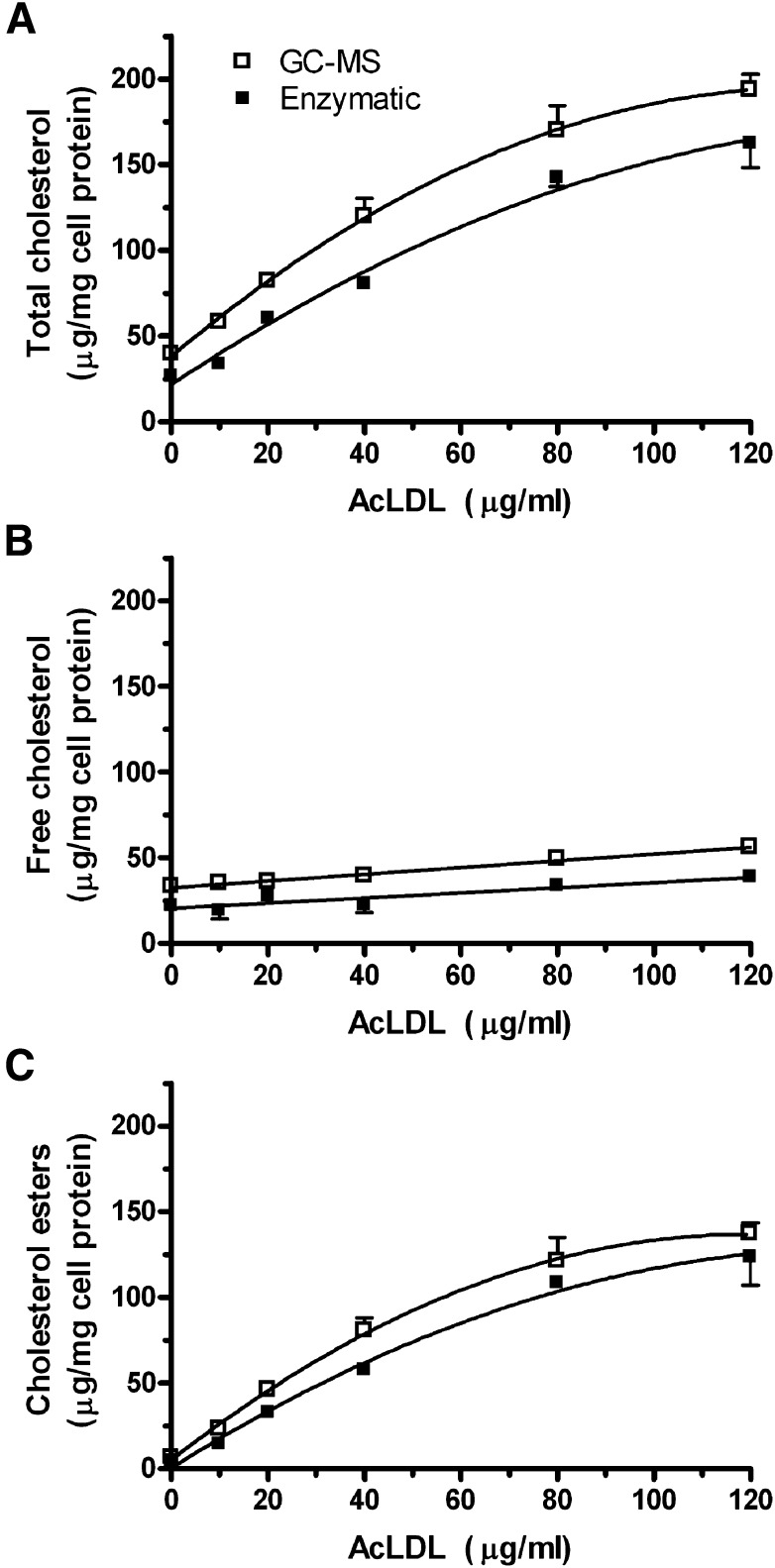
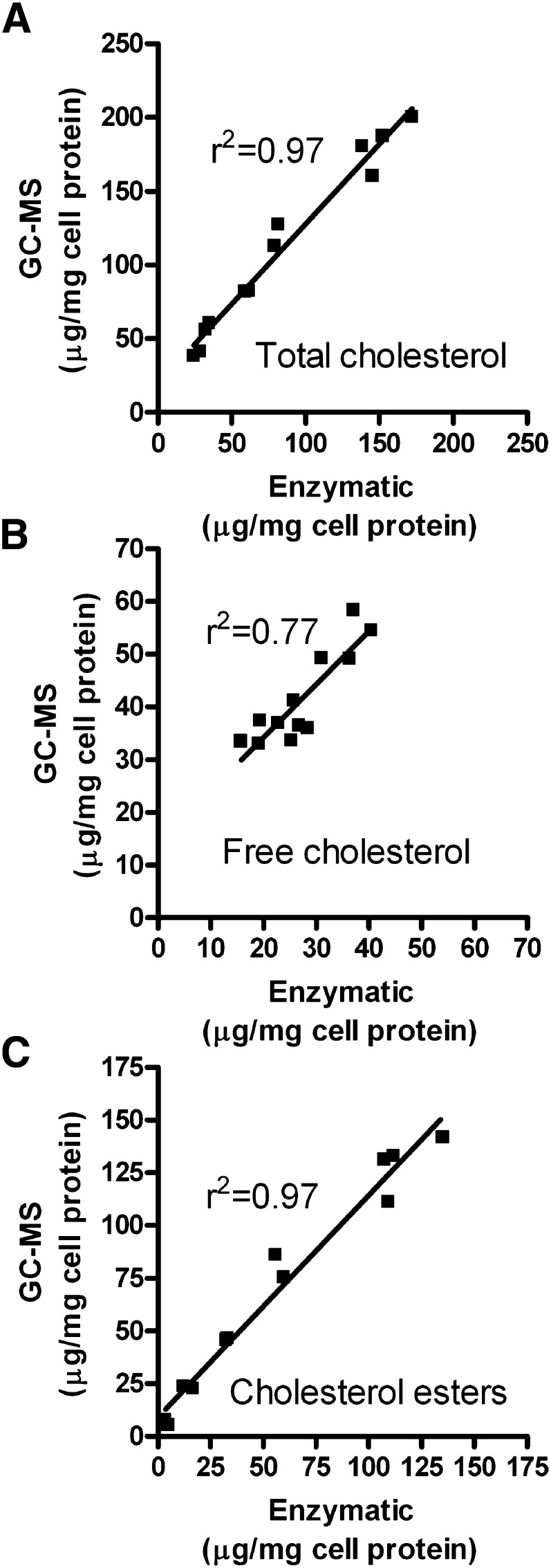
In order to compare the enzymatic and GC-MS assays over a broad range of cholesterol loading, we incubated RAW 264.7 cells with 0, 10, 20, 40, 80, or 120 µg/ml AcLDL in duplicate. Using two internal standards for recovery, as described in the Methods section, each well was assayed for total and free cholesterol by both the enzymatic and GC-MS methods. AcLDL incubations led to dose dependent increases in cellular total cholesterol that are mainly due to increases in cholesterol esters (Fig. 3). In this experiment, the GC-MS assay yielded higher values than the enzymatic assay, with 19% higher total cholesterol levels observed in the cells loaded with 120 µg/ml AcLDL. However, in a separate experiment comparing the two methods using cells loaded with 100 µg/ml AcLDL, we observed the opposite bias, with 8% higher total cholesterol values obtained by the enzymatic assay (193 ± 17 µg total cholesterol/mg cell protein) compared with the GC-MS method (178 ± 5 μg total cholesterol/mg cell protein). In the cells treated with varying doses of AcLDL, we plotted the values of total, free, and esterified cholesterol for each well determined by the enzymatic versus GC-MS methods (Fig. 4). The total cholesterol levels were highly correlated (r2 = 0.97, P < 0.0001), with a slope of 1.08 ± 0.06 (Fig. 4A). The bias in this assay was due to the y-intercept, which was 19.3, indicative of a systematic error. The range of free cholesterol levels did not vary as much but they were still well correlated between the two methods (r2 = 0.77, P = 0.0002, slope = 1.01 ± 0.17, y-intercept = 13.8, Fig. 4B). The cholesterol esters levels were highly correlated between these two methods (r2 = 0.97, P < 0.0001, slope = 1.05 ± 0.06, y-intercept = 9.0, Fig. 4C).
Fig. 3.
Comparison of the enzymatic and GC-MS methods at varying levels of cholesterol loading in RAW 264.7 mouse macrophages. Cells were incubated with increasing concentrations of AcLDL and their total cholesterol (A), free cholesterol (B), and cholesterol esters (C) levels were measured by both the enzymatic (filled squares) and the GC-MS (open squares) methods, using the dual internal standard strategy described in the Methods section. Each symbol represents the mean ± SD of two wells.
Fig. 4.
Correlation analysis of cholesterol values obtained by the enzymatic and GC-MS methods. The data plotted are derived from the 12 independent wells of cells that contain a range of cholesterol levels as described in the legend of Fig. 3. A: Total cholesterol levels (r2 = 0.97, slope = 1.08 ± 0.06; y-intercept = 19.3). B: Free cholesterol levels (r2 = 0.77, slope = 1.01 ± 0.17; y-intercept = 13.8). C: Cholesterol esters levels (r2 = 0.97, slope = 1.05 ± 0.06; y-intercept = 9.0).
DISCUSSION
In this report, we describe a sensitive and easy method to quantify total and free cholesterol content in cultured cells, which does not require chemical saponification or specialized equipment other than a fluorescence plate reader. We demonstrated efficient extraction of cellular sterols and good recovery by redissolving in an isopropanol:NP40 solution that is compatible with the enzymatic assay. We also found that pretreatment of the samples and standards with catalase to remove endogenous peroxides in the isopropanol:NP40 solution was beneficial in reducing the background and increasing sensitivity. We determined that there was no need to specifically inactivate the catalase before the enzymatic cholesterol quantification, which is dependent upon hydrogen peroxide generation. A prior study also used catalase treatment before measurement of cholesterol esters by an enzymatic assay, and these investigators also did not need to inactivate the catalase (10). The reason that catalase does not need to be inactivated is most likely due to catalase's instability at low concentrations at 37°C (11, 12), and the >1000-fold higher affinity of HRP versus catalase for hydrogen peroxide (13, 14).
To evaluate the accuracy of the enzymatic method, we compared it with stable isotope dilution GC-MS, the gold-standard method for free cholesterol and cholesterol ester quantification. On the whole, the results of the high-throughput enzymatic assay were highly correlated with the GC-MS assay. However, in specific assays we could observe one method yielding higher cholesterol levels than the other method. In three separate experiments with unloaded cells, the two methods on average yielded comparable results (Fig. 2), demonstrating that the bias did not consistently affect one assay over the other. In two separate experiments with cholesterol loaded cells, one yielded higher levels for the enzymatic assay, whereas the other yielded higher levels for the GC-MS assay. In the experiment where varying amounts of AcLDL were used to yield a range of cholesterol loading, our analysis showed that the two methods were highly correlated in detecting total, free, and esterified cholesterol (Fig. 4). However, in this study the GC-MS method yielded higher values than the enzymatic method, which was reflected in the y-intercept having values >0, whereas the slope was still ∼1. Thus, both methods could comparably detect increasing amounts of total and free cholesterol. We speculate that the y-intercept difference may be due to a systematic error introduced by use of different cholesterol standards, different operators using different pipettors, and different types of calculations used to determine the cholesterol mass. For example, the enzymatic assay uses a cholesterol standard curve in the linear range to calculate the cholesterol mass in unknown samples. However, the GC-MS results are calculated using an internal deuterated cholesterol standard in each sample that is present at <1% of the sample cholesterol level. Thus, a small variation in the integration of the internal standard peak could lead to a systematic variation in the calculated cholesterol content.
In conclusion, we described a sensitive, simple, and rapid method to quantify cholesterol in cultured macrophages. This method allowed the detection of cholesterol in 80,000 cells prior to cholesterol loading. Combined with the use of 96-well plates, this assay can process a large number of samples per day, which may further be augmented through assay automation. This assay may be widely applicable to all types of cells and tissues after determination that the isopropanol:NP40 mixture fully dissolves all cholesterol in the solvent extract.
Footnotes
Abbreviations:
- AcLDL
- acetylated low density lipoprotein
- ADHP
- 10-acetyl-3,7-dihydroxyphenoxazine
- BCA
- bicinchoninic acid
- NP40
- nonidet P-40
This work was supported by National Institutes of Health grants R01 HL066082 and P01 HL098055 to J.D.S and P01 HL076491 and P01 HL098055 to S.L.H. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Glass C. K., Witztum J. L. 2001. Atherosclerosis. The road ahead. Cell. 104: 503–516. [DOI] [PubMed] [Google Scholar]
- 2.Klansek J. J., Yancey P., St Clair R. W., Fischer R. T., Johnson W. J., Glick J. M. 1995. Cholesterol quantitation by GLC: artifactual formation of short-chain steryl esters. J. Lipid Res. 36: 2261–2266. [PubMed] [Google Scholar]
- 3.Cullen P., Fobker M., Tegelkamp K., Meyer K., Kannenberg F., Cignarella A., Benninghoven A., Assmann G. 1997. An improved method for quantification of cholesterol and cholesteryl esters in human monocyte-derived macrophages by high performance liquid chromatography with identification of unassigned cholesteryl ester species by means of secondary ion mass spectrometry. J. Lipid Res. 38: 401–409. [PubMed] [Google Scholar]
- 4.Paik M. J., Yu J., Hu M. B., Kim S. J., Kim K. R., Ahn Y. H., Choi S., Lee G. 2008. Gas chromatographic-mass spectrometric analyses of cholesterol and its precursors in rat plasma as tert-butyldimethylsilyl derivatives. Clin. Chim. Acta. 396: 62–65. [DOI] [PubMed] [Google Scholar]
- 5.Gamble W., Vaughan M., Kruth H. S., Avigan J. 1978. Procedure for determination of free and total cholesterol in micro- or nanogram amounts suitable for studies with cultured cells. J. Lipid Res. 19: 1068–1070. [PubMed] [Google Scholar]
- 6.Heider J. G., Boyett R. L. 1978. The picomole determination of free and total cholesterol in cells in culture. J. Lipid Res. 19: 514–518. [PubMed] [Google Scholar]
- 7.Amundson D. M., Zhou M. 1999. Fluorometric method for the enzymatic determination of cholesterol. J. Biochem. Biophys. Methods. 38: 43–52. [DOI] [PubMed] [Google Scholar]
- 8.Cullen P., Tegelkamp K., Fobker M., Kannenberg F., Assmann G. 1997. Measuring cholesterol in macrophages: comparison of high-performance liquid chromatography and gas-liquid chromatography with enzymatic fluorometry. Anal. Biochem. 251: 39–44. [DOI] [PubMed] [Google Scholar]
- 9.Basu S. K., Goldstein J. L., Anderson G. W., Brown M. S. 1976. Degradation of cationized low density lipoprotein and regulation of cholesterol metabolism in homozygous familial hypercholesterolemia fibroblasts. Proc. Natl. Acad. Sci. USA. 73: 3178–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizoguchi T., Edano T., Koshi T. 2004. A method of direct measurement for the enzymatic determination of cholesteryl esters. J. Lipid Res. 45: 396–401. [DOI] [PubMed] [Google Scholar]
- 11.Aebi H., Suter H., Feinstein R. N. 1968. Activity and stability of catalase in blood and tissues of normal and acatalasemic mice. Biochem. Genet. 2: 245–251. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimoto M., Sakamoto H., Shirakami H. 2009. Covalent conjugation of tetrameric bovine liver catalase to liposome membranes for stabilization of the enzyme tertiary and quaternary structures. Colloids Surf. B Biointerfaces. 69: 281–287. [DOI] [PubMed] [Google Scholar]
- 13.Li J., Xiao L. T., Liu X. M., Zeng G. M., Huang G. H., Shen G. L., Yu R. Q. 2003. Amperometric biosensor with HRP immobilized on a sandwiched nano-Au / polymerized m-phenylenediamine film and ferrocene mediator. Anal. Bioanal. Chem. 376: 902–907. [DOI] [PubMed] [Google Scholar]
- 14.Nicholls P., Loewen P., Fita I. 2001. Enzymology and structure of catalases. Heme-Fe Proteins, Sykes A. G., Mauk G., Academic Press, San Diego, CA: 52–106. [Google Scholar]