Abstract
Background
Association of genetic-variants in the FADS1-FADS2-gene-cluster with fatty-acid-composition in blood of adult-populations is well established. We analyze this genetic-association in two children-cohort-studies. In addition, the association between variants in the FADS-gene-cluster and blood-fatty-acid-composition with eczema was studied.
Methods and Principal Findings
Data of two population-based-birth-cohorts in the Netherlands and Germany (KOALA, LISA) were pooled (n = 879) and analyzed by (logistic) regression regarding the mutual influence of single-nucleotide-polymorphisms (SNPs) in the FADS-gene-cluster (rs174545, rs174546, rs174556, rs174561, rs3834458), on polyunsaturated fatty acids (PUFA) in blood and parent-reported eczema until the age of 2 years. All SNPs were highly significantly associated with all PUFAs except for alpha-linolenic-acid and eicosapentaenoic-acid, also after correction for multiple-testing. All tested SNPs showed associations with eczema in the LISA-study, but not in the KOALA-study. None of the PUFAs was significantly associated with eczema neither in the pooled nor in the analyses stratified by study-cohort.
Conclusions and Significance
PUFA-composition in young children's blood is under strong control of the FADS-gene-cluster. Inconsistent results were found for a link between these genetic-variants with eczema. PUFA in blood was not associated with eczema. Thus the hypothesis of an inflammatory-link between PUFA and eczema by the metabolic-pathway of LC-PUFAs as precursors for inflammatory prostaglandins and leukotrienes could not be confirmed by these data.
Introduction
It has been well established that genetic variants in the fatty acid desaturase genes (FADS1 and FADS2) associate with the fatty acid composition in adult populations [1]. Several studies have shown that the Δ-5 and the Δ-6 desaturase enzymes are involved in fatty acid metabolism in adults and that these enzymes are genetically regulated by variants of the FADS1 and FADS2 genes, respectively [2]–[8].
Empirical and theoretical evidence exist that fatty acid metabolism may be involved in atopic eczema [9]–[14]. Some studies found, that dietary intake of certain fatty acids can contribute to the development of allergic diseases. Kompauer et al showed a positive association between hay fever and arachidonic acid (AA) intake, and allergic sensitisation and oleic acid intake in German adults [15].
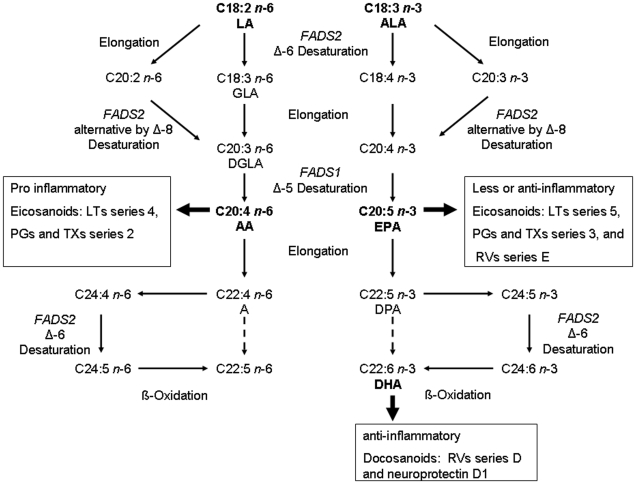
Long chain polyunsaturated fatty acids (LC-PUFA) can influence inflammatory responses, as they are precursors of eicosanoids and docosanoids [16]. The link between PUFA and inflammatory processes are the eicosanoids with arachidonic acid (AA) as their main precursor. Eicosanoids derived from AA (e.g. leukotrienes LTB4, prostaglandins PGE2, PGI2 or thromboxanes TXA2) have mainly pro-inflammatory effects [16]. According to findings of several studies a defect in enzyme activity of Δ-6 desaturase, encoded by the FADS2 gene, leads to enhanced blood levels of the n-6 and n-3 parent fatty acids linoleic (LA) and alpha-linolenic acid (ALA), respectively, and decreased levels of AA and eicosapentaenoic acid (EPA), whereas docosahexaenoic acid (DHA) levels are not influenced [5], [6]. Schaeffer et al. showed that the variability of serum fatty acid levels explained by genetic variants in the FADS1 FADS2 gene cluster is highest for AA with 28% [6]. Thus, variants in the FADS1 FADS2 gene cluster may be indirectly associated with inflammatory processes via their influence on endogenous LC-PUFA production, particularly AA production. Figure 1 show these metabolic pathways of n-6 and n-3 fatty acids and pathways of production of pro-inflammatory and less inflammatory eicosanoids and anti-inflammatory docosanoids schematically [12].
Figure 1. Metabolic pathways of n-6 and and n-3 fatty acids.
Dashed arrows indicate pathways absent in human mammals. Figure is based on modified figures form Duchen et al. Glaser et al., and Park et al. [11], [12], [40]. LA = linoleic acid, GLA = gamma linolenic acid, DGLA = Dihomo-gamma-linolenic acid, AA = arachidonoc acid, A = adrenic acid, ALA = alpha-limolenic acid, EPA, = eicospentaenoic acid, DPA = docosapentaenoic acid, DHA = docosahexaenoic acid; LTs = leukotrienes, PGs = prostaglandins, TXs = thromboxanes, RVs = Resolvins.
No study has empirically confirmed the genetic link between the variants in the FADS1 FADS2 gene cluster with polyunsaturated fatty acids (PUFA) in children. Schaeffer et al found an association of rarer FADS haplotypes with a reduced eczema risk in adults [6], but so far, no study has been published relating both genotypes and fatty acids to eczema development in children. Therefore, we analyzed this genetic association in a children cohort study pooled from two population based birth cohorts and investigate the association between variants in the FADS gene cluster, blood fatty acid composition and eczema up to the age of two years.
Methods
Ethics statement
Approval by the respective local Ethics Committees (Maastricht University/University Hospital of Maastricht, Bavarian General Medical Council) and written informed consent from participants' families (parents) were obtained in both KOALA and LISA studies.
Study design and population
The KOALA Birth Cohort Study (“Kind, Ouders en gezondheid: Aandacht voor Leefstijl en Aanleg”) is a prospective cohort study of 2834 mother-infant pairs in the Netherlands. Aim of the KOALA study was to investigate factors that influence the clinical expression of atopic disease with a main focus on lifestyle factors (e.g. anthroposophy, dietary habits, breastfeeding, intestinal micro flora, and gene-environment-interactions). Enrollment started in October 2000. Details of the study design have been described elsewhere [17], [18].
The LISA study (“Influences of Lifestyle related Factors on the Immune System and the Development of Allergies in Childhood”) is an ongoing population-based birth cohort study of unselected 3097 newborns. The study was designed to assess ‘Influences of Lifestyle related Factors on the Immune System and the Development of Allergies in Childhood’. Between November 1997 and January 1999, n = 3097 healthy full-term newborns (gestational age≥37 weeks) were recruited from 14 obstetrical clinics in Munich (n = 1467), Leipzig (n = 976), Wesel (n = 306), and Bad Honnef (n = 348). Details on study design are published elsewhere [19], [20].
The studied analysis population with information on genotypes, fatty acids and eczema at 2 years of age comprised 546 children in the KOALA birth cohort study and 333 in the LISA birth cohort study.
For details on both studies (see Supporting Information Appendix S1).
Fatty acid analysis
In the KOALA study blood was collected in EDTA-tubes during a home visit to the child around age 2 years, by trained nurses according to a standardized protocol. The EDTA-plasma was used for the analysis of the fatty acid status and was, after centrifugation, stored in cryovials at −80°C. The EDTA-plasma was deproteinated and the precipitate was removed by centrifugation. Then the sample was applied to an aminopropyl-solid-phase column to selectively elute the phospholipid fraction [21], [22].
In the LISA study venous blood samples were collected in serum-separator-tubes. Samples were centrifuged and serum was frozen in plastic vials and stored at −80°C until analysis. The analysis of plasma-glycerophospholipids composition was performed by a sensitive and precise high-throughput method as described recently [23].
For details (see Supporting Information Appendix S2).
Genotyping
In KOALA, genomic DNA was extracted from buccal swabs using standard methods as described previously [24]. In LISA, genomic DNA was extracted from EDTA blood.
Five variants of the FADS1 FADS2 gene cluster (rs174545, rs174546, rs174556, rs174561, rs3834458) were typed with MALDI-TOF-MS.
For details (see Supporting Information Appendix S3).
Determination of infants' IgE
In the KOALA study total and specific IgE were determined at age 1 (against hen's eggs, cow's milk, and peanuts) and 2 years (against eggs, cow's milk, peanut, birch pollen, grass pollen, cat, dog and house-dust mite) by blood sampling during home visits [25]–[29].
In the LISA study blood samples were collected during physical examination of the infant at age 2 years and analysed for total and specific IgE using the RAST-CAP-FEIA-system (Pharmacia, Freiburg, Germany) as previously described [30]. For details (see Supporting Information Appendix S4).
Definition of outcome variable parental reported eczema
In both the KOALA and LISA study the definition of “parental reported eczema” is based on the questionnaire-reported occurrence of “itchy rash that was coming and going” at any time within the first two years of life [18], [31].
For details (see Supporting Information Appendix S5).
Statistical analysis
Allele frequencies, Fisher's exact test of Hardy-Weinberg-Equilibrium (HWE), the linkage disequilibrium (LD) tests Lewontin's D' and pairwise-squared correlations r2 were calculated for the study population and for both studies separately.
Single SNP linear regression analyses for the relation between FADS variants and the nine continuous outcome variables (fatty acids) were conducted applying an additive coded model. P-Values were corrected for multiple testing by Bonferroni correction.
Logistic regression was applied to evaluate the effect of each single SNP and of each fatty acid separately on the dichotomous coded outcome “parental reported eczema during the first two years of life”. No corrections for multiple testing were applied in logistic regression.
All analyses were performed using the statistical software SAS, version 9.1.3 [32], [33], except calculation of linkage disequilibrium measures (JLIN) [34].
For details (see Supporting Information Appendix S6).
Results
Study characteristics for both study populations separately and combined are listed in Table 1 .
Table 1. Characteristics of study population.
| KOALA-Study | LISA-Study | Total | |||||||
| n | mean or % | 95%-CI | n | mean or % | 95%-CI | n | mean or % | 95%-CI | |
| % boys | 546 | 50.4 | (46.2–54.6) | 333 | 55.6 | (50.2–60.9) | 879 | 52.3 | (49.0–55.6) |
| % high maternal education | 543 | 58.9 | (54.8–63.1) | 330 | 66.7 | (61.6–71.8) | 873 | 61.9 | (58.6–65.1) |
| % parental asthma or allergy | 546 | 57.0 | (52.8–61.1) | 332 | 68.7 | (63.7–73.7) | 878 | 61.4 | (58.2–64.6) |
| %maternal smoking during pregnancy | 546 | 3.1 | (1.7–4.6) | 330 | 10.3 | (7.0–13.6) | 876 | 5.8 | (4.3–7.4) |
| % exclusively breastfed for at least 3 months | 538 | 50.0 | (45.8–54.2) | 331 | 71.6 | (66.7–76.5) | 869 | 58.2 | (54.9–61.5) |
| fatty acids (mean % wt of total FA) | |||||||||
| Linoleic (LA) | 546 | 21.37 | (21.14–21.60) | 333 | 22.22 | (21.92–22.52) | 879 | 21.69 | (21.51–21.88) |
| γ-Linolenic (GLA) | 542 | 0.07 | (0.07–0.08) | 333 | 0.11 | (0.10–0.11) | 875 | 0.09 | (0.08–0.09) |
| ln (GLA)a | 542 | −2.93 | (−3.00–−2.85) | 333 | −2.33 | (−2.37–−2.29) | 875 | −2.70 | (−2.75–−2.65) |
| Dihomo-γ-linolenic (DGLA) | 546 | 2.82 | (2.77–2.87) | 333 | 2.91 | (2.84–2.97) | 879 | 2.85 | (2.81–2.89) |
| Arachidonic (AA) | 546 | 9.06 | (8.95–9.16) | 333 | 9.33 | (9.19–9.47) | 879 | 9.16 | (9.08–9.25) |
| Adrenic (A) | 545 | 0.47 | (0.46–0.48) | 333 | 0.40 | (0.39–0.41) | 878 | 0.44 | (0.44–0.45) |
| α-Linolenic (ALA) | 544 | 0.21 | (0.21–0.22) | 333 | 0.24 | (0.23–0.25) | 877 | 0.22 | (0.22–0.23) |
| ln (ALA)a | 544 | −1.61 | (−1.64–−1.58) | 333 | −1.49 | (−1.52–−1.45) | 877 | −1.56 | (−1.59–−1.54) |
| Eicosapentaenoic (EPA) | 546 | 1.00 | (0.98–1.03) | 333 | 0.55 | (0.52–0.58) | 879 | 0.83 | (0.81–0.86) |
| ln (EPA)a | 546 | −0.05 | (−0.09–−0.02) | 333 | −0.68 | (−0.72–−0.63) | 879 | −0.29 | (−0.32–−0.26) |
| n-3 Docosapentaenoic (DPA) | 546 | 0.92 | (0.91–0.94) | 333 | 0.97 | (0.95–0.99) | 879 | 0.94 | (0.93–0.95) |
| Docosahexaenoic (DHA) | 546 | 2.67 | (2.61–2.74) | 333 | 3.07 | (2.99–3.15) | 879 | 2.82 | (2.77–2.88) |
| ln (DHA)a | 546 | 0.95 | (0.93–0.97) | 333 | 1.09 | (1.07–1.12) | 879 | 1.00 | (0.99–1.02) |
| % parental reported eczema of child in first 2 years of life | 542 | 30.6 | (26.7–34.5) | 333 | 14.1 | (10.4–17.9) | 875 | 24.3 | (21.5–27.2) |
| IgE at 2 years of life | |||||||||
| mean total IgE (IU/ml) | 544 | 59.3 | (34.9–83.7) | 327 | 56.3 | (44.3–68.3) | 871 | 58.2 | (42.3–74.0) |
| % specifc IgE (egg) | 544 | 5.5 | (3.6–7.4) | 333 | 4.8 | (2.5–7.1) | 877 | 5.2 | (3.8–6.7) |
| % specifc IgE (milk) | 544 | 16.9 | (13.8–20.1) | 333 | 4.2 | (2.0–6.4) | 877 | 12.1 | (9.9–14.2) |
| % specifc IgE (peanut) | 544 | 5.0 | (3.1–6.8) | 333 | 1.2 | (0.0–2.4) | 877 | 3.5 | (2.3–4.8) |
| % specifc IgE (birch)b | 535 | 0.4 | (−0.1–0.9) | - | - | - | - | - | - |
| % specifc IgE (gras)b | 538 | 1.9 | (0.7–3.0) | - | - | - | - | - | - |
| % specifc IgE (cat) | 543 | 2.8 | (1.4–4.1) | 333 | 0.6 | (−0.2–1.4) | 876 | 1.9 | (1.0–2.9) |
| % specifc IgE (dog)c | 543 | 2.6 | (1.2–3.9) | - | - | - | - | - | - |
| % specifc IgE (dust mite) | 544 | 6.8 | (4.7–8.9) | 333 | 0.6 | (−0.2–1.4) | 877 | 4.7 | (3.3–6.1) |
Mean levels of GLA, ALA, EPA and DHA are naturally logged means to account for the severely skewed distributions of these fatty acids; see methods section.
In the LISA study % specific IgE for sensitization to birch and gras was not determined separately but for a mix of grass, trees and herbs only (0.9% (95%-CI −0.1–1.9)) at age 2.
In the LISA study % specific IgE for sensitization to dog was not determined at age 2.
There are some differences between both studies in the percentages of boys, high maternal education, family history of asthma or allergy, proportion of maternal smoking during pregnancy and exclusive breastfeeding. Mean percentage contributions of fatty acids to plasma phospholipids in the KOALA cohort and to plasma glycerophospholipids in the LISA cohort are similar, except for an almost twofold higher percentage value of EPA in the KOALA study, which might be related to the different population studied with a potentially higher fish consumption in the Netherlands, as well as the different analytical method used.
Information regarding position, possible functional region and genotyping frequencies for the five analyzed SNPs of the FADS1 FADS2 gene cluster (see Supporting Information Table S1). Minimum P-value of Fisher's exact test for violation of Hardy-Weinberg-Equilibrium (HWE) for any of the five SNPs was 0.48 (rs174546) in the whole study population and 0.62 and 0.61 for the KOALA and LISA study populations separately.
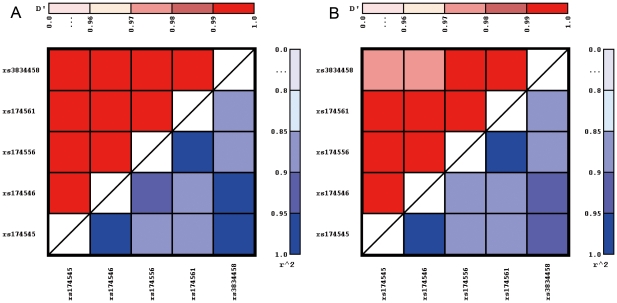
Lewontin's D' and pairwise-squared correlations r2 for the KOALA study and for the LISA study are depicted in Figure 2 . In the KOALA study Lewontin's D' ranged between 0.99 and 1.0 and the pairwise-squared correlations r2 ranged between 0.89 and 0.99. For the LISA study D' ranged between 0.97 and 1.0 and r2 between 0.87 and 1.0. Both measures confirm that all five SNPs are in high linkage disequilibrium (LD).
Figure 2. Pairwise linkage disequilibrium measured by Lewontin's D' and r2 for the common five single nucleotide polymorphisms (SNP) in the KOALA (Panel A) and the LISA (Panel B) birth cohort studies.
Single SNP associations with fatty acids
Mean levels and standard deviations for each fatty acid by genotype of each SNP (rs174545, rs174546, rs174556, rs174561, rs3834458) for the combined study population and for the KOALA and LISA studies separately are listed in in the Online Repository (see Supporting Information Table S2, Table S3 and Table S4). Note that mean levels of GLA, ALA, EPA and DHA are naturally logged means to account for the severely skewed distributions of these fatty acids.
Single SNP regression analyses of each additive coded SNP (0/1/2) with each fatty acid in the combined study population of the KOALA and LISA studies at age 2 years revealed that all SNPs are highly significantly associated with all fatty acids except for ALA and EPA (natural log scale, Table 2 ). This is true even though all P-values have been conservatively corrected for multiple testing according to Bonferroni's method. With respect to ALA, corrected significance at the 5% level was only reached for rs174545 and rs3834458 but not for the other SNPs. Regarding EPA, regression coefficients were in the same direction and the same order of magnitude as for DPA and DHA, but corrected P-values were only between 0.05 and 0.09. Separate analyses for both study populations revealed that in the LISA study associations of the SNPs were significant only with LA, GLA, DGLA, AA and EPA after correction for multiple testing (see Supporting Information Table S5 and Table S6). In the KOALA study all SNPs showed a significant association with all n-6 and n-3 PUFAs except EPA. However, regression coefficients of SNPs were in the same direction for all PUFAs so the lack of significance regarding n-3 PUFA in the LISA study may just reflect a loss of power due to the smaller sample size of the LISA study compared to that of the KOALA study.
Table 2. Associations of the five analyzed variants in the FADS1 FADS 2 gene region with fatty acids.
| Fatty acid | LA | GLA | DGLA | AA | A | ALA | EPA | DPA | DHA | |
| SNP | C18_2n_6 | ln(C18_3n_6) | C20_3n_6 | C20_4n_6 | C22_4n_6 | ln(C18_3n_3) | ln(C20_5n_3) | C22_5n_3 | ln(C22_6n_3) | |
| rs174545 | ||||||||||
| P-value (corrected) | 4.1E-10 | 1.3E-04 | 6.5E-13 | 5.4E-38 | 0.003 | 0.039 | 0.071 | 1.7E-05 | 3.2E-04 | |
| ß-coefficient | 0.964 | −0.191 | 0.224 | −0.818 | −0.021 | 0.057 | −0.080 | −0.046 | −0.059 | |
| R2 (explained var.) | 5.3% | 2.5% | 6.7% | 18.3% | 1.8% | 2.5% | 1.2% | 3.0% | 3.3% | |
| rs174546 | ||||||||||
| P-value (corrected) | 3.7E-10 | 7.9E-05 | 1.4E-12 | 2.9E-39 | 0.002 | 0.066 | 0.066 | 5.9E-06 | 1.5E-04 | |
| ß-coefficient | 0.962 | −0.194 | 0.221 | −0.828 | −0.022 | 0.055 | −0.080 | −0.048 | −0.061 | |
| R2 (explained var.) | 5.3% | 2.6% | 6.5% | 18.8% | 2.0% | 2.6% | 1.2% | 3.2% | 2.5% | |
| rs174556 | ||||||||||
| P-value (corrected) | 1.1E-09 | 0.002 | 2.3E-13 | 3.9E-37 | 0.009 | 0.218 | 0.054 | 1.4E-06 | 6.3E-05 | |
| ß-coefficient | 0.975 | −0.170 | 0.235 | −0.836 | −0.021 | 0.050 | −0.085 | −0.052 | −0.065 | |
| R2 (explained var.) | 5.1% | 1.9% | 6.7% | 17.9% | 1.6% | 1.9% | 1.2% | 3.5% | 2.7% | |
| rs174561 | ||||||||||
| P-value (corrected) | 1.2E-09 | 0.002 | 5.0E-13 | 2.1E-37 | 0.005 | 0.384 | 0.051 | 9.8E-07 | 6.3E-05 | |
| ß-coefficient | 0.971 | −0.174 | 0.232 | −0.836 | −0.021 | 0.047 | −0.085 | −0.053 | −0.065 | |
| R2 (explained var.) | 5.0% | 2.0% | 6.7% | 18.0% | 1.7% | 2.0% | 1.2% | 3.6% | 2.7% | |
| rs3834458 | ||||||||||
| P-value (corrected) | 3.9E-10 | 1.1E-04 | 3.8E-14 | 5.7E-38 | 0.005 | 0.018 | 0.092 | 5.9E-06 | 6.0E-05 | |
| ß-coefficient | 0.970 | −0.193 | 0.236 | −0.823 | −0.019 | 0.061 | −0.079 | −0.049 | −0.064 | |
| R2 (explained var.) | 5.3% | 2.6% | 7.3% | 18.3% | 1.7% | 2.6% | 1.1% | 3.2% | 2.6% | |
Note: Corrected P-values to account for multiple testing are obtained by dividing not corrected P-values by 45 (5 single SNPanalyses of 9 outcomes); ß-coeffcient is the respective regression estimate of the additive coded SNP (0/1/2) for the respective outcome (PUFA) in a simple regression model.
Associations of single SNPs with parental reported eczema
Of all analyzed indicator coded SNPs of the FADS1 FADS2 gene cluster none showed a statistically significant association with the dichotomous outcome parental reported eczema in the first 2 years of life ( Table 3 ) in both unadjusted and adjusted analyses (see methods). Odds ratios (OR) for carriers of one or two minor alleles in comparison to non-carriers range from 1.3 to 1.5 in unadjusted, and 1.3 to 1.4 in adjusted analyses, respectively. However, 95%-confidence limits always include the null-effect of an OR of 1.0. Moreover, tests for multiplicative allelic trend (genotypes coded 0, 1, 2) also showed that none was significant at the 5% level with, even though no correction for multiple testing was applied.
Table 3. Association of FADS1 FADS 2 variants with parental reported eczema.
| SNP | n | OR | 95%-CI | P-value Trend | n | OR | 95%-CI | P-value Trend | |
| unadjusted | adjusted | ||||||||
| rs174545 | C/C | 399 | 1.00 | - | 0.07 | 392 | 1.00 | - | 0.13 |
| C/G | 364 | 1.32 | 0.95–1.85 | 357 | 1.32 | 0.94–1.87 | |||
| G/G | 93 | 1.43 | 0.86–2.39 | 92 | 1.34 | 0.79–2.28 | |||
| rs174546 | C/C | 401 | 1.00 | - | 0.09 | 394 | 1.00 | - | 0.16 |
| C/T | 363 | 1.30 | 0.93–1.81 | 356 | 1.30 | 0.92–1.83 | |||
| T/T | 94 | 1.40 | 0.84–2.33 | 93 | 1.30 | 0.77–2.21 | |||
| rs174556 | C/C | 424 | 1.00 | - | 0.05 | 417 | 1.00 | - | 0.12 |
| C/T | 358 | 1.34 | 0.96–1.86 | 351 | 1.33 | 0.95–1.87 | |||
| T/T | 76 | 1.49 | 0.86–2.58 | 75 | 1.33 | 0.75–2.35 | |||
| rs174561 | T/T | 427 | 1.00 | - | 0.07 | 420 | 1.00 | - | 0.15 |
| T/C | 356 | 1.31 | 0.94–1.82 | 349 | 1.31 | 0.93–1.84 | |||
| C/C | 77 | 1.46 | 0.84–2.51 | 76 | 1.30 | 0.74–2.29 | |||
| rs3834458 | T/T | 396 | 1.00 | - | 0.06 | 389 | 1.00 | - | 0.10 |
| T/Z | 370 | 1.28 | 0.92–1.79 | 363 | 1.28 | 0.91–1.81 | |||
| Z/Z | 91 | 1.54 | 0.93–2.57 | 90 | 1.42 | 0.84–2.42 | |||
Note: Odds ratios (OR) of indicator coded SNPs (reference is homozygous major alleles genotype) on eczema are estimated by logistic regression. Adjustment comprises study cohort (indicator coded dummy variables: KOALA study conventional, KOALA study alternative recruitment group vs. LISA study as reference), sex, maternal education, maternal smoking during pregnancy and exclusive breastfeeding for at least 3 months.
Separate analyses for both study populations revealed that in the LISA-study all SNPs of the FADS1 FADS2 gene cluster showed significant associations with the dichotomous outcome parental reported eczema in the first 2 years of life in both unadjusted and adjusted analyses, with ORs of about 2 and 4 for heterzygous and homozygous minor allele carriers, which is in good agreement with multiplicative increase of risks (confirmed by P-values for multiplicative trend between 0.003 and 0.005 without correction for multiple testing (see Supporting Information Table S7). By contrast, no associations were found within the KOALA study, where all ORs were between 0.9 and 1.2, whereas upper boundaries of the 95% confidence intervals were below 2.0.
No significant association of any analyzed SNP of the FADS gene with total IgE level or specific IgE at the age of one or two years of the child could be established, neither in the KOALA nor in the LISA study (results not presented).
Associations of fatty acids with parental reported eczema
No single PUFA was significantly associated with parental reported eczema of the child in the first 2 years of life after adjustment (see methods), neither in analyses of the combined study population ( Table 4 ), nor in separate analyses for the KOALA and LISA studies (see Supporting Information Table S8).
Table 4. Association of fatty acids with parental reported eczema.
| Fatty Acid | n | OR | 95%-CI | P-value | n | OR | 95%-CI | P-value | |
| unadjusted | adjusted | ||||||||
| LA | C18_2n_6 | 875 | 0.99 | 0.94–1.05 | 0.85 | 860 | 1.03 | 0.97–1.09 | 0.39 |
| GLA | ln(C18_3n_6) | 871 | 0.00 | 0.00–0.08 | 0.00 | 856 | 0.88 | 0.72–1.07 | 0.19 |
| DGLA | C20_3n_6 | 875 | 0.92 | 0.71–1.21 | 0.57 | 860 | 0.95 | 0.72–1.25 | 0.71 |
| AA | C20_4n_6 | 875 | 1.00 | 0.89–1.13 | 0.97 | 860 | 1.02 | 0.90–1.15 | 0.80 |
| A | C22_4n_6 | 874 | 3.41 | 0.82–14.16 | 0.09 | 859 | 0.85 | 0.17–4.15 | 0.84 |
| ALA | ln(C18_3n_3) | 873 | 0.79 | 0.50–1.25 | 0.32 | 858 | 0.99 | 0.61–1.61 | 0.97 |
| EPA | ln(C20_5n_3) | 875 | 1.53 | 1.09–2.15 | 0.01 | 860 | 0.82 | 0.55–1.21 | 0.31 |
| DPA | C22_5n_3 | 875 | 0.46 | 0.19–1.11 | 0.08 | 860 | 0.60 | 0.24–1.50 | 0.27 |
| DHA | ln(C22_6n_3) | 875 | 0.87 | 0.48–1.59 | 0.66 | 860 | 1.53 | 0.80–2.95 | 0.20 |
Note: Odds ratios (OR) of measured PUFA on eczema are estimated by logistic regression. Adjustment comprises study cohort (indicator coded dummy variables: KOALA study conventional, KOALA study alternative recruitment group vs. LISA study as reference), sex, maternal education, maternal smoking during pregnancy and exclusive breastfeeding for at least 3 months.
Discussion
This is the first study presenting associations between the genetic variants in the FADS1 FADS2 gene cluster with fatty acid composition in a population based sample of children.
We found that all five analyzed variants of the FADS1 FADS2 gene cluster are associated with polyunsaturated fatty acids LA, GLA, DGLA, AA, A, ALA, EPA, DPA and DHA, in particular with AA. Except for the n-3 fatty acids ALA and EPA these associations are highly significant even after conservative Bonferroni correction for multiple testing. For most PUFAs this is in line with previous reports showing these associations in adult populations [4]–[8]. However, in contrast to previous studies in adults, all SNPs in the present study in children are also highly significantly associated with DHA in one of the study populations (KOALA study). In the same population, Moltó et al found a relation between DHA in blood of the KOALA mothers during pregnancy and the mothers' FADS variants [35].
Our study confirms previous reports that carriers of the minor alleles showed higher levels of n-6 precursor fatty acids LA, DGLA and n-3 precursor fatty acids ALA and decreased product levels of the n-6 fatty acids GLA, AA, A and n-3 fatty acids EPA, DPA and DHA. Thus carriers of the minor alleles show enhanced desaturase substrate levels (substrate accumulation) and decreased desaturase product levels indicative of a lower desaturase activity, in agreement with previous studies [5], [6]. Thus, our results confirm that the FADS1 FADS2 gene cluster modulates the PUFA metabolism and demonstrate that this is also the case in children at the age of 2 years. A functional basis for such a regulation was recently shown by Lattka et al. in a study investigating the FADS2 variants rs3834458 and rs968567 [36]. According to this study both SNPs are potential promoter polymorphisms and located in a region important for transcription regulation. The minor alleles of SNP rs968567, but not rs3834458 (studied also in our study), showed a statistical significant increased effect on promoter activity and binding activity to two protein complexes activating the transcription factor ELK1 in that study. However, one of the rare other functional studies on FADS2 found a decreased promoter activity for rs3834458, suggesting the need for many more functional studies [37].
A previous study in adults found protective associations of carriers of minor alleles of the FADS1 FADS2 gene cluster with allergic rhinitis and atopic eczema [6]. In our analyses we could confirm such an association between FADS variants and eczema only for the German study population, but clearly not within the larger sample of children of the Netherlands. Whether these inconsistent results suggest that in children less common variants of the FADS1 FADS2 gene cluster are actually related to the development of eczema within the first 2 years of life requires further investigations with further independent study populations.
In contrast to some previous studies summarized in several reviews [9]–[11], we found no evidence for a direct link between the analyzed blood PUFA levels and eczema. This was true for analyses based on the combined study population as well as for separate analyses of the KOALA and LISA cohorts. Apparently, the development of disease cannot be simplified to one underlying pathway on fatty acid metabolism. On the other hand, larger sample sizes and analyses in further study populations may be necessary to draw any final conclusion on the role of fatty acid composition in blood and eczema. Also, studies of gene-diet interactions may be needed to resolve these inconsistencies, since populations vary widely in the intake n-3 PUFAs from fish products.
Strengths and Limitations
With data on variants of the FADS1 FADS2 gene cluster, PUFA and information on eczema status within the first 2 years of life for more than 800 children this is a large study despite the potential lack of power to detect small effects regarding eczema.
Genotyping for both study populations (KOALA, LISA) were done in the same lab at the Helmholtz Center Munich.
Fatty acids were derived from phospholipids in the KOALA study and from serum glycerophospholipids in the LISA study and analyzed at two different labs by somewhat different methods, but we do not think that this compromised our results. Both labs (Laboratory for Health Protection Research, National Institute for Public Health and the Environment, Bilthoven, the Netherlands and Division of Metabolic Diseases and Nutritional Medicine, Dr. von Hauner Children's Hospital, Ludwigs-Maximilians-University of Munich) have a long lasting experience in fatty acid analysis. Moreover, Glaser et al. have recently shown that fatty acid analysis of serum glycerophospholipids by high throughput analysis results in only minor deviations in values of fatty acids compared to analysis of serum phospholipid fatty acids [23]. Indeed, the fatty acid composition data was quite similar for both study populations, except for EPA, which might reflect different dietary habits with higher consumption levels of fish, which is the prime dietary source of EPA, in the Netherlands than in Bavaria (cf. Table 1).
We used a similar definition of the outcome “parental reported eczema of child within the first 2 years of life”, but slightly different wording of the questions and slightly different timing of questionnaires, covering the entire first two years of life in the two cohorts. Despite these similarities we found more than double the percentage in parental reported eczema in the children of the KOALA study (30.6%) than in the sample of the German LISA study (14.1%), even though in KOALA parents the prevalence of asthma and allergies was lower (57.0%) than in LISA parents (68.7%). This raises the question of whether lifestyle differences are responsible for the marked difference in % eczema in children in the two cohorts. There are two additional elements which further press this question: First, the FADS1 and FADS2 polymorphism prevalence is remarkably similar in the two populations, suggesting similar genetic background in the two populations. Second, in the LISA study, 71% of children are exclusively breastfed vs. only 50% in the KOALA study. To compare, the reported prevalence on eczema defined by symptoms in young children in two other birth cohort studies are 25% in children of age 4 years in the Netherlands and 16% for the first 2 years of life years in Germany [38], [39]. However, we do not think that any differences in eczema between the LISA and KOALA study affected the results to an appreciable extent, since we conducted, in addition to unadjusted analyses, also analyses adjusted for study cohort. The higher prevalence of eczema in KOALA could point to a higher proportion of non-atopic eczema, and if the FADS effect would be confined to atopic (IgE-mediated) eczema, it could have been diluted by non-atopic cases in the KOALA study. However, the absence of a FADS effect on total and specific IgE in both cohorts makes this explanation unlikely.
At present we have no convincing explanation, why we found a significant association between variants of the FADS gene and eczema only in the German LISA study, but not in the Netherland's KOALA study. Since Moltó et al found that the DHA deficit in the homozygous minor allele carriers could be overcome by intake of fish at the recommended level of 2 portions a week [35] we speculate that the difference between LISA and KOALA may reflect a lower intake of n-3 long chain PUFAs in LISA, as a consequence of which the genetic effect is manifest in LISA but not in KOALA. Based on this, we recommend that further studies are done in populations with low n-3 PUFA intake.
In conclusion, this is the first study that confirms in children of age 2 years the previously found associations of genetic variants in the FADS1 FADS2 gene cluster with fatty acid composition in serum phospholipids or glycerophospholipids and that also analyzed the potential influence of FADS1 FADS2 genotypes and PUFAs to eczema.
Variants of the FADS1 FADS2 gene cluster clearly do regulate the metabolism of PUFA in young children. Inconsistent results were found for a link between these genetic variants with eczema. In the German LISA study all SNPs were significantly associated with eczema. In the Netherland's KOALA study this was clearly not the case. In both study populations PUFA was not associated with eczema. Thus the hypothesis of an inflammatory link between PUFA and eczema by the metabolic pathway of LC-PUFAs as precursors for inflammatory prostaglandins and leukotrienes could not be confirmed by these data. This would suggest either that this pathway is not or only marginally involved in eczema development, that other risk factors for early childhood eczema may be more important, or that eczema may have heterogeneous etiology with only a small segment of the population susceptible for effects of endogenous fatty acids metabolism or gene-diet-interaction; or a combination of these arguments.
Supporting Information
Study design and population
(0.04 MB DOC)
Fatty acid analysis
(0.04 MB DOC)
Genotyping
(0.04 MB DOC)
Determination of infants' IgE
(0.04 MB DOC)
Definition of outcome variable parental reported eczema
(0.04 MB DOC)
Statistical analysis
(0.04 MB DOC)
Characteristics of the five analyzed variants in the FADS1 FADS 2 gene region
(0.37 MB DOC)
Mean levels of fatty acids (PUFA) by genotype of five variants in the FADS1 FADS 2 gene region (KOALA & LISA study combined)
(0.39 MB DOC)
Mean levels of fatty acids (PUFA) by genotype of five variants in the FADS1 FADS 2 gene region in the KOALA study
(0.39 MB DOC)
Mean levels of fatty acids (PUFA) by genotype of five variants in the FADS1 FADS 2 gene region in the LISA study
(0.39 MB DOC)
Associations of the five analyzed variants in the FADS1 FADS 2 gene region with fatty acids in the KOALA study
(0.39 MB DOC)
Associations of the five analyzed variants in the FADS1 FADS 2 gene region with fatty acids in the LISA study
(0.39 MB DOC)
Association of FADS1 FADS 2 variants with parental reported eczema
(0.38 MB DOC)
Association of fatty acids with parental reported eczema
(0.38 MB DOC)
Acknowledgments
We thank the families for participation in the KOALA and LISA birth cohort studies; the obstetric units for allowing recruitment, the study teams for excellent work. We also like to acknowledge Hans Cremers (Laboratory for Health Protection Research. National Institute for Public Health and the Environment, Bilthoven, the Netherland) for excellent contribution in the analyses of fatty acid composition and Franziska Scharl (Helmholtz Zentrum München, German Research Center for Environmental Health, Institute of Epidemiology, Neuherberg, Germany) for excellent technical assistance in DNA extraction.
LISA Study Group:
Institute of Epidemiology, Helmholtz Zentrum Muenchen - German Research Center for Environmental Health, Neuherberg (Wichmann HE, Heinrich J, Schoetzau A, Franke K, Laubereau B); Schaaf B, Bad Honnef, Department of Pediatrics, University of Leipzig (Borte M, Schulz R, Sierig G); Institute for Clinical Immunology and Transfusion Medicine (Sack U, Emmrich F); Department of Pediatrics, Marien-Hospital, Wesel (von Berg A, Bollrath C); Department of Human Exposure-Research and Epidemiology, UFZ-Center for Environmental Research Leipzig-Halle (Herbarth O, Diez U); Department of Environmental Immunology, UFZ-center for Environmental Research Leipzig-Halle (Lehmann I, Herberth G).
Footnotes
Competing Interests: Royal Friesland Foods has financially contributed to the infrastructure of the KOALA study as a co-sponsor. Data and intellectual property rights are wholly owned by the researchers. None of the authors have financial, professional or personal interests related to this sponsoring. Royal Friesland Foods had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The KOALA study with follow-up until age 2 years was co-sponsored by the Netherlands Organisation for Health Research and Development (ZonMW grants no. 2100.0090 and 912-03-031), the Netherlands Asthma Foundation (grant no. 3.2.03.48), Royal Friesland Foods, Triodos Foundation, Phoenix Foundation, Raphae Foundation, Iona Foundation, Foundation for the Advancement of Heilpedagogie, and the Netherlands Ministry of Public Health, Welfare and Sport, the Netherlands and the Netherlands Organisation for Scientific Research (NWO Spinoza award of Prof. D.S. Postma). The LISA study was funded by grants of the Federal Ministry for Education, Science, Research and Technology (Grant No. 01 EG 9705/2 and 01EG9732). This work was partly supported by the “Kompetenznetz Adipositas (Competence Network Obesity)” and NGFNplus funded by the Federal Ministry of Education and Research (FKZ: 01GI0826, 01GI0823, 01GS0820). Personal and financial support by the Munich Center of Health Sciences (MCHEALTH) as part of the Ludwig-Maximilians University Munich LMU innovative is gratefully acknowledged. Dirkje S. Postma received the NWO Spinoza award from the Netherlands Organisation for Scientific Research. Berthold Koletzko is the recipient of a Freedom to Discover Award of the Bristol-Myers-Squibb Foundation, New York, NY, USA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lattka E, Illig T, Koletzko B, Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol; 2010;21:64–69. doi: 10.1097/MOL.0b013e3283327ca8. [DOI] [PubMed] [Google Scholar]
- 2.Cho HP, Nakamura MT, Clarke SD. Cloning, expression, and nutritional regulation of the mammalian Delta-6 desaturase. J Biol Chem; 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 3.Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J Biol Chem; 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 4.Malerba G, Schaeffer L, Xumerle L, Klopp N, Trabetti E, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids; 2008;43:289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 5.Rzehak P, Heinrich J, Klopp N, Schaeffer L, Hoff S, et al. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br J Nutr; 2009;101:20–26. doi: 10.1017/S0007114508992564. [DOI] [PubMed] [Google Scholar]
- 6.Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet; 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet; 2009;5 doi: 10.1371/journal.pgen.1000338. e1000338-doi:10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie L, Innis SM. Genetic Variants of the FADS1 FADS2 Gene Cluster Are Associated with Altered (n-6) and (n-3) Essential Fatty Acids in Plasma and Erythrocyte Phospholipids in Women during Pregnancy and in Breast Milk during Lactation. Journal of Nutrition; 2008;138:2222–2228. doi: 10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
- 9.Calder PC. Polyunsaturated fatty acids and inflammation. Biochem Soc Trans; 2005;33:423–427. doi: 10.1042/BST0330423. [DOI] [PubMed] [Google Scholar]
- 10.Calder PC. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am J Clin Nutr; 2006;83:1505S–1519S. doi: 10.1093/ajcn/83.6.1505S. [DOI] [PubMed] [Google Scholar]
- 11.Duchen K, Bjorksten B. Polyunsaturated n-3 fatty acids and the development of atopic disease. Lipids; 2001;36:1033–1042. doi: 10.1007/s11745-001-0814-5. [DOI] [PubMed] [Google Scholar]
- 12.Glaser C, Heinrich J, Koletzko B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism; 2010;59:993–999. doi: 10.1016/j.metabol.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Lattka E, Illig T, Heinrich J, Koletzko B. FADS gene cluster polymorphisms: important modulators of fatty acid levels and their impact on atopic diseases. J Nutrigenet Nutrigenomics; 2009;2:119–128. doi: 10.1159/000235559. [DOI] [PubMed] [Google Scholar]
- 14.Sausenthaler S, Koletzko B, Heinrich J. Dietary Fat Intake and Allergic Diseases. Current Nutrition & Food Science; 2006;2:351–359. [Google Scholar]
- 15.Kompauer I, Demmelmair H, Koletzko B, Bolte G, Linseisen J, et al. Association of fatty acids in serum phospholipids with hay fever, specific and total immunoglobulin E. Br J Nutr; 2005;93:529–535. doi: 10.1079/bjn20041387. [DOI] [PubMed] [Google Scholar]
- 16.Calder PC. Polyunsaturated fatty acids, inflammatory processes and inflammatory bowel diseases. Mol Nutr Food Res; 2008;52:885–897. doi: 10.1002/mnfr.200700289. [DOI] [PubMed] [Google Scholar]
- 17.Kummeling I, Thijs C, Penders J, Snijders BE, Stelma F, et al. Etiology of atopy in infancy: the KOALA Birth Cohort Study. Pediatr Allergy Immunol; 2005;16:679–684. doi: 10.1111/j.1399-3038.2005.00333.x. [DOI] [PubMed] [Google Scholar]
- 18.Snijders BE, Thijs C, van Ree R, van den Brandt PA. Age at first introduction of cow milk products and other food products in relation to infant atopic manifestations in the first 2 years of life: the KOALA Birth Cohort Study. Pediatrics; 2008;122:e115–e122. doi: 10.1542/peds.2007-1651. [DOI] [PubMed] [Google Scholar]
- 19.Heinrich J, Bolte G, Holscher B, Douwes J, Lehmann I, et al. Allergens and endotoxin on mothers' mattresses and total immunoglobulin E in cord blood of neonates. Eur Respir J; 2002;20:617–623. doi: 10.1183/09031936.02.02322001. [DOI] [PubMed] [Google Scholar]
- 20.Sausenthaler S, Koletzko S, Schaaf B, Lehmann I, Borte M, et al. Maternal diet during pregnancy in relation to eczema and allergic sensitization in the offspring at 2 y of age. Am J Clin Nutr; 2007;85:530–537. doi: 10.1093/ajcn/85.2.530. [DOI] [PubMed] [Google Scholar]
- 21.Agren JJ, Julkunen A, Penttila I. Rapid separation of serum lipids for fatty acid analysis by a single aminopropyl column. J Lipid Res; 1992;33:1871–1876. [PubMed] [Google Scholar]
- 22.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem; 1957;226:497–509. [PubMed] [Google Scholar]
- 23.Glaser C, Demmelmair H, Koletzko B. High-throughput analysis of fatty acid composition of plasma glycerophospholipids. J Lipid Res; 2010;51:216–221. doi: 10.1194/jlr.D000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bottema RW, Reijmerink NE, Kerkhof M, Koppelman GH, Stelma FF, et al. Interleukin 13, CD14, pet and tobacco smoke influence atopy in three Dutch cohorts: the allergenic study. Eur Respir J; 2008;32:593–602. doi: 10.1183/09031936.00162407. [DOI] [PubMed] [Google Scholar]
- 25.Reimerink J, Stelma F, Rockx B, Brouwer D, Stobberingh E, et al. Early-life rotavirus and norovirus infections in relation to development of atopic manifestation in infants. Clin Exp Allergy; 2009;39:254–260. doi: 10.1111/j.1365-2222.2008.03128.x. [DOI] [PubMed] [Google Scholar]
- 26.Aalberse RC, Koshte V, Clemens JG. Immunoglobulin E antibodies that crossreact with vegetable foods, pollen, and Hymenoptera venom. J Allergy Clin Immunol; 1981;68:356–364. doi: 10.1016/0091-6749(81)90133-0. [DOI] [PubMed] [Google Scholar]
- 27.Stallman PJ, Aalberse RC. Estimation of basophil-bound IgE by quantitative immunofluorescence microscopy. Int Arch Allergy Appl Immunol; 1977;54:9–18. doi: 10.1159/000231803. [DOI] [PubMed] [Google Scholar]
- 28.Stapel SO, Eysink PE, Vrieze J, Aalberse RC. IgE testing in capillary blood. Pediatr Allergy Immunol; 2004;15:230–233. doi: 10.1111/j.1399-3038.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 29.Akkerdaas JH, Wensing M, Asero R, Fernandez RM, Knulst AC, et al. IgE binding to pepsin-digested food extracts. Int Arch Allergy Immunol; 2005;138:203–208. doi: 10.1159/000088720. [DOI] [PubMed] [Google Scholar]
- 30.Chen CM, Rzehak P, Zutavern A, Fahlbusch B, Bischof W, et al. Longitudinal study on cat allergen exposure and the development of allergy in young children. J Allergy Clin Immunol; 2007;119:1148–1155. doi: 10.1016/j.jaci.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Chen CM, Sausenthaler S, Bischof W, Herbarth O, Borte M, et al. Perinatal exposure to endotoxin and the development of eczema during the first 6 years of life. Clin Exp Dermatol; 2010;35:238–244. doi: 10.1111/j.1365-2230.2009.03460.x. [DOI] [PubMed] [Google Scholar]
- 32.SAS Institute Inc. SAS/STAT 9.1 User's Guide. SAS Institute Inc., Cary, NC, USA; 2004. [Google Scholar]
- 33.SAS Institute Inc. SAS/Genetics 9.1.3 User's Guide. SAS Institute Inc., Cary, NC, USA; 2005. [Google Scholar]
- 34.Carter KW, McCaskie PA, Palmer LJ. JLIN: a java based linkage disequilibrium plotter. BMC Bioinformatics; 2006;7:60. doi: 10.1186/1471-2105-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molto-Puigmarti C, Plat J, Mensink RP, Muller A, Jansen E, et al. FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. Am J Clin Nutr; 2010;91:1368–1376. doi: 10.3945/ajcn.2009.28789. [DOI] [PubMed] [Google Scholar]
- 36.Lattka E, Eggers S, Moeller G, Heim K, Weber M, et al. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res; 2010;51:182–191. doi: 10.1194/jlr.M900289-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nwankwo JO, Spector AA, Domann FE. A nucleotide insertion in the transcriptional regulatory region of FADS2 gives rise to human fatty acid delta-6-desaturase deficiency. J Lipid Res; 2003;44:2311–2319. doi: 10.1194/jlr.M300273-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Brauer M, Hoek G, Smit HA, de Jongste JC, Gerritsen J, et al. Air pollution and development of asthma, allergy and infections in a birth cohort. Eur Respir J; 2007;29:879–888. doi: 10.1183/09031936.00083406. [DOI] [PubMed] [Google Scholar]
- 39.Filipiak B, Zutavern A, Koletzko S, von Berg A, Brockow I, et al. Solid food introduction in relation to eczema: results from a four-year prospective birth cohort study. J Pediatr; 2007;151:352–358. doi: 10.1016/j.jpeds.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 40.Park WJ, Kothapalli KSD, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product {Delta}8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res; 2009;50:1195–1202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study design and population
(0.04 MB DOC)
Fatty acid analysis
(0.04 MB DOC)
Genotyping
(0.04 MB DOC)
Determination of infants' IgE
(0.04 MB DOC)
Definition of outcome variable parental reported eczema
(0.04 MB DOC)
Statistical analysis
(0.04 MB DOC)
Characteristics of the five analyzed variants in the FADS1 FADS 2 gene region
(0.37 MB DOC)
Mean levels of fatty acids (PUFA) by genotype of five variants in the FADS1 FADS 2 gene region (KOALA & LISA study combined)
(0.39 MB DOC)
Mean levels of fatty acids (PUFA) by genotype of five variants in the FADS1 FADS 2 gene region in the KOALA study
(0.39 MB DOC)
Mean levels of fatty acids (PUFA) by genotype of five variants in the FADS1 FADS 2 gene region in the LISA study
(0.39 MB DOC)
Associations of the five analyzed variants in the FADS1 FADS 2 gene region with fatty acids in the KOALA study
(0.39 MB DOC)
Associations of the five analyzed variants in the FADS1 FADS 2 gene region with fatty acids in the LISA study
(0.39 MB DOC)
Association of FADS1 FADS 2 variants with parental reported eczema
(0.38 MB DOC)
Association of fatty acids with parental reported eczema
(0.38 MB DOC)