Abstract
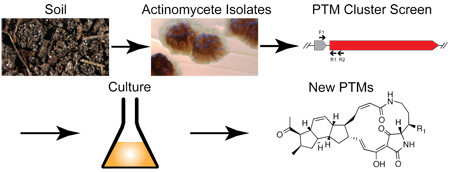
A targeted PCR-based screening approach was used to identify candidate polycyclic tetramate macrolactam (PTM) biosynthetic gene clusters in environmental Streptomyces isolates. Isolation and characterization of the small molecules produced by one of the strains confirmed the production of two new PTMs (clifednamides A, 4 and B, 5), and more generally, the utility of using a targeted approach for the discovery of new members of this interesting class.
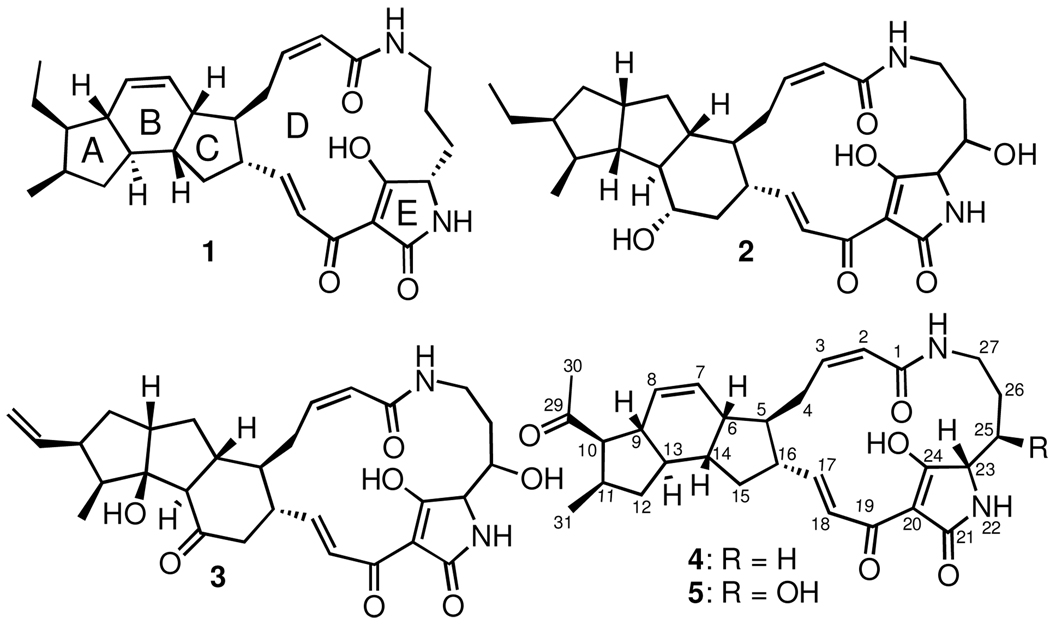
Ikarugamycin (1), dihydromaltophilin (2), and frontalamide A (3) comprise the founding, the most widely encountered, and the most recently discovered members of a related group of polycyclic tetramate macrolactams (PTMs).1 As the name suggests, this family of structurally complex and biologically active small molecules shares a macrocyclic lactam ring with an embedded tetramic acid ring along with a variable set of stereochemically rich carbocyclic rings. Ikarugamycin (1) has a 5-6-5 set of carbocyclic rings, but all other well-characterized PTMs have 5-5-61b, 1c, 2 or 5-53 ring arrangements. Ikarugamycin (1) was originally reported in 1972 as an antiprotozal agent,4 and subsequent studies identified other family members in a desultory fashion. Bacteria from diverse genera – Streptomyces, Alteromonas, Stenotrophomonas, and Lysobacter – produce PTMs.1–4
Many PTMs display antifungal activity, and a dihydromaltophilin (2) producer (Lysobacter enzymogenes C3) has been widely studied as a biocontrol agent for fungal infestations of agricultural crops.5 In addition to the antiprotozoal and antifungal activities of PTMs, a recent study using expression-based pathway signature analysis identified ikarugamycin (1) and related compounds as “promising leads” for anticancer therapeutics.6
The recent identification of the frontalamide (3) biosynthetic locus enabled systematic searches for similar loci.1d Analyses of sequenced bacterial genomes revealed surprisingly large numbers of PTM biosynthetic loci, particularly within the Streptomyces genus. Strikingly, almost none of the identified bacteria were previously recognized as PTM producers. Further, a PCR-based screen to detect PTM biosynthetic loci was used to probe environmental isolates, leading to numerous isolates harboring putative PTM clusters. Together these studies suggested that a targeted approach to PTM discovery would lead to new family members with potentially valuable properties. This communication outlines the implementation of such a PCR-guided search strategy to discover two new PTMs (4 and 5) with the unusual 5-6-5 set of carbocyclic rings found in 1. These results corroborate previous sequence-driven PTM studies and illustrate the general utility of a targeted strategy to specifically discover new PTMs.
Streptomyces sp. JV178 produced putative PTM compounds (HPLC retention time, UV, and MS) after growth for five to six days at 30 °C on ISP4 agar. A crude extract of the culture plates was prepared by soaking the agar, with adhering mycelia intact, in ethyl acetate. After removing the solvent under vacuum, the dry crude material was dissolved in methanol before C-18 SPE treatment. Eluates containing 4 and 5 were separated by C-18 HPLC to afford the pure compounds, which we have named the clifednamides A and B.
Clifednamides A (4) and B (5) were obtained as colorless powders. Initial LC/MS analyses indicated that 4 had a molecular weight of 492 Dalton and a strong UV absorption at 327 nm. Compound 5 showed a similar UV absorption, but a molecular weight of 508. The hydrocarbon skeletons of 4 and 5 were characterized with 1D (1H, 13C) and 2D NMR methods.
The molecular formula of 4 was established as C29H36N2O5 by high-resolution electrospray ionization mass spectrometry (HR-ESI-MS). The 1H NMR and HSQC spectra of 4 in 90% pyridine-d5/CD3OD showed the presence of six olefinic protons (δH 5.75, br d, J = 9.6 Hz; 5.87, br d, J = 9.6 Hz; 6.06, td, J = 11.1, 2.4 Hz; 6.32, d, J = 11.1 Hz; 6.86, dd, J = 15.6, 10.2 Hz; 8.05, d, J = 15.6 Hz), nine methine, six methylenes, and two methyl groups (δH 2.21, s; 0.88 d, J = 7.2 Hz), and the 13C NMR spectrum exhibited twenty nine signals, including four carbonyls (δC 167.6, 174.3, 197.6, 209.9), four double bonds (δC 103.2, 124.0, 125.8, 129.7, 130.5, 139.9, 147.5, 178.0), and seventeen sp3 carbons from δC 19.3 to 61.4 ppm. In the COSY spectrum of 4, three coupling systems from H-23 (δH 4.05, m) to H2-27 (δH 3.91, m; 2.85, m) through H2-25 (δH 2.24, m; 2.06, m) and H2-26 (δH 1.82, m; 1.60, m), from H-2 (δH 6.32, d, J = 11.1 Hz) to H-5 (δH 1.49, m) through H-3 (δH 6.06, td, J = 11.1, 2.4 Hz) and H-4 (δH 4.05, m; 2.60, m), and from H-18 (δH 8.05, d, J = 15.6 Hz) to H-16 (δH 2.45, m) through H-17 (δH 6.86, dd, J = 15.6, 10.2 Hz) were clearly demonstrated. The HMBC correlations from H-2, H-3, and H2-27 to C-1 (δC 167.6), from H-18 to C-19 (δC 178.0), C-20 (δC 103.2), and from H-23 to C-20, C-21 (δC 174.3), C-24 (δC 197.6), C-25 (δC 28.3) and C-26 (δC 22.3) established rings D and E of compound 4 to be a macrolactam with a tetramic acid unit as shown. The following key HMBC correlations were also observed: H-6 (δH 2.50, m) to C-8 (δC 130.5) and C-13 (δC 48.2); H-9 (δH 2.50, m) to C-7 (δC 129.7) and C-14 (δC 41.8); H3-30 (δH 2.21, s) to C-10 (δC 59.4) and C-29 (δC 209.9); H3-31 (δH 0.88 d, J = 7.2 Hz) to C-10, C-11 (δC 34.2) and C-12 (δC 39.3); H-16 (δH 2.45, m) to C-6 (δC 43.2) and C-14 (δC 41.8). Based on these HMBC correlations, together with both COSY and TOCSY information, rings A–C of 4 were determined as a 5-6-5 tricyclic unit. In the ROESY spectrum of 4, correlations between H-14 and H-6/H-9, H-10 and H-13, H3-31 and H-9, H-16 and H-6/H-14 indicated trans-cis-trans A/B-B/C-C/D ring fusions. Hence the structure of 4 (clifednamide A) was determined as shown.
The molecular formula for clifednamide B (5) was determined as C29H36N2O6 by HR-ESI-MS and 13C NMR spectroscopy. The 1H and 13C NMR signals indicated rings A-E of 5 were very similar to those of 1 and 4. The only observed difference between 4 and 5 was at the C-25-position, and the chemical shifts (δH 4.94, m; δC 73.1) of this position in 5 indicated an oxygenated methine. Given the likelihood that PTMs incorporate L-ornithine during their biosynthesis, H-23, which was originally the α-proton of L-ornithine, should have a β-orientation. Molecular modeling (Chem3D-Ultra 9.0 MM2) predicted a dihedral angle of 78° (J = ~0.1 Hz) for an α-oriented H-25 (H-23–C-23–C-25–H-25) compared to 38° (J = ~5.0 Hz) for a β-oriented H-25. H-23 (δH 4.74) appeared as a broad singlet, indicating that the dihedral angle must be close to 78°, that the 25-OH had a β-orientation, and that the structure of clifednamide B (5) is as shown.
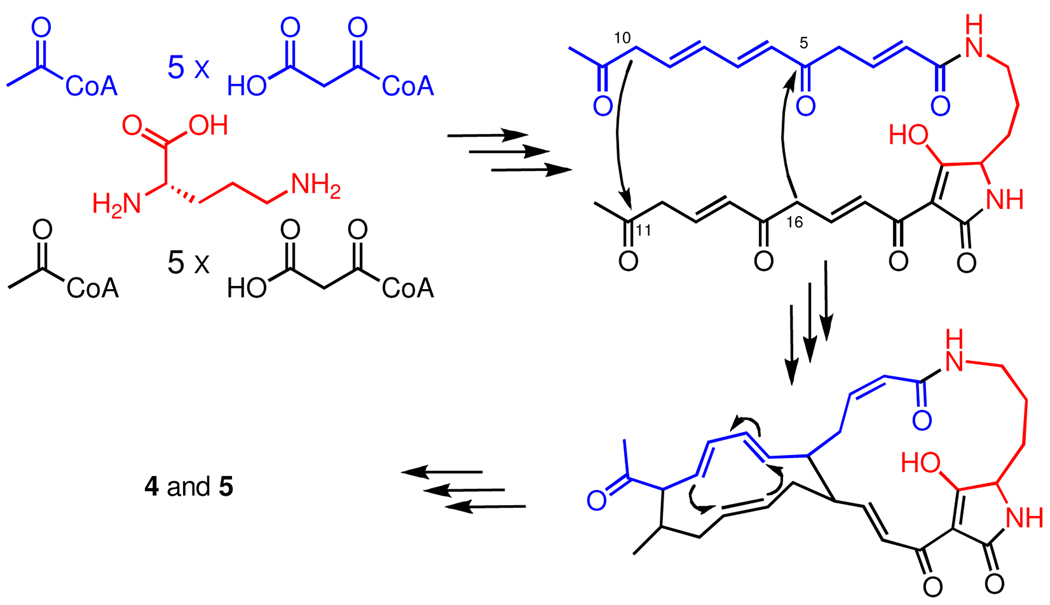
Based on the structure of 1, Ito and Hirata proposed that its biosynthesis involved coupling three fragments: one derived from ornithine and two twelve-carbon chains derived from six acetate units1a – what today would be described as a hybrid PKS-NRPS pathway. The authors also proposed that the 5-6-5 ring system of 1 could, in principle, be formed by two aldol condensations and a formal Diels-Alder reaction (Figure 2) between appropriately functionalized PKS chains. By extension, the same might apply to the clifednamides. The later discovered and more common PTMs containing 5-5-6 and 5-5 systems obviously require different ring forming reactions.
Figure 2.
Hypothetical pathway1a leading to the production of 4 and 5
Identifying conserved PTM loci in sequenced bacterial genomes and environmental isolates underscores the pathway’s evolutionary success. As a result, we undertook the targeted discovery of what are likely the many additional small molecules made by PTM pathways to explore their chemical diversity, biological potential and ultimately, the basis of the pathway’s high prevalence. The PTM cluster screen detects a small, but highly conserved (~800 bp in JV178) region common to all PTM loci identified thus far, with PCR primers annealing to portions of two conserved biosynthetic genes (Figure 3).1d One, ftdA, encodes a hydroxylase, and the other ftdB, encodes a putative iterative hybrid PKS-NRPS synthetase. Based on the biosynthesis of 3, it is likely that the JV178 FtdA homolog installs the hydroxyl at C-25 in 5 and the FtdB hybrid PKS-NPRS forms the mixed polyketide/peptide backbone common to 4 and 5.
Figure 3.
PTM biosynthetic locus for compound 3, consisting of genes ftdA-ftdF.1d PCR priming sites F1 and R1/R2 used to detect PTM clusters are shown for clarity. Primers not shown to scale.
The molecular diversity of the family therefore almost certainly lies in the pathway’s other genes, those that code for enzymes to direct the multiple links between the two PKS-derived chains or perform tailoring reactions. Currently we know little about the reactions carried out by these genes, and a more detailed understanding will require connecting fully sequenced pathways with the small molecules they produce. Since nearly 50% of sequenced Streptomyces genomes contain apparent PTM biosynthetic loci,1d there are plenty of opportunities.
Supplementary Material
Figure 1.
Structures of compounds 1–5. Note the rings A–E of ikarugamycin (1) are identical to those of the clifednamides (4 and 5).
Acknowledgment
This project was supported by NIH GM086258 (JC), Harvard NERCE AI057159 (JC), CA024487 (JC) and a Harvard Microbial Sciences Initiative Postdoctoral Fellowship to JB.
Footnotes
Supporting Information Available Experimental methods; NMR spectra, data tables, and supporting chemical data for clifednamides A and B (4 and 5). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Ito S, Hirata Y. Bull. Chem. Soc. 1977;50:1813–1820. [Google Scholar]; (b) Graupner PR, Thornburgh S, Mathieson JT, Chapin EL, Kemmitt GM, Brown JM, Snipes CE. J. Antibiot. 1997;50:1014–1019. doi: 10.7164/antibiotics.50.1014. [DOI] [PubMed] [Google Scholar]; (c) Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. Antimicrob. Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Blodgett JAV, Oh DC, Cao S, Currie CR, Kolter R, Clardy J. Proc. Natl. Acad. Sci. U S A. 2010;107:11692–11697. doi: 10.1073/pnas.1001513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Jakobi M, Winkelmann G, Kaiser D, Kempler C, Jung G, Berg G, Bahl H. J. Antibiot. 1996;49:1101–1104. doi: 10.7164/antibiotics.49.1101. [DOI] [PubMed] [Google Scholar]; (b) Gunasekera SP, Gunasekera M, McCarthy P. J. Org. Chem. 1991;56:4830–4833. [Google Scholar]
- 3.(a) Bae MA, Yamada K, Uemura D, Seu JH, Kim YH. J. Microbiol. Biotechn. 1998;8:455–460. [Google Scholar]; (b) Shigemori H, Bae MA, Yazawa K, Sasaki T, Kobayashi J. J. Org. Chem. 1992;57:4317–4320. [Google Scholar]; (c) Kanazawa S, Fusetani N, Matsunaga S. Tetrahedron Lett. 1993;34:1065–1068. [Google Scholar]
- 4.Jomon K, Kuroda Y, Ajisaka M, Sakai H. J. Antibiot. 1972;25:271–280. doi: 10.7164/antibiotics.25.271. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Jochum CC, Yu F, Zaleta-Rivera K, Du L, Harris SD, Yuen GY. Phytopathology. 2008;98:695–701. doi: 10.1094/PHYTO-98-6-0695. [DOI] [PubMed] [Google Scholar]
- 6.Tenenbaum JD, Walker MG, Utz PJ, Butte AJ. BMC Med. Genomics. 2008;1:51. doi: 10.1186/1755-8794-1-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.