Abstract
The loss of expression of the fragile X mental retardation protein (FMRP) leads to fragile X syndrome. FMRP has two types of RNA binding domains, two K-homology domains and an arginine-glycine-glycine box domain, and it is propose to act as a translation regulator of specific messenger RNA. The interest to produce sufficient quantities of pure recombinant FMRP for biochemical and biophysical studies is high. However, the recombinant bacterial expression of FMRP has had limited success, and subsequent recombinant eukaryotic and in vitro expression has also resulted in limited success. In addition, the in vitro and eukaryotic expression systems may produce FMRP which is posttranslationally modified, as phosphorylation and arginine methylation have been shown to occur on FMRP. In this study, we have successfully isolated the conditions for recombinant expression, purification and long term storage of FMRP using Escherichia coli, with a high yield. The expression of FMRP using E. coli renders the protein devoid of the posttranslational modifications of phosphorylation and arginine methylation, allowing the study of the direct effects of these modifications individually and simultaneously. In order to assure that FMRP retained activity throughout the process, we used fluorescence spectroscopy to assay the binding activity of the FMRP arginine-glycine-glycine box for the Semaphorin 3F mRNA and confirmed that FMRP remained active.
Keywords: FMRP, Fragile X Syndrome, protein purification
Introduction
Fragile X syndrome, the most prevalent inheritable mental retardation which affects ~1 in 4000 males and ~1 in 8000 females, is caused by the lack of fragile X mental retardation protein (FMRP) expression [1]. The absence of FMRP is the result of transcriptional inactivation of the fragile X mental retardation-1 (FMR1) gene by the unstable expansion of a cytosine-guanine-guanine (CGG) trinucleotide repeat in its 5′-untranslated region [2, 3]. Upon exceeding ~200 CGG repeats, the cytosines of the repeats are hypermethylated leading to the transcriptional silencing of FMR1 and loss of FMRP expression. In addition to a nuclear localization signal near its N-terminus and a nuclear export signal near its C-terminus, FMRP carries two types of RNA binding domains: two K-homology domains and one arginine-glycine-glycine box (RGG box), suggesting that the protein exerts its function via RNA binding (Fig 1) [4-6]. Despite extensive studies of the FMRP function, the mechanism by which its loss of expression leads to mental retardation is still not fully understood. FMRP has demonstrated RNA chaperone properties and has been found localized to actively translating ribosomes, leading to the postulation that it serves as a translational regulator for specific messenger RNA (mRNA) targets [7-9]. FMRP is expressed in most tissues, being found abundantly in the dendritic spines of neurons [10-12].
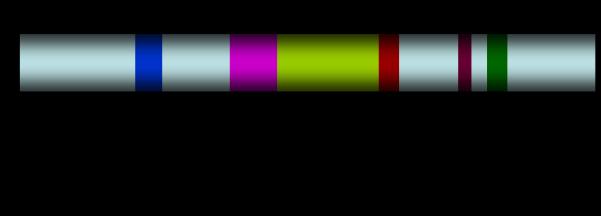
Figure 1.
(A) Schematic of full length FMRP showing the nuclear localization signal (NLS), K-homology domains (KH1, KH2), nuclear export signal (NES), region of phosphorylation (P), and the RGG box (RGG) domain. (B) Sequence of the phosphorylation region and the RGG box domain of full-length ISO1. The sequence shown represents splice site acceptor 15a of FMR1 mRNA. The RGG repeats of the RGG box domain are underlined; the phosphorylation sites are indicated by a double dagger (‡); the arginine methylation sites, located within the RGG box domain, are indicated by an arrow (↓). Also shown is the sequence of the synthetic RGG box peptide used in the previous studies of binding with the S3F-sh_8AP mRNA [42]. Adapted from [28].
The FMR1 gene has 17 exons and it can undergo alternative splicing that involves the exons 12 and 14 and the choice of acceptor sites in exons 15 and 17 [4, 13-15]. These splicing patterns could potentially result in 12 FMRP isoforms, however, so far only five isoforms have been detected in various tissues [15, 16]. The largest molecular weight FMRP species corresponds to the isoform 1 (ISO1) (Fig 1) [13].
Previous attempts towards FMRP recombinant bacterial expression, purification and long-term storage in sufficient quantities for biochemical and biophysical studies resulted in limited success, which was not greatly improved upon the subsequent utilization of eukaryotic cells or in vitro systems [4, 12, 15, 17-30]. Following an extensive study, we have isolated the experimental conditions for expression, purification and storage of active recombinant ISO1, representing full-length FMRP, in Escherichia coli. Besides improving yield, the bacterial expression of FMRP ISO1 has the advantage that it renders the protein product devoid of posttranslational modifications, such as phosphorylation and arginine methylation. This is particularly important, as it has been proposed that phosphorylation and arginine methylation might be relevant for the regulation of FMRP function. Mammalian FMRP was found to be phosphorylated in a region N-terminal to its RGG box [31]. The initial suggestion that this modification modulates the FMRP translation regulator function came from the findings that unphosphorylated FMRP associates with actively translating polyribosomes, whereas phosphorylated FMRP associates with stalled polyribosomes [30]. Recently, two enzymes implicated in the phosphorylation and dephosphorylation of FMRP were identified as the ribosomal protein S6 kinase and the protein phosphatase 2A, respectively [32, 33]. It has been suggested that FMRP phosphorylation is coupled to translational suppression, whereas the FMRP dephosphorylation releases the target mRNAs for translation [32-34]. However, it is not clear if these phosphorylation and dephosphorylation events regulate the FMRP translational regulator function by affecting its RNA binding properties or its interactions with other protein partners. In addition, it has been proposed that the phosphorylation state of FMRP determines its inclusion with the microRNA pathway by regulating its association with Dicer and Dicer-containing complexes [35].
It has also been shown that FMRP is methylated within its RGG box domain and that its binding affinity for homoribopolymers (poly (rG), poly (rC), poly (rA) or poly (rU)) changes when the protein is produced in the presence of protein methyltransferases [19, 36-38]. Moreover, FMRP methylation reduced its ability to associate with Sc1 RNA, a G quadruplex forming synthetic RNA [19]. These results suggest that protein arginine methylation is important in defining the interactions of FMRP with its G quadruplex forming mRNA targets.
As stated previously, the expression of FMRP ISO1 using E. coli renders the protein devoid of posttranslational modifications. Thus, this method allows for the future analysis of the explicit effects of each posttranslational modification, phosphorylation and arginine methylation, individually and simultaneously. The ability to study the effects of these modifications as potential regulators of the mRNA binding activity of FMRP is particularly important due to their locations relative to the mRNA-binding RGG box domain, as the four proposed sites of phosphorylation occur N-terminal, but in close proximity to the RGG box domain (Fig 1B). Moreover, of the arginine methylation sites, two occur on the arginine residues of the RGG repeats within the RGG box, whereas the other two sites occur at the only two arginines found between these repeats. Hence, the probability that the posttranslational modifications of arginine methylation and phosphorylation acting as a regulatory mechanism for the mRNA binding activity of the RGG box domain is high.
Thus, the successful purification of a significant yield of active recombinant FMRP ISO 1 expressed in E. coli, devoid of posttranslational modifications, is significant in that it will allow us to investigate to what extent FMRP posttranslational modifications, such as phosphorylation and arginine methylation, modulate its translational regulator function.
Materials and Methods
Expression of recombinant FMRP ISO1
The recombinant plasmid pET21a-FMRP, encoding ISO1 fused with a C-terminal 6x histidine tag, was a gift kindly provided by Dr. Bernhard Laggerbauer [17]. The plasmid was transformed into and cultured using Rosetta 2(DE3) pLysS E. coli cells (Novagen). The cells were cultured at 37 °C, 250 rpm in Luria-Bertani (LB; Fisher Scientific) media containing 200 μg/mL ampicillin (Amp; MP Biomedical) and 15 μg/mL chloramphenicol (Chl; MP Biomedical) and then mixed 1:1 with glycerol and frozen at −80 °C.
Cells were spread onto an LB + Amp + Chl agar plate using 4-way spread technique and incubated 12 h at 37 °C. Single colonies were picked and cultured (culture 1) in 250 mL LB + Amp + Chl media in a 1 L flask at 37 °C, 250 rpm, 12 h. Eight 2 L flasks (culture 2) were prepared with 500 mL LB + Amp + Chl and 25 mL culture 1 was added to each flask, followed by incubation at 37 °C, 250 rpm until OD600 reached 0.8 – 1.0. The expression of FMRP ISO1 was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM to culture 2 which was then transferred to a separate incubator at 25 °C, 250 rpm for 12 h.
The eight 500 mL culture 2 flasks were equally combined into six 1 L centrifuge flasks and cells were pelleted at 5000 g, 4 °C, 10 min. Pellets were weighed and transferred to individual 50 mL round-bottom polycarbonate centrifuge tubes (Nalgene). A 1:1 mixture of B-PER Bacterial Protein Extraction Reagent (Thermo Scientific) and 2x lysis buffer (20 mM HEPES, pH 7.5, 600 mM LiCl, 10 mM β-mercaptoethanol, 40 mM imidazole, 10% glycerol) was added at 10 mL buffer/g cells to each pellet, followed by the re-suspension of the pellets by vortexing. Cells were incubated at −20 °C until further purification.
Purification of recombinant FMRP ISO1
Resuspended cell pellets were thawed in a 25 °C water bath and then incubated on ice for 30 min to allow lysozyme action encoded by the pLysS cell line. Cells were then sonicated (Branson Sonifier Cell Disruptor 185) three times at a power of 6 for 30 sec with 2 min on ice between bursts. The crude lysate was then centrifuged at 40,000 g, 4 °C, 30 min. A 10 cm × 1.5 cm diameter column was loaded with 10 mL Ni-NTA Superflow resin slurry (Qiagen) at 4 °C and equilibrated with 5 column volumes of equilibration buffer 1 (10 mM HEPES, pH 8.0, 300 mM LiCl, 5 mM β-mercaptoethanol, 5% glycerol, 20 mM imidazole) at 2 mL/min using the BioRad BioLogic LP system. The entire clarified crude supernatant was loaded onto the column at a flow rate of 0.5 mL/min, followed by washes performed at a rate of 2 mL/min: 5 column volumes of equilibration buffer 1, 10 column volumes of equilibration buffer 2 (same components as equilibration buffer 1 except at pH 7.0), and 10 column volumes of wash buffer 1 (10 mM HEPES, pH 7.5, 300 mM LiCl, 5 mM β-mercaptoethanol, 5% glycerol, 100 mM imidazole). An additional wash step was performed, at a flow rate of 1 mL/min with wash buffer 2 (10 mM HEPES, pH 7.5, 300 mM LiCl, 5 mM β-mercaptoethanol, 5% glycerol, 250 mM imidazole). Pure FMRP ISO1 was eluted at a flow rate of 1 mL/min with elution buffer (10 mM HEPES, pH 7.5, 300 mM LiCl, 5 mM β-mercaptoethanol, 5% glycerol, 500 mM imidazole), collecting 15 3 mL fractions. Immediately after elution, ethylenediaminetetraacetic acid (EDTA) was added to each collected fraction to a final concentration of 1 mM and the fractions were stored at 4 °C until dialysis. Fractions from the elution were analyzed via 10% tris-glycine SDS-PAGE and visualized by Coomassie blue stain to assure purity (Fig 2A).
Figure 2.
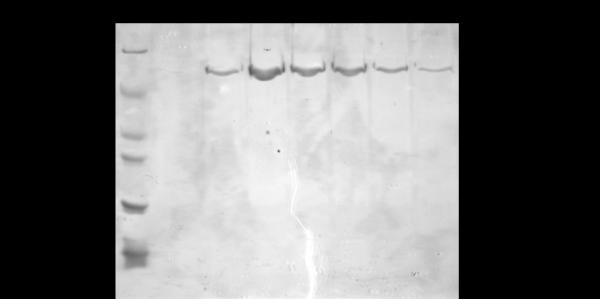
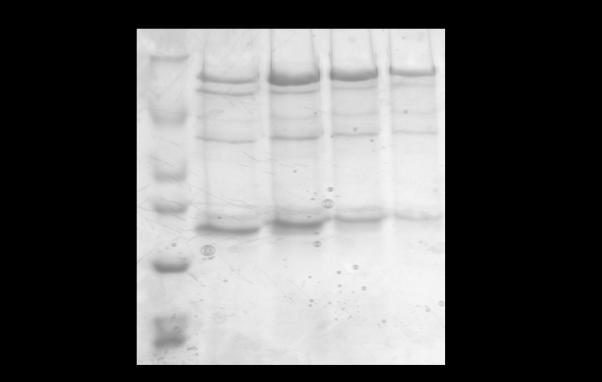
Tris-glycine SDS-PAGE (10%) analysis of FMRP ISO1 purity (A) after the Ni-NTA affinity chromatography method developed herein and (B) prior to the optimization of the purification protocol. FMRP ISO1 is not expressed strongly enough in the presence of IPTG to be visibly distinguished from other protein bands in the cleared crude supernatant hence that sample is not shown. (A) Lane 1: EZ-Run protein marker (Fisher Scientific). Lanes 2-8: eluted pure FMRP ISO1 fractions 4-10, respectively. (B) Lane 1: EZ-Run protein marker. Lanes 2-5: eluted impure FMRP ISO1 fractions 5-8, respectively.
Concentration of FMRP ISO1
The concentration protocol was adapted from [39]. A sterile 50 mL beaker was pre-chilled on ice and elution fractions 5 – 8, containing the highest concentration of FMRP ISO1, were pooled therein. Dialysis tubing of 3500 MWCO, 29.3 mm diameter (Fisher Scientific) was washed in sterile dH2O and filled with polyethylene glycol 20,000 (J.T. Baker) such that the polyethylene glycol consumed approximately 30% of the available inner volume to allow for expansion. The filled tubing was immersed completely into the beaker containing FMRP ISO1. After pooling FMRP ISO1, the volume for concentrating was 12 mL and the protein was concentrated to a final volume of approximately 3 mL.
Dialysis of ISO1
For the purpose of the mRNA binding experiments that were monitored by fluorescence spectroscopy, FMRP ISO1 had to be devoid of imidazole. All dialysis buffers were prepared two days in advance of use, without adjusting the pH, and allowed equilibrate overnight at 4 °C. The pH adjustments occurred one day prior to use and allowed to finish equilibration over night at 4 °C. Dialysis tubing of 10,000 MWCO, 7.5 mm diameter (Spectra/Por) was washed with sterile dH2O and the concentrated FMRP ISO1 was added. All dialysis buffers were, volume-wise, approximately 667:1 to FMRP ISO1 and incubations in the respective dialysis buffer occurred for 12 h at 4 °C. Each dialysis buffer (2L) contained 10 mM HEPES, pH 7.5, 300 mM LiCl, 5 mM β-mercaptoethanol, 5% glycerol, 1 mM EDTA and variable imidazole concentrations, as follows: the first dialysis buffer contained 200 mM imidazole, the second contained 100 mM imidazole, the third contained 50 mM imidazole and the fourth contained no imidazole. After dialysis, the concentration of FMRP ISO1 was determined to be 50 μM by absorbance at 280 nm using an extinction coefficient 46370 M−1 cm−1 adapted from [40, 41]. The purity and retention of the protein were analyzed by 10% tris-glycine SDS-PAGE and visualized by Coomassie blue stain (Fig 3).
Figure 3.
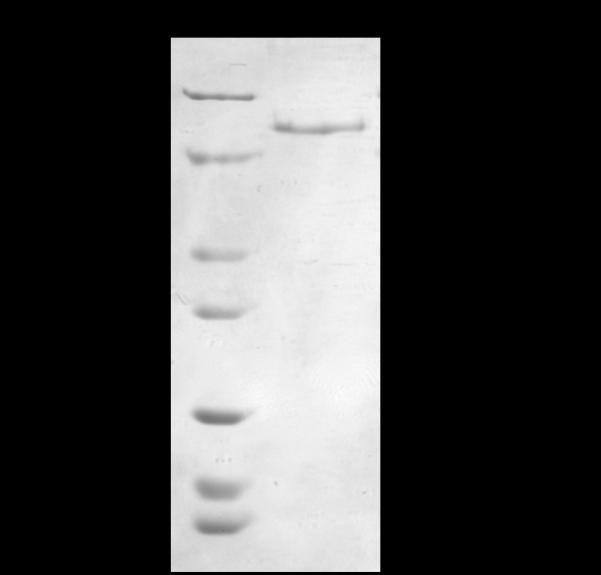
Tris-glycine SDS-PAGE (10%) analysis of FMRP ISO1 after the purification, concentration and dialysis processes and ready for binding analysis with G quadruplex mRNAs. Lane 1: EZ-Run protein marker. Lane 2: dialyzed FMRP ISO1.
Mass spectrometry analysis of FMRP ISO1
The identity of FMRP ISO1 was confirmed using peptide mass fingerprinting (Genomics and Proteomics Core Laboratories, University of Pittsburgh) (Table 1). Briefly, FMRP ISO1 from an SDS-PAGE gel was excised and trypsin digested followed by analysis via matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS). The five MALDI-TOF-MS characterized amino acid sequences matched that found in FMRP ISO1.
Table 1.
Results from the MALDI-TOF-MS sequence analysis of trypsin digested FMRP ISO1
| Peptide Sequence |
Observed Mass |
Expected Mass |
Δ Mass | Protein Identified |
|---|---|---|---|---|
| (K)LIQEIVDK(S) | 958.13403 | 958.1400 | −0.00597 | FMRP |
| (K)NVPQEEEIM*PPNSLPSNNSR(V) | 2269.43498 | 2269.4425 | −0.00752 | FMRP |
| (R)VLVASSVVAGESQKPELK(A) | 1842.12671 | 1842.1375 | −0.01079 | FMRP |
| (R)EDLM*GLAIGTHGANIQQAR(K) | 2012.23703 | 2012.2433 | −0.00627 | FMRP |
| (K)AWQGM*VPFVFVGTK(D) | 1583.87812 | 1583.8837 | −0.00558 | FMRP |
indicates oxidized methionine
Analysis of FMRP ISO1 Activity
In order to ensure that FMRP ISO1 maintained mRNA binding activity after the aforementioned processes, we performed a fluorescence RNA binding assay according to [5, 42]. Briefly, the G quadruplex forming semaphorin 3F mRNA (S3F-sh) fragment was labeled by the highly fluorescent purine analog 2-aminopurine (AP) at position 8, producing S3F-sh_8AP (Dharmacon, Inc) (Fig 4). The replacement of adenine with 2-aminopurine has been shown to not disrupt G quadruplex formation and binding interactions of various mRNAs [5, 6, 42]. The fluorescence spectroscopy experiments were performed on a J.Y. Horiba Fluoromax-3 equipped with a variable temperature control in the sample chamber and a 3 mm path-length quartz cuvette (Starna Cells) in a 150 μL final volume. The excitation wavelength was set at 310 nm and the emission spectrum was recorded in the range of 330 – 450 nm with a bandpass of 5 mm for both the excitation and emission monochromators.
Figure 4.
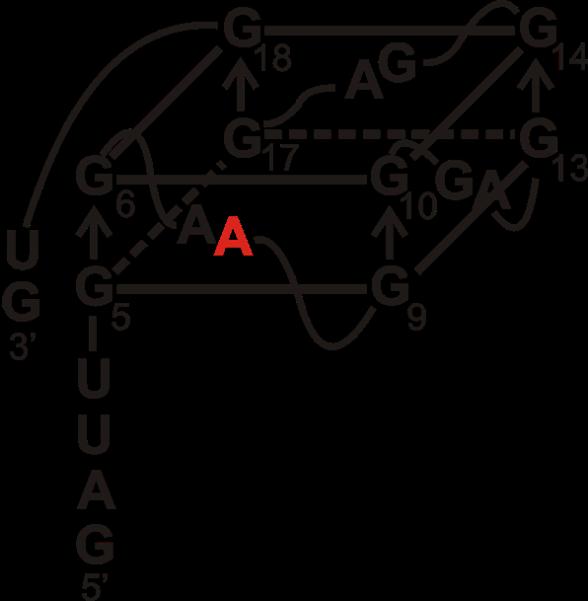
Proposed G quadruplex structure of S3F-sh mRNA [42]. The adenine at position 8 replaced with fluorescent 2-aminopurine tag, to form S3F-sh_8AP, is shown in red.
The binding was measured by titrating FMRP ISO1 in 30 nM aliquots to a fixed concentration of 150 nM S3F-sh_8AP mRNA (in 10 mM cacodylic acid, pH 6.5, 150 mM KCl) at 25 °C. The samples were incubated for 7 min after each addition of FMRP ISO1. The binding dissociation constant, Kd, was determined by plotting the normalized S3F-sh_8AP mRNA steady state fluorescence intensity at 371 nm as a function of the FMRP ISO1 concentration and fitting the resultant binding curves to the equation [42]:
| (1) |
where IB and IF are the steady state fluorescence intensity of the bound and free S3F-sh_8AP mRNA, [RNA]t is the total concentration of S3F-sh_8AP mRNA, and [P]t is the total FMRP ISO1 concentration. The best-fit curve and subsequent Kd and R2 values for FMRP ISO1 binding S3F-sh_8AP mRNA were generated using Origin 7.0 (Fig 5). The experiment was repeated in triplicate and bovine serum albumin (BSA) was also similarly titrated as a negative control (Fig 5).
Figure 5.
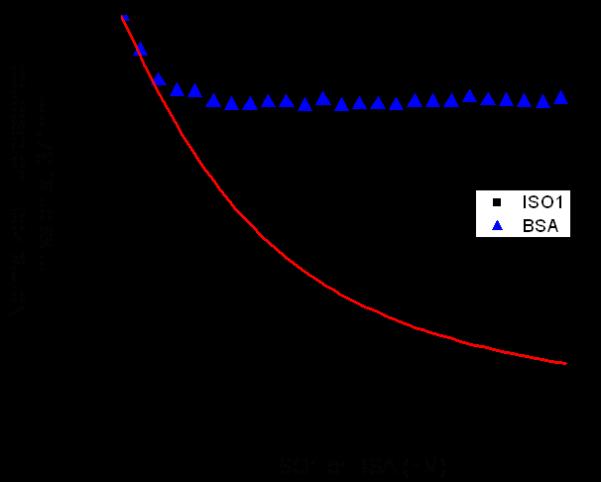
Fluorescence spectroscopy assay where FMRP ISO1 (black squares) is titrated into a fixed concentration of S3F-sh_8AP mRNA resulting in a Kd of 104 ± 11 nM and R2 value of 0.995. The Kd value was obtained by curve-fitting (red curve) the titration data points with equation 1. As a negative control, BSA (blue triangles) was similarly titrated and showed no significant binding to S3F-sh_8AP mRNA.
Results and Discussion
Since the discovery of the FMR1 gene, the recombinant expression of FMRP in E. coli has been continuously pursued, however the results of these efforts have been only satisfactory, as we and others have experienced problems involving low levels of FMRP expression and yield, as well as protein precipitation [4, 15, 17-19, 21, 22, 24, 25, 27, 28]. In trying to circumvent these problems, other studies have employed eukaryotic cell lines and in vitro transcription and translation systems, but they also had limited success [12, 18, 20-26, 28-30]. Furthermore, FMRP is known to undergo phosphorylation and arginine methylation, and such posttranslational modifications might occur when using eukaryotic and in vitro systems of expression, as the enzymes involved in these pathways are present in such systems [19, 30-38]. Thus, to elucidate the effects of these posttranslational modifications on FMRP activity, the expression of FMRP in the prokaryotic E. coli is vital.
Expression and purification of recombinant FMRP ISO1
Initially, we attempted the FMRP ISO1 expression using the plasmid pET21a-FMRP (a generous gift from Dr. Bernhard Laggerbauer), which was transformed in the E. coli BL21 Gold (DE3) cell line; however, these experiments were not successful due to the low expression of the protein. One cause for this poor FMRP ISO1 expression could be codon bias, which involves seven human codons, as the sequence analysis of FMR1 reveals that all seven biased codons are present in the gene a total of 72 times. To overcome this potential problem of codon bias associated with the expression of eukaryotic genes in prokaryotic E. coli cells, we transformed the FMR1 containing vector into the Rosetta 2(DE3) pLysS E. coli cell line. In addition to compensating for codon bias, the utilization of the pLysS cell line allows the cells to endogenously express lysozyme, eliminating the need to add exogenous lysozyme and reducing the time necessary for the protein purification procedure. Based on the tendency of FMRP to precipitate, the endogenous lysozyme also enables a gentler lysis of the cells during the purification. The transformed cells were grown at 37 °C until OD600 was 0.8 – 1.0 and following induction with IPTG, the cultures were grown for an additional 12 h at 25 °C. Following cell harvesting and lysis, we employed Ni-NTA affinity chromatography for the purification process, eluting FMRP in the presence of 500 mM imidazole. The fraction containing FMRP also contained other protein impurities, which we attempted to remove using additional steps, such as gel filtration chromatography, but this was unsuccessful as protein precipitation occurred (Fig 2B). We then proceeded to optimize the conditions for FMRP purification, by varying the buffer components and the concentration of imidazole in the wash steps during the Ni-NTA affinity chromatography, and were successful in producing pure FMRP ISO1 using only the Ni-NTA resin column (Fig 2A). Utilizing single-column affinity chromatography reduces the necessary amount of time for purification, allowing analyses of FMRP sooner; hence a longer period of time available for analyses before precipitation occurs. Furthermore, we have noted that long term storage of FMRP ISO1 at 4 °C in the presence of the elution buffer containing 500 mM imidazole and 1 mM EDTA, followed by the dialysis and concentration steps described below maintains its activity and reduces precipitation. MALDI-TOF-MS analysis was employed to confirm the identity of trypsin digested FMRP ISO1, and as summarized in Table 1, the five peptide fragments detected were characteristic to FMRP based on matches within the Xcalibur mass spectrometry database.
Concentration and dialysis of recombinant FMRP ISO1
After obtaining pure FMRP ISO1, we attempted to concentrate the protein using centrifugal means with a regenerated cellulose membrane, and subsequently with a polyethersulfone membrane. These efforts were unsuccessful as a considerable amount of FMRP ISO1 adsorbed to either membrane, and visible precipitation was present in solution. We then used a dialysis tubing concentration method adapted from [39], which drastically reduced the FMRP ISO1 precipitation and protein loss. Furthermore, concentration of the protein in the presence of hydrogen bonding-capable agents, such as glycerol and imidazole, inhibited precipitation. Based on this, FMRP ISO1 was concentrated while in the elution buffer containing 500 mM imidazole and 1 mM EDTA.
During the development of the dialysis procedure we noted that the presence of hydrogen bonding buffer components, such as imidazole, and EDTA reduced FMRP ISO1 precipitation. The presence of imidazole is not compatible with the fluorescence spectroscopy mRNA binding assay, yet the presence of imidazole reduces FMRP ISO1 loss. Hence, we developed the dialysis conditions for the gradual removal of imidazole which resulted in minimal loss through precipitation. Initially, the concentrated FMRP ISO1 is in elution buffer containing 500 mM imidazole and 1 mM EDTA. Every dialysis step was performed at 4 °C with 2 L of buffer that contained 10 mM HEPES, pH 7.5, 300 mM LiCl, 5 mM β-mercaptoethanol, 5% glycerol, 1 mM EDTA and varying concentrations of imidazole. The approximate 3 mL of concentrated FMRP ISO1, in elution buffer with 500 mM imidazole, was transferred to a dialysis buffer containing 200 mM imidazole for 12 h, followed by 100 mM imidazole for 12 h, then 50 mM for 12 h and finally zero imidazole for 12 h. Following the concentration and imidazole removal, analysis by SDS-PAGE indicates that FMRP ISO1 remains pure and free of proteolytic degradation (Fig 3). After purification and dialysis, the concentration of FMRP ISO1 was typically 50 μM in a volume of approximately 3mL.
G quadruplex mRNA binding activity of FMRP ISO1
To confirm that the recombinant FMRP ISO1 is active, we have evaluated its ability to bind to G quadruplex RNA [5, 6, 27, 42, 43].
Depending on the particular RNA sequence, the formation of the G quadruplex RNA structure requires K+ ions in different concentrations. Based on the variability of the required K+ concentration, the binding assay buffer must have K+ concentration flexibility; hence FMRP ISO1 must be in a buffer devoid of K+. Moreover, the G quadruplex can form in the presence of Na+ ions but this is thermodynamically less stable relative to when K+ is utilized [44]. Thus, Li+ was employed throughout the purification and dialysis processes in place of the routinely utilized K+ and Na+ buffer counterions.
The RNA we employed in the FMRP ISO1 fluorescence spectroscopy binding assays (S3F-sh_8AP) was derived from the semaphorin 3F mRNA, which encodes for the Sema 3F protein (Fig 4). Sema 3F is expressed in cerebellar granule cells and plays an important role in brain development by functioning as a chemo repellant to axon extension, neuronal migration and growth cone guidance [45, 46]. Previous studies have shown that the S3F mRNA is an in vivo neuronal binding target of FMRP and that S3f-sh_8AP mRNA folds into a G quadruplex structure which is bound with high affinity and specificity by a synthetic FMRP RGG box peptide [5, 27, 42, 47]. S3F-sh_8AP was labeled at position 8 by the highly fluorescent purine analog 2-amino purine (2AP), and increasing amounts of FMRP ISO1 were titrated into a fixed concentration of the labeled RNA, monitoring the steady-state fluorescence changes of the 2AP reporter. By fitting the resulting binding curve with Equation 1 (Materials and Methods) a dissociation constant, Kd, of 104 ± 11 nM was obtained, with an R2 value of 0.995 (Fig 5). This dissociation constant is in the nM range, being comparable, but not identical, to that previously reported for the synthetic FMRP RGG box peptide (Fig 1B) with this mRNA, at 1.0 ± 0.4 nM [42]. Additionally, the Kd of FMRP ISO1 for the G quadruplex forming S3F-sh_8AP mRNA is also in the same range with the Kd of the synthetic RGG box peptide for the G quadruplex forming MAP1B mRNA, at 20.1 ± 6.4 nM [6]. As a negative binding control, BSA was titrated into S3F-sh_8AP and it showed a minimal change (less than 10%) in the steady state fluorescence of the 2AP reporter (Fig 5).
Thus, following an extensive optimization of the parameters for purification, concentration and dialysis, we have successfully developed a method for the expression and purification of recombinant FMRP ISO1 in E.coli. This purified recombinant FMRP ISO1 is devoid of the posttranslational modifications of phosphorylation and arginine methylation, allowing for further studies of the direct effects of these posttranslational modifications upon the protein function. The yield obtained through this method is higher than that obtained from employing various other recombinant expression systems and methods. We have also shown that the FMRP ISO1 binding activity for G quadruplex forming mRNAs has been preserved throughout this process and shown that the Kd for binding the S3F-sh_8AP mRNA is comparable to that previously reported using the synthetic RGG box peptide.
In summary, the method herein allows the production of pure, active full-length FMRP ISO1 and, since the expression occurred in E. coli, opens the door for the analyses of the direct effects of posttranslational modifications on the RGG box binding activity for G quadruplex forming mRNAs.
Acknowledgments
We thank Dr. Bernhard Laggerbauer (Department of Biochemistry, Theodor Boveri Institute) for the plasmid pET21a-FMRP encoding for FMRP ISO1, and Dr. Stephanie Ceman (Department of Cell and Developmental Biology, University of Illinois, Urbana-Champaign) and Dr. Robert Denman (Department of Molecular Biology, Biochemical Molecular Neurobiology Laboratory, New York State Institute for Basic Research in Developmental Disabilities) for their helpful suggestions in the initial stages of this study. This work was supported by the N.I.H. grant 2 R15 GM074660-02A1 to M.R.M.
Abbreviations used
- FMRP
fragile X mental retardation protein
- RGG box
arginine-glycine-glycine box
- ISO1
full-length isoform 1 of FMRP
- S3F-sh_8AP
semaphorin 3F mRNA fragment containing 2-aminopurine
- LB
Luria-Bertani
- Amp
ampicillin
- Chl
chloramphenicol
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- EDTA
ethylenediaminetetraacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crawford DC, Acuna JM, Sherman SL. FMR1 and the fragile X syndrome: human genome epidemiology review. Genet Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin P, Warren ST. Understanding the molecular basis of fragile X syndrome. Human molecular genetics. 2000;9:901–908. doi: 10.1093/hmg/9.6.901. [DOI] [PubMed] [Google Scholar]
- 3.O’Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annual review of neuroscience. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 5.Menon L, Mihailescu MR. Interactions of the G quartet forming semaphorin 3F RNA with the RGG box domain of the fragile X protein family. Nucleic acids research. 2007;35:5379–5392. doi: 10.1093/nar/gkm581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menon L, Mader SA, Mihailescu MR. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA (New York, N.Y. 2008;14:1644–1655. doi: 10.1261/rna.1100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antar LN, Bassell GJ. Sunrise at the synapse: the FMRP mRNP shaping the synaptic interface. Neuron. 2003;37:555–558. doi: 10.1016/s0896-6273(03)00090-4. [DOI] [PubMed] [Google Scholar]
- 8.Jin P, Warren ST. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends in biochemical sciences. 2003;28:152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- 9.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic acids research. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, Bellini M, Ceman S. Fragile X mental retardation protein FMRP binds mRNAs in the nucleus. Molecular and cellular biology. 2009;29:214–228. doi: 10.1128/MCB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamanini F, Bontekoe C, Bakker CE, van Unen L, Anar B, Willemsen R, Yoshida M, Galjaard H, Oostra BA, Hoogeveen AT. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Human molecular genetics. 1999;8:863–869. doi: 10.1093/hmg/8.5.863. [DOI] [PubMed] [Google Scholar]
- 12.Sittler A, Devys D, Weber C, Mandel JL. Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Human molecular genetics. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 13.Ashley CT, Sutcliffe JS, Kunst CB, Leiner HA, Eichler EE, Nelson DL, Warren ST. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nature genetics. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 14.Verkerk AJMH, de Graaf E, De Boulle K, Eichler EE, Konecki DS, Reyners E, Manca A, Poustka A, Willems PJ, Nelson DL, Oostra B. Alternative splicing in the fragile X gene FMR1. Human molecular genetics. 1993;2:399–404. doi: 10.1093/hmg/2.4.399. [DOI] [PubMed] [Google Scholar]
- 15.Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nature genetics. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 16.Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile x syndrome. Annual review of genomics and human genetics. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 17.Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Human molecular genetics. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 18.Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson KD, Warren ST. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. The Journal of biological chemistry. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 19.Stetler A, Winograd C, Sayegh J, Cheever A, Patton E, Zhang X, Clarke S, Ceman S. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Human molecular genetics. 2006;15:87–96. doi: 10.1093/hmg/ddi429. [DOI] [PubMed] [Google Scholar]
- 20.Bardoni B, Schenck A, Mandel JL. A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Human molecular genetics. 1999;8:2557–2566. doi: 10.1093/hmg/8.13.2557. [DOI] [PubMed] [Google Scholar]
- 21.Mazroui R, Huot ME, Tremblay S, Boilard N, Labelle Y, Khandjian EW. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Human molecular genetics. 2003;12:3087–3096. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- 22.Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The fragile X mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. The Journal of biological chemistry. 2003;278:15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- 23.Tamanini F, Van Unen L, Bakker C, Sacchi N, Galjaard H, Oostra BA, Hoogeveen AT. Oligomerization properties of fragile-X mental-retardation protein (FMRP) and the fragile-X-related proteins FXR1P and FXR2P. The Biochemical journal. 1999;343(Pt 3):517–523. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, O’Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. The EMBO journal. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis SA, Willemsen R, van Unen L, Hoogeveen AT, Oostra BA. Prospects of TAT-mediated protein therapy for fragile X syndrome. Journal of molecular histology. 2004;35:389–395. doi: 10.1023/b:hijo.0000039841.22959.3c. [DOI] [PubMed] [Google Scholar]
- 26.Ceman S, Brown V, Warren ST. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Molecular and cellular biology. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 28.Denman RB, Sung YJ. Species-specific and isoform-specific RNA binding of human and mouse fragile X mental retardation proteins. Biochemical and biophysical research communications. 2002;292:1063–1069. doi: 10.1006/bbrc.2002.6768. [DOI] [PubMed] [Google Scholar]
- 29.Sung YJ, Conti J, Currie JR, Brown WT, Denman RB. RNAs that interact with the fragile X syndrome RNA binding protein FMRP. Biochemical and biophysical research communications. 2000;275:973–980. doi: 10.1006/bbrc.2000.3405. [DOI] [PubMed] [Google Scholar]
- 30.Ceman S, O’Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Human molecular genetics. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 31.Siomi MC, Higashijima K, Ishizuka A, Siomi H. Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties. Molecular and cellular biology. 2002;22:8438–8447. doi: 10.1128/MCB.22.24.8438-8447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayanan U, Nalavadi V, Nakamoto M, Thomas G, Ceman S, Bassell GJ, Warren ST. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. The Journal of biological chemistry. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. RNA (New York, N.Y. 2009;15:362–366. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denman RB. Methylation of the arginine-glycine-rich region in the fragile X mental retardation protein FMRP differentially affects RNA binding. Cellular & molecular biology letters. 2002;7:877–883. [PubMed] [Google Scholar]
- 37.Dolzhanskaya N, Merz G, Aletta JM, Denman RB. Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. Journal of cell science. 2006;119:1933–1946. doi: 10.1242/jcs.02882. [DOI] [PubMed] [Google Scholar]
- 38.Dolzhanskaya N, Merz G, Denman RB. Alternative splicing modulates protein arginine methyltransferase-dependent methylation of fragile X syndrome mental retardation protein. Biochemistry. 2006;45:10385–10393. doi: 10.1021/bi0525019. [DOI] [PubMed] [Google Scholar]
- 39.Degerli N, Akpinar MA. A novel concentration method for concentrating solutions of protein extracts based on dialysis techniques. Analytical biochemistry. 2001;297:192–194. doi: 10.1006/abio.2001.5335. [DOI] [PubMed] [Google Scholar]
- 40.Valverde R, Pozdnyakova I, Kajander T, Venkatraman J, Regan L. Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure. 2007;15:1090–1098. doi: 10.1016/j.str.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 41.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic acids research. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bole M, Menon L, Mihailescu MR. Fragile X Mental Retardation Protein Recognition of G quadruplex Structure per se is Sufficient for High Affinity Binding to RNA. Molecular BioSystems. 2008;4:1212–1219. doi: 10.1039/b812537f. [DOI] [PubMed] [Google Scholar]
- 43.Zanotti KJ, Lackey PE, Evans GL, Mihailescu MR. Thermodynamics of the fragile X mental retardation protein RGG box interactions with G quartet forming RNA. Biochemistry. 2006;45:8319–8330. doi: 10.1021/bi060209a. [DOI] [PubMed] [Google Scholar]
- 44.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers. 2000;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 45.Nasarre P, Constantin B, Rouhaud L, Harnois T, Raymond G, Drabkin HA, Bourmeyster N, Roche J. Semaphorin SEMA3F and VEGF have opposing effects on cell attachment and spreading. Neoplasia (New York, N.Y. 2003;5:83–92. doi: 10.1016/s1476-5586(03)80020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan F, Villegas G, Teichman J, Mundel P, Tufro A. Autocrine class 3 semaphorin system regulates slit diaphragm proteins and podocyte survival. Kidney international. 2006;69:1564–1569. doi: 10.1038/sj.ki.5000313. [DOI] [PubMed] [Google Scholar]
- 47.Rackham O, Brown CM. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. The EMBO journal. 2004;23:3346–3355. doi: 10.1038/sj.emboj.7600341. [DOI] [PMC free article] [PubMed] [Google Scholar]