Summary
Heme Nitric oxide/OXygen (H-NOX) domains are a family of hemoprotein sensors that are widespread in bacterial genomes, but limited information is available on their function. Legionella pneumophila is the only prokaryote found thus far to encode two H-NOX proteins. This paper presents data supporting a role for one of the Legionella pneumophila H-NOXs in the regulation of biofilm formation. In summary: (i) unmarked deletions in the hnox1 gene do not affect growth rate in liquid culture or replication in permissive macrophages; (ii) theΔhnox1 strain displays a hyper-biofilm phenotype; (iii) the gene adjacent to hnox1 is a GGDEF-EAL protein, lpg1057, and overexpression in Legionella pneumophila of this protein, or the well-studied diguanylate cyclase, vca0956, results in a hyper-biofilm phenotype; (iv) the Lpg1057 protein displays diguanylate cyclase activity in vitro and this activity is inhibited by the Hnox1 protein in the Fe(II)-NO ligation state, but not the Fe(II) unligated state; (v) consistent with the Hnox1 regulation of Lpg1057, unmarked deletions of lpg1057 in theΔhnox1 background results in reversion of the hyper-biofilm phenotype back to wild-type biofilm levels. Taken together, these results suggest a role for hnox1 in regulating c-di-GMP production by lpg1057 and biofilm formation in response to NO.
Keywords: H-NOX, Heme-Nitric oxide/OXygen Binding, Legionella pneumophila, Biofilm, c-di-GMP, Nitric Oxide
Introduction
The Heme-Nitric oxide/OXygen binding (H-NOX) domain is best known as the heme domain of soluble guanylate cyclase (sGC), a mammalian nitric oxide receptor (Derbyshire and Marletta, 2009). However, the recent proliferation of sequenced prokaryotic genomes has revealed a widespread distribution of H-NOX domains in bacteria (Boon et al., 2006, Karow et al., 2004, Iyer et al., 2003). All bacterial H-NOX domains expressed and characterized thus far are relatively stable in the ferrous oxidation state, bind diatomic gases, and are found adjacent to or fused to signaling proteins (Boon et al., 2006, Karow et al., 2004, Iyer et al., 2003). These bacterial H-NOX proteins fall into at least two distinct classes based on their ligand binding characteristics and genomic context. In obligate anaerobes, H-NOX domains are fused to methyl accepting chemotaxis proteins. In facultative aerobic bacteria, H-NOX domains are stand-alone proteins and are typically found adjacent to histidine kinases or proteins involved in c-di-GMP metabolism, such as those with GGDEF, EAL or HD-GYP domains (Iyer et al., 2003). Biochemical characterization of prokaryotic H-NOXs has shown that the H-NOX domains from obligate anaerobes are able to bind O2, a property conferred upon them by the presence of a distal pocket tyrosine (Karow et al., 2004, Boon et al., 2006). H-NOX proteins from facultative aerobes lack this tyrosine and do not bind O2 (Karow et al., 2004, Boon et al., 2006, Boon et al., 2005).
To date, all characterized H-NOX domains bind NO tightly with a koff of ~10−3 s-1 (Boon et al., 2006). Assuming a diffusion limited on-rate kon of ~108 M−1 s−1, which has been observed for sGC and is likely to be conserved in the prokaryotic H-NOX proteins (Zhao et al., 1999, Stone and Marletta, 1996), the H-NOX family binds NO with picomolar affinity, Kd ~ 10 × 10−12M. Such tight binding suggests a potential biological function for the H-NOX as an NO receptor. If these H-NOX domains do indeed function as NO receptors, then they are tuned to sense concentrations several orders of magnitude below other characterized prokaryotic NO receptors, such as NorR, which has a nanomolar Kd for NO (D’Autreaux et al., 2008) or NsrR, which regulates gene transcription in response to low micromolar levels of NO (Heurlier et al., 2008). Furthermore, the ability of the H-NOX proteins from facultative aerobes to not bind O2 distinguishes them from other bacterial diatomic gas sensing heme proteins that have this function (Wan et al., 2009, Tuckerman et al., 2009). Thus, a reasonable hypothesis is that prokaryotic H-NOX proteins from facultative aerobes are specific, high-affinity NO receptors that sense picomolar levels of NO and regulate cellular processes. H-NOX mediated NO sensing pathways may allow bacterial cells to adjust or prepare for aspects of the environment that are signified by low concentrations of NO. A hypothetical analogy may be drawn to the NO signaling paradigm of mammalian vascular nitric oxide signaling; a NO generating endothelial or neuronal cell produces a non-toxic, picomolar concentration of NO which is sensed by soluble guanylate cyclase in smooth muscle cells to cause vasodilation (Derbyshire and Marletta, 2009).
Previous work suggests that the H-NOX proteins in facultative aerobic bacteria are indeed involved in the regulation of signaling pathways. In Shewanella oneidensis, Price et al. showed that the H-NOX inhibits the activity of an adjacent histidine kinase in the NO ligated state, but not in the ferrous unligated state (Price et al., 2007). A similar mechanism of regulation has been observed for regulation of a phosphotransfer pathway by Hnox2 in Legionella pneumophila (H. K. Carlson and M. A. Marletta, unpublished results).
In Vibrio fischeri, the H-NOX regulates a phosphotransfer pathway that modulates the colonization efficiency of the bacteria in their consortial symbiotic association with the squid host, Euprymna scolopes. H-NOX mutant strains of Vibrio fischeri hyper-colonize squid hosts 10-fold better than wild-type bacteria. This striking result may be explained by the upregulation of iron uptake genes in the H-NOX mutants, which gives the mutants access to parts of the squid crypts that are iron-limited and limit the growth of wild-type bacteria (Wang et al., 2010). It may be that the V. fischeri H-NOX represses iron-uptake genes because the combination of NO and iron is toxic to the bacterium. In this way, the H-NOX responds to low levels of NO to prime the bacterial cell for higher, more toxic levels of NO.
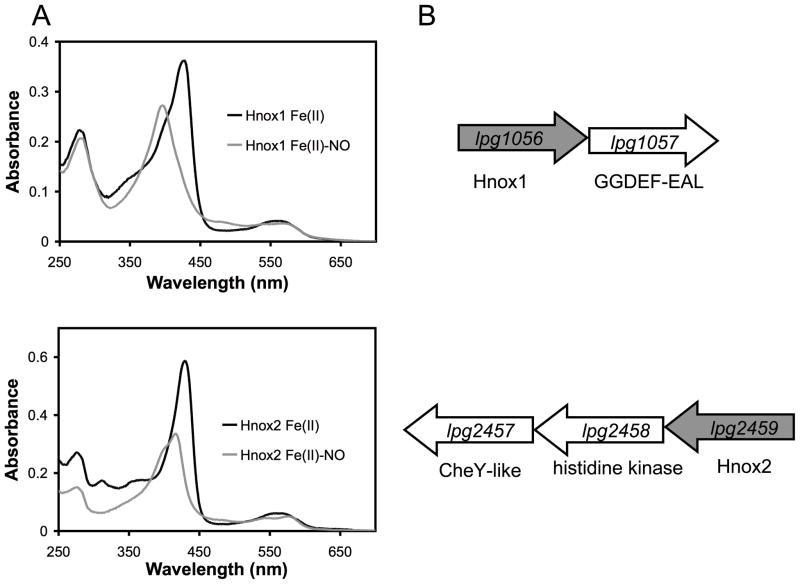
In this report we have focused on Legionella pneumophila and utilized phenotypic experiments to elucidate the role of H-NOX proteins in this organism. As L. pneumophila has two genes coding for H-NOX proteins, it provides a unique opportunity to study the roles of H-NOX proteins in regulating different types of signaling pathways (Figure 1). Both H-NOX proteins in L. pneumophila do not bind O2, but do bind NO (Boon et al., 2006) (and Figure 1A). One of the L. pneumophila H-NOX genes, lpg2459 (hnox2), is adjacent to a histidine kinase, lpg2458, and a CheY-like response regulator, lpg2457. The other H-NOX gene, lpg1056 (hnox1), is adjacent to a GGDEF-EAL protein, lpg1057 (Figure 1B). In the absence of clear phenotypic data, it is difficult to assign a function to CheY-like response regulators (Jenal and Galperin, 2009). These response regulator proteins are known to affect protein localization, function as phosphate sinks, or control chemotactic or chemokinetic responses (Jenal and Galperin, 2009). GGDEF-EAL proteins have a more clearly defined role. These proteins are involved in the metabolism of the bacterial second messenger, c-di-GMP, regulating biofilm formation in a number of bacterial systems (Hengge, 2009). There may be spatial, temporal or functional distribution within the family of GGDEF-EAL proteins, but the processes that they regulate generally lead to an effect on the transition between the biofilm-associated state of a bacterial cell and the planktonic or virulent state (Tamayo et al., 2007, Hengge, 2009).
Figure 1.
(A) Electronic absorption spectra of the Fe(II) and Fe(II)-NO complexes for the two H-NOX proteins from L. pneumophila, showing the Soret and α/β region. Heme concentration was ~5 μm. (B) The predicted H-NOX operons from L. pneumophila. The gene coding for Hnox1 is found adjacent to a diguanylate cyclase gene. The gene coding for Hnox2 is found adjacent to a histidine kinase and single domain response regulator.
From a public health perspective, the mixed species biofilms that form in anthropogenic water systems are well known and important environmental reservoirs for Legionella pneumophila growth (Declerck et al., 2009), and some recent studies have provided insights into biofilm formation by L. pneumophila. It has been shown that L. pneumophila biofilm formation is more robust in rich media compared to minimal media (Mampel et al., 2006), and adheres to surfaces at 25 °C better than at 37 °C. At higher temperatures, L. pneumophila forms longer mycelial mat-like filaments in static cultures (Piao et al., 2006). In the environment, L. pneumophila is found in biofilms in association with other bacteria, as well as protozoa. It has been demonstrated that these interactions with other bacteria and protozoa facilitate the persistence of L. pneumophila in mixed-species biofilms (Mampel et al., 2006). Thus far, the only genes clearly implicated in biofilm formation by L. pneumophila are fliA (Mampel et al., 2006) and the tatB/C genes (De Buck et al., 2005). Genes involved in c-di-GMP metabolism have not been characterized in L. pneumophila.
In this paper, we present evidence that an unmarked, in-frame deletion of the L. pneumophila hnox1 gene results in a hyper-biofilm phenotype, but that neither hnox1 nor hnox2 deletions affect the virulence of L. pneumophila in either amoebae or mouse macrophage infections. Results from the biofilm assays lead us to propose that the H-NOX proteins are likely to be NO bound due to the presence of low nanomolar concentrations of NO present in the rich media used for L. pneumophila growth. In addition, we show that overexpression in L. pneumophila of Vca0956, a well-studied diguanylate cyclase, and overexpression of Lpg1057 both result in hyper-biofilm phenotypes. We also demonstrate that the Lpg1057 protein has diguanylate cyclase activity in vitro that is inhibited by the presence of the H-NOX in the NO ligated state. Finally, we confirm the regulation of Lpg1057 by Hnox1 in vivo through deletion of lpg1057 in the Δhnox1 background and showing that the double mutant Δhnox1 Δlpg1057 displays wild-type levels of biofilm formation. Taken together, these results suggest a role for Hnox1 as a sensitive switch, responsive to picomolar levels of NO, that regulates c-di-GMP metabolism and biofilm formation in Legionella pneumophila.
Results
H-NOX proteins are not required for growth in rich media (BYE), mouse macrophages or Acanthamoeba castellanii
In order to determine the functional role of the H-NOX proteins in L. pneumophila, deletions of both hnox1 (lpg1056) and hnox2 (lpg2459) as well as the double mutant,Δhnox1Δhnox2 were made as described in the Experimental section. The growth kinetics of the H-NOX mutant strains were identical to that of wild-type LP02 in rich media (BYE) (Figure S1), mouse macrophages (Figure S2) or Acanthamoeba castellanii (Figure S3). All strains became motile between an OD of 3.7 and 4.1, and produced the same amount of pyomelanin pigment in late post-exponential phase (Figure S1).
Δhnox1 strains display a hyper-biofilm phenotype in BYE media
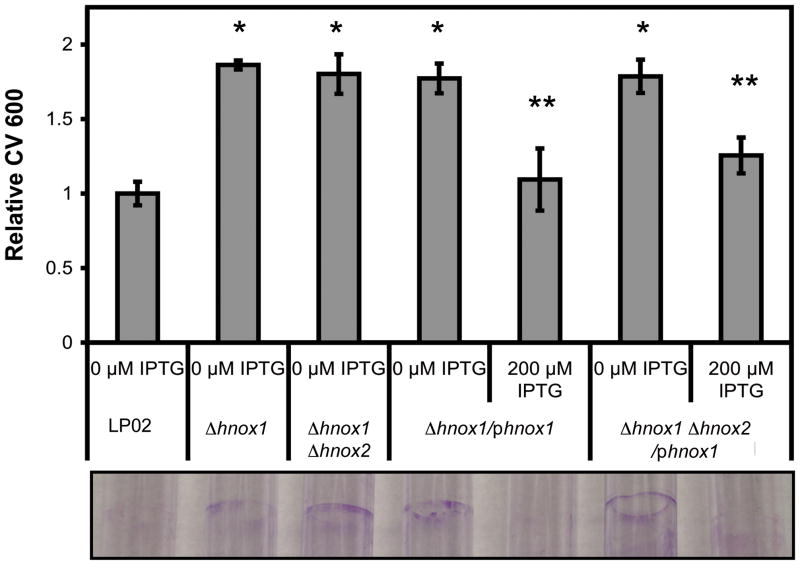
A logical hypothesis for the function of the Hnox1 protein is that it regulates the activity of Lpg1057, the GGDEF-EAL protein adjacent to it in the L. pneumophila genome. GGDEF-EAL proteins are implicated in c-di-GMP metabolism and the regulation of biofilm formation in a number of prokaryotes (Tamayo et al., 2007, Hengge, 2009). In general, bacterial cells with higher levels of c-di-GMP produce more exopolysaccharides, adhere to solid surfaces more efficiently, form thicker biofilms, and are less motile and less virulent (Tamayo et al., 2007, Hengge, 2009). However, in L. pneumophila, only three genes have been reported to regulate biofilm formation, fliA (Mampel et al., 2006), tatB and tatC (De Buck et al., 2005). We compared the growth and biofilm formation of L. pneumophila wild-type and H-NOX mutant strains in static cultures in BYE media. Initial observations of the strains revealed that the Δhnox1 strains consistently formed thicker biofilms. The hyper-biofilm phenotype was quantified using crystal violet staining, and this assay was used to show that strains lacking the hnox1 gene produced ~40% more biofilm mass than wild-type bacteria (Figure 2). Complementation of the Δhnox1 strains by re-introduction of the Δhnox1 gene on a plasmid resulted in a recovery of wild-type levels of biofilm formation upon induction with IPTG (Figure 3). These results clearly implicate hnox1 in the regulation of biofilm formation.
Figure 2.
Biofilm formation by LP02 and H-NOX mutants. A) Optical density was measured at 600 nm (OD 600) to quantify the growth of H-NOX mutant strains and LP02 in static culture in 96 well polystyrene microtiter plates. B) Biofilms were stained with crystal violet and resuspended in 95% ethanol before the absorbance at 600 nm (CV 600) was measured to quantify biofilm production for hnox1 mutant and LP02 in 96 well polystyrene microtiter plates. C) Relative biofilm formation (relative CV 600) at 5 days post-inoculation for all strains in 96 well polystyrene microtiter plates, and D) L. pneumophila was grown without shaking for 5 days in 5 mL polystyrene culture tubes, and then adherent cells (biofilms) were stained with crystal violet. *p-values<0.05 for comparison of Δhnox1 and LP02. **p-values<0.05 for comparison of Δhnox1 Δhnox2 and LP02.
Figure 3.
Complementation of hyper-biofilm phenotype in hnox1 mutants. Biofilms were stained with crystal violet then resuspended in 95% ethanol and the absorbance at 600 nm (CV 600) was measured. Relative CV 600 staining is shown for wild type and mutant strains harboring the pMMB206-GENT vector and the phnox1 vector with and without IPTG induction. Biofilm formation was quantified in 96 well polystyrene microtiter plates (bars) and shown visually in 5 mL polystyrene culture tubes (lower image) at 3 days post-inoculation. *p-values<0.05 for comparison of Δhnox1 or Δhnox1 Δhnox2/pΔhnox1 and LP02. **p-values<0.05 for comparison of uninduced and induced Δhnox1/pΔhnox1 or Δhnox1 Δhnox2/pΔhnox1. Induction of the phnox1 plasmid in LP02 had no effect on biofilm formation (data not shown).
Furthermore, the H-NOX proteins are likely to be NO bound due to the presence of millimolar concentrations of NO3− and low nanomolar concentrations of NO in the rich media used to grow L. pneumophila (Figure S4A). The chemistry responsible for the production of NO in BYE involves ferric iron and L-cysteine that are commonly added as supplements to support L. pneumophila growth (Figure S4B). Direct measurement of NO in growing static cultures of L. pneumophila was not possible. However, it is likely that the oxygen concentration in the static cultures decreases greatly below the ring of growing biofilms at the air-liquid interface. Sealed Hungate culture tubes were used to mimic the microaerobic to anaerobic environment present in static bacterial cultures, thereby providing a closed environment to trap NO. Under these conditions low nanomolar concentrations of NO were produced (Figure S4). Given that the H-NOX proteins likely have picomolar Kds for NO, the absence of the Hnox1 Fe(II)-NO species is likely responsible for the hyper-biofilm phenotype observed in the mutant strains.
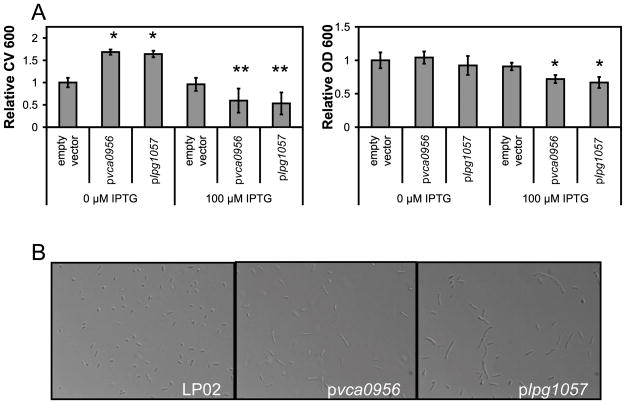
Plasmid expression of diguanylate cyclase proteins in L. pneumophila produces a hyper-biofilm phenotype, but high levels of these proteins inhibits growth
To further investigate the role of c-di-GMP in biofilm formation in Legionella pneumophila, the vector pMMB206-Gent (Hammer and Swanson, 1999, Molofsky and Swanson, 2003) was used to express two diguanylate cyclase proteins in L. pneumophila, Vca0956 (Tischler and Camilli, 2004) and Lpg1057. pMMB206-Gent has a Ptac promoter and is known to have leaky expression when used in L. pneumophila (Molofsky and Swanson, 2003). Therefore, it was not surprising to find that both the pvca0956 and plpg1057 strains displayed hyper-biofilm phenotypes in the absence of IPTG (Figure 4A). Induction with IPTG inhibited growth of the strains, suggesting that higher levels of c-di-GMP are toxic or severely inhibitory on L. pneumophila growth (Figure 4A). In shaken cultures, IPTG induction of pvca0956 and plpg1057 produced more filamentous cells in early post-exponential phase than wild-type (Figure 4B). Also, nucleotide extracts from cultures over-expressing pvca0956 or plpg1057 had higher concentrations of c-di-GMP than vector controls (Figure S5). Filamentous L. pneumophila are observed in late PE phase, or in static cultures grown at 37 °C in the form of mycelial mat-like biofilms (Piao et al., 2006). These observations further support the role of higher c-di-GMP levels in biofilm formation in L. pneumophila, and support the hypothesis that Lpg1057 has dominant diguanylate cyclase activity in vivo unless it is inhibited by the Hnox1 protein.
Figure 4.
Effect of diguanylate cyclase protein expression on L. pneumophila biofilm formation. Growth of static cultures was monitored by measuring the optical density at 600 nm (OD 600). Biofilms were stained with crystal violet and resuspended in 95% ethanol before the absorbance at 600 nm (CV 600) was measured. A) The relative CV 600 and relative OD 600 of static cultures in 96 well polystyrene microtiter plates at 5 days post-inoculation for LP02 harboring pMMB206-GENT pvca0956 or plpg1057. B) 40x magnification images of post-exponential (OD 600 = 4.0) L. pneumophila grown in liquid cultures with shaking and 100 μM IPTG induction. LP02 harboring the pMMB206-GENT vector forms coccoid, motile cells in post-exponential phase, but Vca0956 and Lpg1057 induce filamentous morphology. **p-values<0.05 for CV 600 comparison of LP02 harboring the empty pMMB-206-GENT vector with LP02 harboring pvca0956 or plpg1057. *p-values<0.05 for OD 600 comparison of LP02 harboring the empty pMMB-206-GENT vector with LP02 harboring pvca0956 or plpg1057.
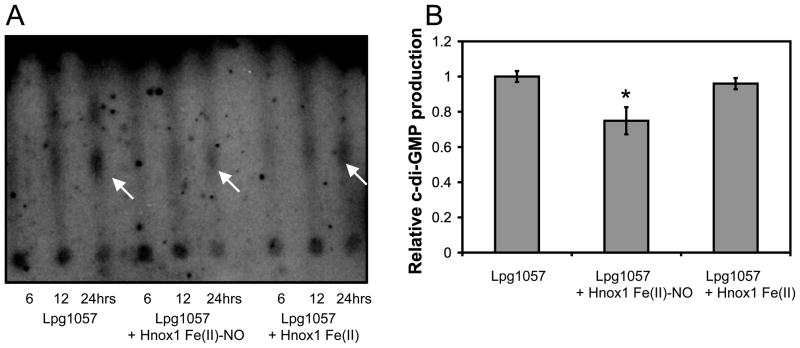
Lpg1057 is an active diguanylate cyclase and is inhibited by Hnox1 Fe(II)-NO
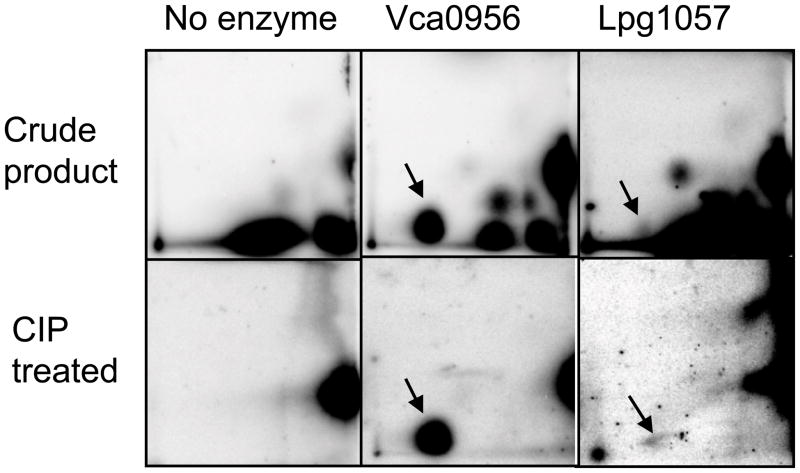
To better understand the role of the Hnox1 protein in the regulation of biofilm formation, Hnox1 and Lpg1057 proteins and Vca0956, an active GGDEF domain containing protein from V. cholerae were expressed and purified (Thormann et al., 2006, Tischler and Camilli, 2004). We found that Lpg1057 was an active diguanylate cyclase, though over 100-fold less active than Vca0956 (Figure 5). The specific activity of the Lpg1057 protein preparation was estimated to be 0.04 ± 0.02 nmol/min/mg based on the quantification of the 24-hour time point, but this value is likely to be a lower limit since the protein concentration had to be estimated (~ 50% pure). If Hnox1 is acting as a NO responsive switch to regulate Lpg1057 in vivo, then it should alter the production of c-di-GMP by the Lpg1057 protein in vitro. Hnox1 Fe(II)-NO inhibited the formation of c-di-GMP by Lpg1057, but Hnox1 Fe(II) unligated had no effect (Figure 6). Unfortunately, due to the low activity of the Lpg1057 protein, 24-hour reaction times were required to see quantifiable c-di-GMP in these assays. The low diguanylate cyclase activity of Lpg1057 may be due to the presence of a c-di-GMP phosphodiesterase, EAL, domain. However, phosphodiesterase assays did not reveal any measurable phosphodiesterase activity (data not shown). Furthermore, the Lpg1057 EAL domain has non-conservative mutations in a conserved active site loop that may render this domain catalytically inactive (Rao et al., 2009). The low activity of Lpg1057 limits further conclusions. However, given the slow growth rate of Legionella pneumophila and the potential for c-di-GMP signaling pathways to be spatially or temporally located (Hengge, 2009), this low activity may be sufficient to explain the hyper-biofilm phenotype of the Δhnox1 strains. Furthermore, the comparison with the highly active Vca0956 may be misleading. Nucleotide extracts from L. pneumophila strains overexpressing Vca0956 had higher concentrations of c-di-GMP compared to strains overexpressing Lpg1057 (Figure S5).
Figure 5.
2-D TLC analysis of GTP turnover to c-di-GMP by Vca0956 and Lpg1057. The top row shows the crude mixture of products after 24 hours at 25 °C for no enzyme, Vca0956 (~1 μM), or Lpg1057 (~ 1 μM) incubated with 500 μM GTP and 10 μCi GTPα32P. Arrows indicate the c-di-GMP spot. The bottom row shows the same reactions after treatment with CIP (Calf intestine alkaline phosphatase). c-di-GMP is resistant to CIP treatment and remains, while other nucleotides are degraded.
Figure 6.
Effect of Hnox1 Fe(II) and Hnox1 Fe(II)-NO on c-di-GMP production by Lpg1057 in vitro. A) 1-D TLC analysis of GTP turnover to c-di-GMP by Lpg1057. Lpg1057 (~ 1 μM) in the presence of Hnox1 (50 μM) in the Fe(II)-NO or Fe(II) unligated states was incubated in sealed anaerobic vials with 500 μM GTP and 10 μCi GTPα32P for 24 hours at 25 °C and then treated with CIP. Arrows indicate the c-di-GMP spot. Spectral analysis of identical reactions without radioactive GTP revealed that less than 10% of the Fe(II) protein was oxidized after 24 hours in the sealed vials, and the Fe(II)-NO complex was stable. B) Relative c-di-GMP production for the Lpg1057 protein alone or in the presence of the Hnox1 Fe(II)-NO or Hnox1 Fe(II). The intensity of the c-di-GMP spots in the phosphorimage were quantified using ImageQuant software (Molecular Dynamics). The error bars represent average values obtained from three independent experiments. *p-values<0.05 for comparison of c-di-GMP production of Lpg1057 with Lpg1057 + Hnox1 Fe(II)NO.
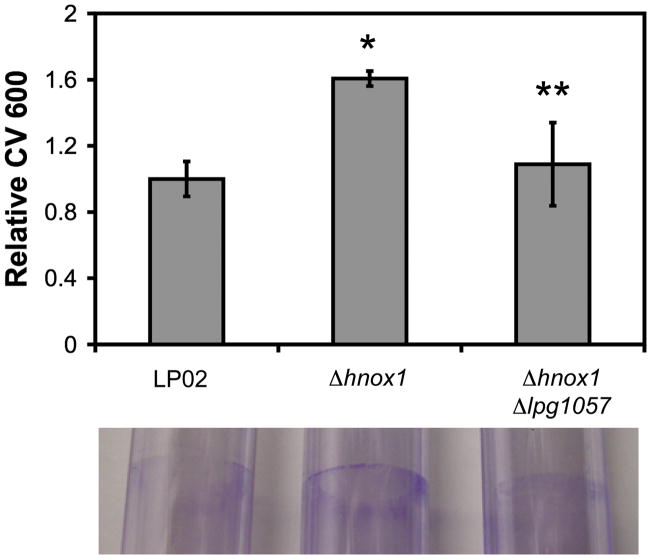
Δlpg1057 strains display wild-type biofilm growth
We hypothesized that the hyper-biofilm phenotype observed in the Δhnox1 strains was due to a lack of inhibition of the diguanylate cyclase activity of Lpg1057 by the H-NOX. To gather support for this hypothesis, deletions of the lpg1057 gene in the Δhnox1 background were made. We found, as expected, that the deletion of the lpg1057 gene reversed the hyper-biofilm phenotype of the hnox1 mutants (Figure 7). This finding clearly demonstrates that Lpg1057 is part of the Hnox1 signaling pathway.
Figure 7.
Biofilm formation by LP02, Δhnox1, and Δhnox1 Δlpg1057 strains. Biofilms were stained with crystal violet then resuspended in 95% ethanol and the absorbance at 600 nm (CV 600) was measured. Relative CV 600 was quantified for 96 well polystyrene microtiter plates (top) and shown visually for crystal violet stained 5 mL polystyrene culture tubes for wild-type LP02 and mutants in the hnox1 signaling pathway at 5 days post-inoculation (bottom). *p-values<0.05 for comparison of LP02 with Δhnox1. **p-values<0.05 for comparison of Δhnox1 with Δhnox1 Δlpg1057.
Discussion
Legionella H-NOX proteins are highly-sensitive, specific NO sensors
Previous studies have found that the H-NOX domains from facultative aerobic bacteria do not form an O2 complex, including the two H-NOX proteins from Legionella pneumophila (Karow et al., 2004, Boon et al., 2006). In contrast, the H-NOX domains from obligate anaerobes form stable and tight complexes with O2, a property conferred upon them by the presence of a distal pocket tyrosine (Boon et al., 2005). The non O2 binding H-NOX domains are distinctive among heme proteins in their ability to bind NO with high affinity in the presence of much higher concentrations of oxygen. The L. pneumophila Hnox1 protein involved in the biofilm phenotype reported here has a koff of ~10−3 s−1 at 20 °C(Boon et al., 2006), and assuming a nearly diffusion limited kon of ~4.5 × 108 M−1 s−1 at 20 °C(Stone and Marletta, 1996, Zhao et al., 1999), the Kd would be in the picomolar range. This high affinity Kd suggests that the H-NOX is likely to be NO bound in the rich media used for L. pneumophila. The media contains millimolar concentrations of NO3-, and low nanomolar concentrations of NO produced by the iron and L-cysteine dependent, heat-catalyzed, inorganic decomposition of the NO3− (Figure S4). Therefore, any phenotypes observed in the mutant strains grown in rich media are likely due to the absence of the H-NOX Fe(II)-NO species. L. pneumophila H-NOX proteins, like all H-NOX domains, also bind CO and can be oxidized. However, in previous studies, CO bound H-NOX domains were found to have a much weaker effect on the regulation of other protein domains compared to NO bound H-NOX (Derbyshire and Marletta, 2009, Price et al., 2007). Furthermore, L. pneumophila does not possess the CO dehydrogenase that has been shown to be responsible for CO production in other bacteria (Roberts et al., 2004). Though the H-NOX domain from S. oneidensis inhibits its associated kinase in the Fe(III) state, it is likely that H-NOX domains are in the Fe(II) state in the reducing environment of the bacterial cell (Price et al., 2007). It is more likely that proteins such as OxyR, which is present L. pneumophila, are responsible for sensing oxidative stress (Christman et al., 1989, LeBlanc et al., 2008).
L. pneumophila could be exposed to NO in a number of ways. Both eukaryotic and bacterial cells produce NO through the action of nitric oxide synthase (NOS) (Agapie et al., 2009, Shatalin et al., 2008, Sudhamsu and Crane, 2009). Denitrifying bacteria also produce NO as an intermediate of this process (Corker and Poole, 2003, Bedmar et al., 2005, Rodionov et al., 2005), but NO3− can also be chemically reduced by Fe(II) in the presence of catalytic amounts of other transition metals, such as Cu(II) (Ottley et al., 1997). The chemical reduction of NO3− is rapid at high temperatures and pressures, and is an important nitrogen sink and likely NO source in extreme environments, such as hydrothermal vents or the early earth (Brandes et al., 1998). In fact, it has been proposed that NO was the first biological terminal electron sink to emerge on the earth before the appearance of molecular oxygen (Ducluzeau et al., 2009). The photolysis of NO3− to NO2− to NO is another an important abiotic source of NO, and this process is at least partly responsible for the halo of low nanomolar NO levels that encircles the planet at the bottom of the photic zone in the oceans (Zafiriou et al., 1980). Bacteria with H-NOX proteins are found in all of these environments, and the H-NOX may be regulating biofilm formation in response to NO in these environments.
Deletions of L. pneumophila hnox genes do not affect growth in liquid culture or phagocytes
A well-studied part of the L. pneumophila cell cycle is the transition from replicative phase during exponential growth in liquid culture or phagocytes to transmissive phase during post-exponential, stationary growth. This transition is regulated by a ppGpp alarmone response to starvation conditions and involves a number of well-studied regulatory genes such as csrA, letA and fliA (Molofsky and Swanson, 2004, Byrne and Swanson, 1998, Hammer and Swanson, 1999). If the H-NOX were regulating any of these genes, it would likely have affected the virulence of the mutant strains, but we did not observe virulence phenotypes for the H-NOX mutants (Figure S2, Figure S3).
A few studies have focused on the potential role for NO in Legionella pneumophila infections. As an intracellular pathogen, L. pneumophila potentially encounters toxic levels of immune-derived NO since iNOS is induced in L. pneumophila infected macrophages (Heath et al., 1996). However, the addition of iNOS inhibitors to permissive A/J macrophages infected with L. pneumophila had little effect on the intracellular growth of the bacteria (Gebran et al., 1994, Yamamoto et al., 1996). In L. pneumophila infected A/J mice, however, the NOS inhibitor, N-methyl-L-arginine, diminished the ability of the mice to clear the bacteria (Brieland et al., 1995). Hence, although NO may not be directly involved in killing intracellular bacteria it may play a role in the immune response to Legionella. NO is clearly toxic to L. pneumophila, and it would be interesting to know if L. pneumophila strains with deletions in the nsrR gene (Rodionov et al., 2005) or the genes coding for NO reductases (Rodionov et al., 2005) are deficient in replication within macrophages or amoebae.
L. pneumophila H-NOX proteins are NO sensitive switches that regulate biofilm formation
Regulation of biofilm attachment, detachment and growth by changes in the concentration of diatomic gases such as NO or O2 has been observed in several bacteria (Barraud et al., 2009, Thormann et al., 2005, Barraud et al., 2006). c-di-GMP metabolism is well established as a regulator of biofilm formation (Hengge, 2009, Tamayo et al., 2007), and it has been proposed that protein domains involved in sensing diatomic gases act as in vivo switches to regulate domains involved in c-di-GMP metabolism (Wan et al., 2009, Tuckerman et al., 2009).
Microarrays show that the L. pneumophila H-NOX genes are upregulated alongside the other proteins in their predicted operons in transmissive phase, as are many other proteins involved in virulence (Bruggemann et al., 2006). Also upregulated in transmissive phase are proteins involved in c-di-GMP metabolism (Bruggemann et al., 2006). These results suggest that more developmental checkpoints may exist in transmissive phase, rather than replicative phase, to determine whether the bacteria will form biofilms or infect phagocytes. In support of this hypothesis, several studies show that transmissive phase L. pneumophila in static cultures adhere to solid surfaces in the initial phase of biofilm attachment better than replicative phase L. pneumophila (Hindre et al., 2008).
Some heme-containing diatomic gas sensors have been shown to regulate diguanylate cyclase and phosphodiesterase domains, but these proteins bind both NO and O2 and their activity is affected by both ligands (Wan et al., 2009, Tuckerman et al., 2009). However, the L. pneumophila H-NOX protein does not bind O2; thus, NO is the most likely physiological ligand. Hence, this study is the first to describe a biofilm phenotype that implicates both a diatomic gas sensor that does not bind O2, the Hnox1, and a protein involved in c-di-GMP metabolism, Lpg1057.
Relatively few L. pneumophila mutant strains are reported to have a biofilm phenotype. To date, only fliA mutants and tatB/C mutants have been shown to have defects in biofilm formation (Hindre et al., 2008, De Buck et al., 2005). Microarrays suggest that genes involved in the response to oxidative stress and iron uptake are up-regulated in biofilms (Hindre et al., 2008), and supplementation of static cultures with excess iron has been shown to inhibit biofilm formation (Hindre et al., 2008). It is highly likely that mutations in many of the genes involved in the metabolism of c-di-GMP will display biofilm phenotypes, but no other studies have analyzed the role of these genes in L. pneumophila.
The results presented in this paper suggest that the Hnox1 protein regulates biofilm formation in response to NO by altering the c-di-GMP production activity of the Lpg1057 protein (Figure 8). This is the first report of biofilm formation being regulated by c-di-GMP in L. pneumophila. Future studies will focus on the role of H-NOX proteins in the regulation of biofilm formation in other bacteria, and how the H-NOX domains may work in concert with other diatomic gas sensor proteins to allow L. pneumophila to persist and survive in environments with varying concentrations of NO.
Figure 8.

A model for the hnox1 pathway in L. pneumophila. The Hnox1 protein inhibits the diguanylate cyclase activity of Lpg1057 in response to NO leading to lower c-di-GMP levels and less biofilm formation. Δhnox1 strains produce more biofilms because c-di-GMP production by Lpg1057 protein is no longer inhibited by Hnox1.
Experimental Procedures
Bacterial strains, culture conditions and reagents
LP02 is a streptomycin-resistant thymidine auxotroph derived from LP01. All mutant strains were made in the LP02 background. L. pneumophila was grown on ACES buffered yeast extract (BYE) charcoal agar plates (BCYE) supplemented with 0.1 g/L thymidine, 0.4 g/L L-cysteine and 0.135 g/L ferric nitrate. Liquid cultures in BYE with supplements were inoculated with patches of L. pneumophila strains. Antibiotics were added to BCYE plates or liquid BYE, when needed at the following concentrations: streptomycin 100 μg/mL, kanamycin, 25 μg/mL, and gentamycin, 10 μg/mL. The optical density at 600 nm (OD 600) of cultures was measured by diluting the cultures 10-fold in BYE in 1 cm plastic cuvettes and reading the OD 600 with an Ultraspec 100 cell density reader (Amersham Biosciences). An OD600 of 3 represents approximately 109 bacteria/mL.
Mutant strain construction
Unmarked in-frame deletions in the genes coding for the two H-NOX proteins and the GGDEF-EAL protein were constructed using previously published methods (Ren et al., 2006, Merriam et al., 1997). Briefly, the flanking regions of the genes to be deleted were cloned into the vector pSR47S and transformed into DH5α λ pir E. coli. For matings, patches of L. pneumophila were mixed with the mating strain containing the knock-out vector and the helper strain 2174. Matings were spread on BCYE plates containing 100 μg/mL streptomycin and 25μg/mL kanamycin to select for the first cross-over event. The second cross-over event was selected for on BCYE plates containing 100 μg/mL streptomycin and 6% sucrose. Colonies that did not grow when patched onto BCYE plates containing 25 μg/mL kanamycin were deletion candidates. These candidates were confirmed by PCR and sequenced with primers flanking the gene of interest to verify the presence of clean in-frame deletions. Multiple isolates of each mutant strain were tested to confirm the phenotypes reported in this paper.
Strains and Plasmids
Strains and plasmids are listed in Table 1. Oligonucleotide primers are listed in Supplemental Table 1. A detailed description of cloning techniques is described in Supplemental Appendix 1.
Table 1.
Strains, plasmids and primers used in this study
| Strains and plasmids | Relevant characteristics | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH5α λ pir | Mating strain | (Merriam et al., 1997, Ren et al., 2006) |
| 2174 | Helper strain | (Merriam et al., 1997, Ren et al., 2006) |
| BL21(DE3) pLysS | Expression strain | (Invitrogen) |
| L. pneumophila | ||
| LP02 | Wild-type L. pneumophila, thymidine auxotroph Smr | |
| Δhnox1 | LP02 Δhnox1 Smr | This work |
| Δhnox2 | LP02 Δhnox2 Smr | This work |
| Δhnox1 Δhnox2 | LP02 Δhnox1 Δhnox2 Smr | This work |
| Δhnox1 Δlpg1057 | LP02 Δhnox1 Δlpg1057 Smr | This work |
| LP02/pJB908 | LP02 pJB908 Smr Ampr | This work |
| Δhnox1/pJB908 | LP02 Δhnox1 pJB908Smr Ampr | This work |
| Δhnox2/pJB908 | LP02 Δhnox2 pJB908Smr Ampr | This work |
| Δhnox1 Δhnox2/pJB908 | LP02 Δhnox1 Δhnox2 pJB908 Smr Ampr | This work |
| LP02/pMMB206-GENT | LP02 pMMB206-GENT Smr Gentr | This work |
| LP02/pvca0956 | LP02 pvca0956 Smr Gentr | This work |
| LP02/plpg1057 | LP02 plpg1057 Smr Gentr | This work |
| Δhnox1/phnox1 | Δhnox1 phnox1 Smr Gentr | This work |
| Plasmids | ||
| pSR47S | sacB Kanr | (Merriam et al., 1997, Ren et al., 2006) |
| pSRhnox1 | psR47S::Δhnox1 sacB Kanr | This work |
| pSRhnox2 | psR47S::Δhnox2 sacB Kanr | This work |
| pSRlpg1057 | psR47S::Δlpg1057 sacB Kanr | This work |
| pJB908 | thymidylate synthetase Ampr | Gift from Prof. Ralph Isberg, Tufts Univ. |
| pMMB206-GENT | pTac promoter Gentr | (Hammer and Swanson, 1999) |
| plpg1057 | pMMB206-GENT::lpg1057 | This work |
| pvca0956 | pMMB206-GENT::vca0956 | This work |
| phnox1 | pMMB206-GENT::hnox1 | This work |
| pET20-b(+) | T7 promoter E. coli expression vector Ampr | Invitrogen |
| pEThnox1 | pET20-b(+)::Hnox1-His6 Ampr | This work |
| pETlpg1057 | pET20-b(+)::Lpg1057-His6 Ampr | This work |
| pAC1758 | pBAD33::Vca0956-His6, Cmr | Gift from Prof. Andrew Camilli, Tufts Univ. |
L. pneumophila Biofilm Formation Assay
L. pneumophila biofilms were generated as described previously with slight modifications (Hindre et al., 2008, Mampel et al., 2006). Overnight cultures of L. pneumophila in exponential phase (OD 600 = 3) were diluted 10x into a final volume of 200 mL fresh BYE in 96-well polystyrene microtiter plates (Costar). Biofilm formation was induced by placing the microtiter plates containing bacteria into a 30 °C incubator and growing static cultures for between 1 and 7 days. Alternatively, 5 mL polystyrene culture tubes were inoculated with 10x dilutions of OD 600 = 3 bacteria and grown for between 1 and 7 days at 30 °C.
Quantification of L. pneumophila Biofilm Formation
L. pneumophila biofilm formation was quantified essentially as described previously (Hindre et al., 2008, Mampel et al., 2006). The optical density of static cultures in the 96 well microplates was determined by reading OD 600 on a SpectraMax M2 optical plate reader (Molecular Dynamics). At the indicated timepoints, the planktonic bacteria were removed and the plate was washed twice with 200 μL BYE per well. Crystal violet (0.3%; 200 μL) was then added to the wells for 10 minutes to stain the biofilms. The excess crystal violet was removed and the wells were washed 3 times with 200 μL water. To quantify biofilm formation, 200 μL of ethanol was added to the wells to resuspend the biofilms and the OD 600 of each well was read on the plate reader. Two-tailed Student’s T-tests were used to analyze the data. P values < 0.05 were considered significant.
Expression and purification of Hnox1, Lpg1057, and Vca0956 proteins
L. pneumophila Hnox1 (Lpg1056) and GGDEF-EAL protein (Lpg1057) were cloned out of genomic DNA and ligated into the pET-20b(+) expression vector (Invitrogen) cut with NcoI and XbaI restriction enzymes (NEB) and then transformed into E. coli DH5a. Positive transformants of all constructs were screened for on LB plates containing 100 μg/mL ampicillin, and the DNA sequences were confirmed by sequencing (Elim Biopharmaceuticals). All proteins were expressed as follows: E. coli BL21(DE3)pLysS cells containing the appropriate plasmid were grown at 37 °C to an OD 600 of 0.6–0.9, induced with 10 μM IPTG, and grown for 16–18 hours at 25 °C. Cells were harvested by centrifugation at 7,000 rpm (6370 xg) for 15 minutes in an Avanti J20I centrifuge with a JLA 8.1 rotor (Beckman), resuspended in lysis buffer (100 mM sodium phosphate, 250 mM NaCl, 5% glycerol, 20 mM imidazole, 1 mM Pefabloc (Roche), pH 7.9) and lysed with a high-pressure homogenizer (Avestin). Lysate was clarified by centrifugation at 42,000 rpm (200,000 xg) for 1 hour in an Optima XL-100K ultracentrifuge with a Ti-45 rotor (Beckman) prior to loading on Ni-NTA agarose resin (Qiagen). The Ni-NTA resin was washed with lysis buffer and eluted with elution buffer (250 mM imidazole in 100 mM sodium phosphate, 250 mM NaCl, 5% glycerol, pH 7.9). For Hnox1, gel filtration chromatography with a S200 26/60 HiLoad Resin column (Pharmacia Biotech) connected to a Biologic HR FPLC was used for further purification. The gel filtration column was equilibrated and run in 50 mM TEA, 50 mM NaCl, 5% glycerol, pH 7.5 buffer. Hnox1 was >95% pure for assays, as determined by SDS-PAGE and Coomassie staining. The diguanylate cyclases used in the assays were approximately 50% pure as assessed by Coomassie staining. Protein concentrations were determined by the Bradford method.
Spectral analysis of Hnox1 and preparation of Fe(II) and Fe(II)-NO
Purified H-NOX protein was desalted into the spectral buffer (50 mM TEA, 50 mM NaCl, pH 7.5) using a PD-10 desalting column (GE Healthcare) in an anaerobic glove bag. The protein was then oxidized with 20-fold molar excess of potassium ferricyanide, and desalted to give the oxidized protein. Oxidized protein was reduced with a 50-fold excess of sodium dithionite, and desalted to give the ferrous unligated species, H-NOX Fe(II). NO was added by providing a 10-fold excess of DEA-NONOate (Cayman Chemical) to the reduced protein preparation, and the protein was desalted to give the ferrous nitrosyl complex, H-NOX Fe(II)-NO. The H-NOX spectra were recorded on a Cary 3E spectrophotometer at 20 °C.
In vitro diguanylate cyclase assays
The activity assay for the GGDEF-EAL protein adjacent to the H-NOX for the formation or degradation of c-di-GMP was done as reported (Tamayo et al., 2005) with some modifications. Due to difficulties in obtaining pure Lpg1057, protein that was approximately 50% pure was used in this assay. For diguanylate cyclase assays, Vca0956 or Lpg1057 (~1 μM) were incubated with 500 μM GTP and 10 μCi GTPα32P for 24 hrs at 25 °C. Products were treated with calf intestine alkaline phosphatase (CIP) (New England Biolabs) for 2 hours. Nucleotides were separated from protein by with a 10K MWCO spin filter and spotted onto PEI-cellulose TLC plates. For 2-D TLCs, the plates were developed first with 0.2 M NH4HCO3, pH 7.5 and then with 10 M KH2PO4, pH 4. For 1-D TLCs, the plates were developed with 10 M KH2PO4, pH 4. Developed TLC plates were exposed to a phosphorimager plates (Molecular Dynamics) and imaged with a Typhoon (Molecular Dynamics) and the intensity of the spots was quantified using ImageQuant software (Molecular Dynamics).
For assays with H-NOX proteins, ~1 μM Lpg1057 and 50 μM Hnox1 were mixed in an anaerobic glove bag (Coy) in sealed vials. A small volume of aerobic GTP was added with a gas-tight syringe (Hamilton) to give a final concentration 500 μM GTP and 10 μCi GTPα32P, and timepoints were removed with the gas-tight syringe between 0 and 24 hrs at 25 °C. Control vials in which no radioactive GTP was added were also prepared, and spectral analysis of the Hnox1 with a Cary 3E spectrophotometer revealed less than 10% of the total Hnox1 Fe(II) was oxidized after overnight reactions, and the Hnox1 Fe(II)-NO complex was completely intact.
Supplementary Material
Acknowledgments
We thank Drs. Michele Swanson (University of Michigan) and Jean-Francois Dubuisson (University of Michigan) for helpful advice, and for conducting early experiments related to this project. We are also grateful to members of the Vance and Marletta labs for helpful advice and critical comments on this manuscript. This work was supported by NIH grants GM070671 (M.A.M.) and AI075039 and AI080749 (R.E.V.) and Investigator Awards from the Burroughs Wellcome Fund and the Cancer Research Institute (R.E.V.).
References
- Agapie T, Suseno S, Woodward JJ, Stoll S, Britt RD, Marletta MA. NO formation by a catalytically self-sufficient bacterial nitric oxide synthase from Sorangium cellulosum. Proc Natl Acad Sci U S A. 2009;106:16221–16226. doi: 10.1073/pnas.0908443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedmar EJ, Robles EF, Delgado MJ. The complete denitrification pathway of the symbiotic, nitrogen-fixing bacterium Bradyrhizobium japonicum. Biochem Soc Trans. 2005;33:141–144. doi: 10.1042/BST0330141. [DOI] [PubMed] [Google Scholar]
- Boon EM, Davis JH, Tran R, Karow DS, Huang SH, Pan D, Miazgowicz MM, Mathies RA, Marletta MA. Nitric oxide binding to prokaryotic homologs of the soluble guanylate cyclase beta1 H-NOX domain. J Biol Chem. 2006;281:21892–21902. doi: 10.1074/jbc.M600557200. [DOI] [PubMed] [Google Scholar]
- Boon EM, Huang SH, Marletta MA. A molecular basis for NO selectivity in soluble guanylate cyclase. Nat Chem Biol. 2005;1:53–59. doi: 10.1038/nchembio704. [DOI] [PubMed] [Google Scholar]
- Brandes JA, Boctor NZ, Cody GD, Cooper BA, Hazen RM, Yoder HS., Jr Abiotic nitrogen reduction on the early Earth. Nature. 1998;395:365–367. doi: 10.1038/26450. [DOI] [PubMed] [Google Scholar]
- Brieland JK, Remick DG, Freeman PT, Hurley MC, Fantone JC, Engleberg NC. In vivo regulation of replicative Legionella pneumophila lung infection by endogenous tumor necrosis factor alpha and nitric oxide. Infect Immun. 1995;63:3253–3258. doi: 10.1128/iai.63.9.3253-3258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006;8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- Byrne B, Swanson MS. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman MF, Storz G, Ames BN. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989;86:3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corker H, Poole RK. Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J Biol Chem. 2003;278:31584–31592. doi: 10.1074/jbc.M303282200. [DOI] [PubMed] [Google Scholar]
- D’Autreaux B, Tucker N, Spiro S, Dixon R. Characterization of the nitric oxide-reactive transcriptional activator NorR. Methods Enzymol. 2008;437:235–251. doi: 10.1016/S0076-6879(07)37013-4. [DOI] [PubMed] [Google Scholar]
- De Buck E, Maes L, Meyen E, Van Mellaert L, Geukens N, Anne J, Lammertyn E. Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem Biophys Res Commun. 2005;331:1413–1420. doi: 10.1016/j.bbrc.2005.04.060. [DOI] [PubMed] [Google Scholar]
- Declerck P, Behets J, Margineanu A, van Hoef V, De Keersmaecker B, Ollevier F. Replication of Legionella pneumophila in biofilms of water distribution pipes. Microbiol Res. 2009;164:593–603. doi: 10.1016/j.micres.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Derbyshire ER, Marletta MA. Biochemistry of soluble guanylate cyclase. Handb Exp Pharmacol. 2009:17–31. doi: 10.1007/978-3-540-68964-5_2. [DOI] [PubMed] [Google Scholar]
- Ducluzeau AL, van Lis R, Duval S, Schoepp-Cothenet B, Russell MJ, Nitschke W. Was nitric oxide the first deep electron sink? Trends Biochem Sci. 2009;34:9–15. doi: 10.1016/j.tibs.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Gebran SJ, Yamamoto Y, Newton C, Klein TW, Friedman H. Inhibition of Legionella pneumophila growth by gamma interferon in permissive A/J mouse macrophages: role of reactive oxygen species, nitric oxide, tryptophan, and iron(III) Infect Immun. 1994;62:3197–3205. doi: 10.1128/iai.62.8.3197-3205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Swanson MS. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- Heath L, Chrisp C, Huffnagle G, LeGendre M, Osawa Y, Hurley M, Engleberg C, Fantone J, Brieland J. Effector mechanisms responsible for gamma interferon-mediated host resistance to Legionella pneumophila lung infection: the role of endogenous nitric oxide differs in susceptible and resistant murine hosts. Infect Immun. 1996;64:5151–5160. doi: 10.1128/iai.64.12.5151-5160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Heurlier K, Thomson MJ, Aziz N, Moir JW. The nitric oxide (NO)-sensing repressor NsrR of Neisseria meningitidis has a compact regulon of genes involved in NO synthesis and detoxification. J Bacteriol. 2008;190:2488–2495. doi: 10.1128/JB.01869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindre T, Bruggemann H, Buchrieser C, Hechard Y. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology. 2008;154:30–41. doi: 10.1099/mic.0.2007/008698-0. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Aravind L. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics. 2003;4:5. doi: 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Galperin MY. Single domain response regulators: molecular switches with emerging roles in cell organization and dynamics. Curr Opin Microbiol. 2009;12:152–160. doi: 10.1016/j.mib.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochemistry. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- LeBlanc JJ, Brassinga AK, Ewann F, Davidson RJ, Hoffman PS. An ortholog of OxyR in Legionella pneumophila is expressed postexponentially and negatively regulates the alkyl hydroperoxide reductase (ahpC2D) operon. J Bacteriol. 2008;190:3444–3455. doi: 10.1128/JB.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mampel J, Spirig T, Weber SS, Haagensen JA, Molin S, Hilbi H. Planktonic replication is essential for biofilm formation by Legionella pneumophila in a complex medium under static and dynamic flow conditions. Appl Environ Microbiol. 2006;72:2885–2895. doi: 10.1128/AEM.72.4.2885-2895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect Immun. 1997;65:2497–2501. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol. 2003;50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- Ottley CJ, Davison W, Edmunds WM. Chemical catalysis of nitrate reduction by iron(II) Geochimica Et Cosmochimica Acta. 1997;61:1819–1828. [Google Scholar]
- Piao Z, Sze CC, Barysheva O, Iida K, Yoshida S. Temperature-regulated formation of mycelial mat-like biofilms by Legionella pneumophila. Appl Environ Microbiol. 2006;72:1613–1622. doi: 10.1128/AEM.72.2.1613-1622.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, Liang ZX. The functional role of a conserved loop in EAL domain-based cyclic di-GMP-specific phosphodiesterase. J Bacteriol. 2009;191:4722–4731. doi: 10.1128/JB.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts GP, Youn H, Kerby RL. CO-sensing mechanisms. Microbiol Mol Biol Rev. 2004;68:453–473. doi: 10.1128/MMBR.68.3.453-473.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, I, Dubchak L, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalin K, Gusarov I, Avetissova E, Shatalina Y, McQuade LE, Lippard SJ, Nudler E. Bacillus anthracis-derived nitric oxide is essential for pathogen virulence and survival in macrophages. Proc Natl Acad Sci U S A. 2008;105:1009–1013. doi: 10.1073/pnas.0710950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JR, Marletta MA. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry. 1996;35:1093–1099. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- Sudhamsu J, Crane BR. Bacterial nitric oxide synthases: what are they good for? Trends Microbiol. 2009;17:212–218. doi: 10.1016/j.tim.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Tischler AD, Camilli A. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem. 2005;280:33324–33330. doi: 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann KM, Saville RM, Shukla S, Spormann AM. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol. 2005;187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol Microbiol. 2004;53:857–869. doi: 10.1111/j.1365-2958.2004.04155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman JR, Gonzalez G, Sousa EH, Wan X, Saito JA, Alam M, Gilles-Gonzalez MA. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry. 2009;48:9764–9774. doi: 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- Wan X, Tuckerman JR, Saito JA, Freitas TA, Newhouse JS, Denery JR, Galperin MY, Gonzalez G, Gilles-Gonzalez MA, Alam M. Globins synthesize the second messenger bis-(3′–5′)-cyclic diguanosine monophosphate in bacteria. J Mol Biol. 2009;388:262–270. doi: 10.1016/j.jmb.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci U S A. 2010;107:8375–8380. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Klein TW, Friedman H. Immunoregulatory role of nitric oxide in Legionella pneumophila-infected macrophages. Cell Immunol. 1996;171:231–239. doi: 10.1006/cimm.1996.0198. [DOI] [PubMed] [Google Scholar]
- Zafiriou OC, McFarland M, Bromund RH. Nitric Oxide in Seawater. Science. 1980;207:637–639. doi: 10.1126/science.207.4431.637. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Brandish PE, Ballou DP, Marletta MA. A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc Natl Acad Sci U S A. 1999;96:14753–14758. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.