Abstract
In this study, we assess on a large scale the possibility of deriving self-inhibitory peptides from protein domains with globular architectures. Such inhibitory peptides would inhibit interactions of their origin domain by mimicking its mode of binding to cognate partners, and could serve as promising leads for rational design of inhibitory drugs. For our large-scale analysis, we analyzed short linear segments that were cut out of protein interfaces in-silico in complex structures of protein-protein docking Benchmark 3.0 and CAPRI targets from rounds 1-19. Our results suggest that more than 50% of these globular interactions are dominated by one short linear segment at the domain interface, which provides more than half of the original interaction energy. Importantly, in many cases the derived peptides show strong energetic preference for their original binding mode independently of the context of their original domain, as we demonstrate by extensive computational peptide docking experiments. As an in-depth case study, we computationally design a candidate peptide to inhibit the EphB4-EphrinB2 interaction based on a short peptide derived from the G-H loop in EphrinB2. Altogether, we provide an elaborate framework for the in-silico selection of candidate inhibitory molecules for protein-protein interactions. Such candidate molecules can be readily subjected to wet-lab experiments and provide highly promising starting points for subsequent drug design.
Keywords: inhibitory peptides, derived peptides, protein-protein interactions, peptide docking, Rosetta FlexPepDock, mimicry peptides
Introduction
Protein-protein interactions (PPIs) mediate and regulate a majority of cellular processes. Due to their central role, these interactions have been the subject of intensive research, providing attractive targets for the inhibition and manipulation of specific pathways. Two modes of interaction are particularly prevalent, namely interactions between two globular domains and interactions between a globular domain and a flexible linear peptide1.
Peptide-protein interactions have been acknowledged as key mediators of PPIs, predominantly in signaling and regulatory networks1. Whereas in globular PPIs the two domains are structured prior to binding, these interactions are characterized by a short flexible peptide that binds to a globular protein receptor, often folding only upon binding1,2. Such peptides may bind independently, or as linear motifs within larger intrinsically disordered domains3. Only recently has sufficient amount of structural data become available to conduct large-scale analyses of peptide-protein interactions4,5 and computational approaches for the modeling of these interactions have emerged (e.g., 6-9; reviewed in 10,11).
In this study, we examine on a large scale the possibility of deriving inhibitory peptides from globular domains as a general mechanism to inhibit their cognate interactions. Such peptides could serve as promising leads for the rational design of inhibitory drugs. There are several advantages in targeting drugs at protein-protein interfaces rather than at enzyme active sites - most importantly, blocking these interactions provides a way to modulate signaling activity without harming the intrinsic catalytic activity of the targets12. However, designing small molecule inhibitors of protein–protein interactions is considered difficult, due to challenges such as the size of protein-protein interfaces (PPIs are large13,14), as well as the lack of well-defined binding pockets (PPIs are relatively flat15). Nevertheless, recent years have seen important progress and several success stories in this field16. The use of peptides as leads for the inhibition of protein-protein interactions has recently gained substantial interest, and inhibitory peptides have been designed for several important therapeutics targets17-20. Some of these therapeutic peptides are no other than linear stretches of amino-acids derived from one of the cognate globular partners in the interactions (e.g. Angiotensin II receptor antagonists mimicking the Angiotensinogen terminus21, and glycoprotein IIb/IIIa receptor blockers mimicking Integrin motifs22). These derived peptides might originate from loops within globular domains, from disordered regions at inter-domain linkers or from protein termini.
While it seems only natural to derive inhibitory peptides from linear motifs within intrinsically disordered regions, deriving peptides from interfaces of globular proteins is possibly more complicated. First, the interface architecture is often formed by several binding modules which are brought together by the tertiary topology of the globular domain23,24. Therefore, it cannot be taken for granted that any single linear segment derived from a globular protein would be able to mimic the binding mode of the origin domain. Moreover, the large size and flat surface13-15 of most globular interfaces may hamper competitive binding by derived inhibitory peptides. On the other hand, protein binding is often mediated by a small number of residues, usually termed “hotspots”, which make the major contribution to binding at the interface25. Therefore, designing inhibitory peptides to target “hotspot” residues might be feasible nevertheless. The derived peptides are more likely to be specific to their target (relative to small molecules taken from molecular screening), since they were derived from the native interface. In addition, the smaller size of the peptide compared to the protein confers advantage in competitive binding thanks to increased effective peptide concentration at the interface, resulting in increased effective affinity16. Peptides derived from native protein sequences were able to inhibit interactions of their origin domain in several examples, including MDM2–p5326, Bcl-xL–Bak27 and MD2-TLR428.
To what extent then could peptides extracted from a globular protein monomer be used to inhibit the interaction to its partner? In order to effectively compete with their origin domains for native interactions, the derived peptides are expected to bind their target with comparable (or increased) affinities, while utilizing a similar binding mode. In this work, we first screened for high-affinity linear segments on interfaces of globular proteins (using the Rosetta modeling framework29), which could be used to derive candidate inhibitory peptides. Subsequently, we used peptide docking experiments and energy funnel analysis, using our recently developed protocol for flexible peptide docking (FlexPepDock6), to assess the binding of these peptides to the same binding site independently of the context of their original domain. The results suggest that most of the peptides will indeed adopt a similar structure even when cut out of their protein context. We conclude with an in-depth case study, where we use our methodology to extract and design a cyclic inhibitory peptide for the EphB4-EphrinB2 interaction, which is involved in pathological forms of angiogenesis and in tumorigenesis30. Altogether, we present here an elaborate framework for in-silico selection of candidate inhibitory molecules for a multitude of protein interactions, which can in turn be readily subjected to wet-lab experiments and subsequent lead optimization.
Results
Interactions between globular proteins are often dominated by a single high-affinity linear peptide segment
As a first step, we examine a large dataset of protein interactions for the prevalence of high-affinity linear peptide segments on globular protein domains that participate in the protein-protein interface. Such peptides would have the potential to compete with their origin domains for the same interactions. Our dataset comprises 27 docking targets from the CAPRI competition rounds 1-1931 (Supp. Table S1), and 124 interactions from the non-redundant docking Benchmark 3.032 (Supp. Table S2; partially overlapping with the CAPRI set). We have created a computational tool that extracts all possible linear peptide segments from the interface of each domain, and evaluates the binding energy of each derived peptide to the other domain, using the Rosetta energy function29 (see Methods). For each interaction in the Benchmark 3.0 dataset and the CAPRI targets from rounds 1-19, we apply this tool to identify a low-energy linear segment, namely the decamer* linear peptide segment with the lowest absolute interaction energy estimate (see Methods). For comparison, we also estimate the energy of the original interaction between the two globular domains.
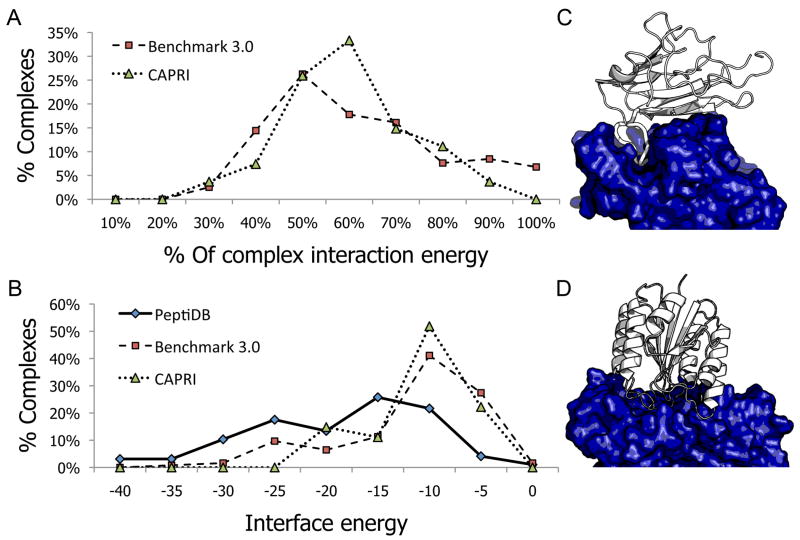
Figure 1A shows the distribution of the relative binding energy of these low-energy linear segment peptides (i.e. the percentage of the interaction binding energy of the decamer, relative to the globular protein-protein interaction binding energy). In 57% of the protein complex structures in Benchmark 3.0, and in 63% of the CAPRI set, a single peptide derived from a linear segment at the interface was estimated to contribute over half of the interaction energy of the entire complex (Average contributions of these high affinity segments (>50%) were 70%±14% and 60%±9% in Benchmark 3.0 and CAPRI, respectively). Figures 1C and 1D illustrate different types of peptide mediated interactions: In Figure 1C, the interaction is mediated by one single linear segment from EphrinB2 that binds to its partner EphB4 with 65% of the total interaction energy (-19.5 Rosetta energy units; REU; pdb: 2HLE30). In contrast, in Figure 1D no single linear segment contributes the majority of the binding energy (the best decamer peptide binds with 43% of the total interaction energy: -8.5 REU; pdb: 1T6B33).
Figure 1. Most protein-protein interactions are mediated by one dominant linear peptide at the interface.
A.The distribution of the relative interaction energy contribution (% of total interaction energy) of the highest-affinity decamer at the interface, for complexes from the docking Benchmark 3.032 (squares) and CAPRI31 (triangles) data sets. For roughly 50% of the complexes, a single linear segment contributes more than half of the interaction energy. See Methods for more details about the estimation of interaction energies. B. The corresponding distribution of absolute interaction energy values of the highest-affinity decamers (in Rosetta energy units; REU). For reference, the distribution of absolute interaction energy is given for a dataset of solved structures of peptide-protein interactions (peptiDB4; diamonds). A large number of the proposed peptides have energetic values that are comparable to known peptide interactions. C and D. Examples of protein interactions that are dominated by a single linear peptide, vs. interactions that are not. C. In the complex of Ephrin type-b receptor 4 kinase (EphB4; dark blue) and EphrinB2 (white) (pdb: 2HLE30), most of the interaction energy is contributed by a linear segment at the interface of EphrinB2 (cartoon, widened). D. In the complex of b. Anthracis protective antigen (dark blue) and human anthrax toxin receptor (white) (pdb: 1T6B33), the interface is composed of several joined loops. No single linear segment contributes a significant fraction of the interaction energy.
Absolute binding energies of derived peptides are comparable with native peptide interactions
A plot of the absolute values of interaction energies (see Methods) indicates that a significant fraction of the peptides derived from linear segments with the highest affinity (both from Benchmark 3.0 and the CAPRI dataset) contribute more than -10, and in some cases even more than -15 REU. Such values overlap with interaction energies typically calculated for native peptide-protein interactions in the peptiDB dataset4 (Figure 1B). In general, this suggests that in many cases the use of a single linear segment extracted from one of the protein partners is a good strategy to obtain binding peptides, which in turn can provide starting points for the design of high affinity competitive inhibitors of globular PPIs. Supp. Tables S1 and S2 list the highest affinity decamers at the interface and their predicted contribution to binding for each of the complexes in the CAPRI and Docking Benchmark 3.0 datasets.
Secondary structure preferences of derived peptides
Analysis of the secondary structures content of high-affinity linear segments from Benchmark 3.0 reveals that 76% of the peptides display no defined secondary structure (coil; see Supp. Figure S1A), 20% form an α-helix, and only 4% complement a β-sheet. This under-representation of β-sheets (as compared to 18% in native peptide-mediated interactions from peptiDB4) might be explained by the geometric constraints enforced by the globular fold of the parent protein. The relative energy contribution to the interface for peptides assuming different secondary structures is similar (Supp. Figure S1B).
High-affinity linear segments are prevalent in different classes of protein interactions
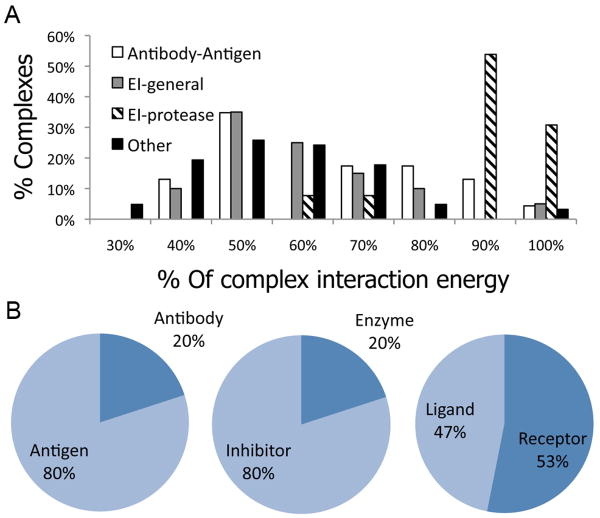
Figure 2A shows the distribution of relative binding energies for the highest-affinity linear segment in the different sub-classes of Benchmark 3.0 (Enzyme-Inhibitor - EI, Antibody-Antigen, and Others). The results indicate that for all classes of interactions, a substantial fraction of complexes obtain the majority of their binding energy by a single linear segment at their interface. The subset of enzyme inhibitors that comprises Trypsin- and Subtilisin–inhibitor complexes (EI-protease; accounting for 35% of the EI class) have a remarkably high tendency to utilize such linear segments for both Benchmark 3.0 (Figure 2A) and CAPRI targets (data not shown), contributing most of the peptides that bind with more than -25 units in absolute energy terms. Examples for additional cases where a derived peptide covers most of the interaction with a predicted strong binding affinity include (1) The interaction between HemK and Release Factor 1: a decamer in RF1, AGGQHVNTTD, provides an estimated binding energy of -21.8 REU, which amounts to 47% of the total estimated binding energy of this interaction (Capri Target 20; pdb: 2BT334), and (2) The interaction between UCL-H3 deubiquitinase and ubiquitin: a decamer in ubiquitin, (TLHLVLRLRG), provides an estimated binding energy of -26.6 REU, which amounts to 59% of the total estimated binding energy (pdb: 1×D335).
Figure 2. Derived peptides in different types of interfaces.
A. The distribution of relative binding energies of the highest affinity decamer at the interface (see Figure 1) is shown for different classes of interactions (see legend). Dominating linear peptides at the interface appear in particular in the subclass of Trypsin- and Subtilisin enzyme-inhibitor complexes (EI-proteases). Nonetheless, significant contributions by one dominant interface peptide are also observed for all other classes of protein interactions. B. Which side contributes the highest-affinity decamer? High-affinity linear segments show an asymmetric distribution in the Antibody-Antigen and Enzyme-Inhibitor classes of interactions, where in 80% of the complexes the high-affinity peptide is derived from the Antigen or Inhibitor, respectively. In the ‘Other’ class in turn, no particular preference for any of the partners is observed.
Preferred location of highest-affinity linear segments in Antibody-Antigen and Enzyme-Inhibitor interactions
In 80% of the Antibody-Antigen and Enzyme-Inhibitor complexes, the linear peptide segment with the predicted highest binding affinity is preferentially located on one particular side of the protein complex, namely on the antigen and inhibitor, respectively (Figure 2B). Only in 20% of the complexes it is derived from the antibody or the enzyme. For Antibody-Antigen complexes, this finding may sound obvious considering that antibodies use three complementarity determining region (CDR) loops to bind the antigen with high affinity and specificity. The antigen in turn can provide various forms of linear structural epitopes that are recognized by antibodies, although interestingly, we could identify dominant linear peptides in several non-continuous epitopes. As for Proteases, these proteins have been designed and selected for binding a protein stretch and to cut a peptide bond. It is therefore but natural that inhibitors would use the same strategy of binding as the protein substrate would. No such asymmetric tendencies are observed among the remaining complexes in the ‘Other’ class (Figure 2B).
Peptide docking experiments: do derived peptides favor their native binding conformation?
In the previous section, we have demonstrated that many protein interactions are dominated by a linear peptide segment, and suggested that such a peptide provides a good starting point for deriving potential inhibitory peptides. However, it is not clear whether such peptides would still assume their original binding mode after being cut out of their origin domain. Namely, inhibitory peptides are expected to lie in the minimum of an energy funnel in their original binding pockets.
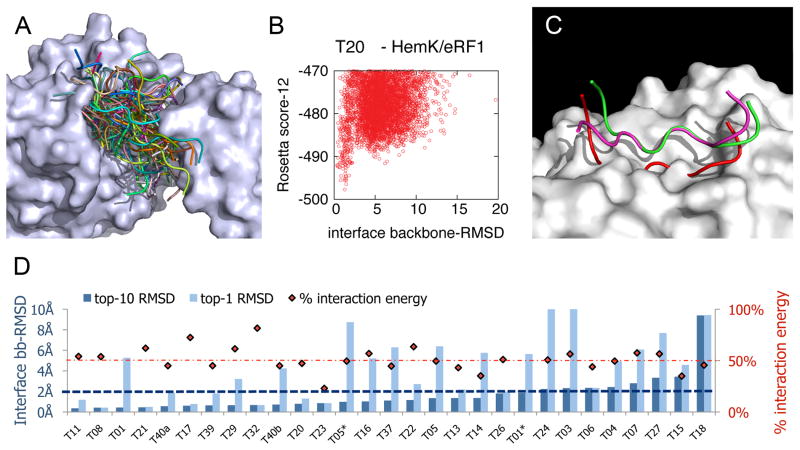
We have recently developed Rosetta FlexPepDock6, a high-resolution peptide modeling protocol that is designed to dock peptides to their receptor with full peptide backbone flexibility and side-chain modeling, starting from an initial description of the complex. We used our modeling protocol to aggressively sample the energy landscape of the derived peptide around the original binding pocket (see Methods). For each derived peptide, we created 9,000 models of its complex with the cognate target of its origin domain. The different models cover a wide range of peptide conformations around the original binding pocket, deviating up to well over 15Å backbone RMSD from the original native structure (Figure 3A). Due to the intensive nature of this test, we have restricted this analysis to 29 peptides derived from the 27 CAPRI interactions. We assume however, that the results are representative, since distribution of the binding energy values of derived peptides are very similar for the CAPRI targets and Benchmark 3.0.
Figure 3. Derived peptides favor the native binding conformation observed in the full protein.
A. Sampling of a wide range of conformations allows accurate mapping of the energy landscape. Example of sampled range of peptide conformations for CAPRI target 20, showing 100 representative models of a docked peptide derived from RF1 (ribbon presentation in different colors; out of the total 9000) bound to HemK (surface representation). B. Energy funnel for RF1-derived peptide - HemK docking. Each model is presented as a dot, indicating its total energy (y-axis) vs. the backbone RMSD of the peptide interface residues to the peptide in its native protein context (x-axis). For clearness, only top-scoring models out of the total 9000 are shown. See Supp. Figure S2 for energy funnel plots of peptides derived from 13 additional CAPRI targets. C. Example of successful docking simulations on peptides derived from the Plasminogen receptor antibody/Plasminogen activator receptor complex (pdb: 2FD6). In this example, docking starts from the unbound conformation of the derived peptide in its native context. More examples can be found in Supp. Figure S3. The native peptide is colored green, the peptide in its unbound form (as part of the unbound monomer) is colored red, and the model created with the FlexPepDock protocol is colored magenta. D. Mapping the energy landscapes of peptides derived from CAPRI targets indicates that derived peptides favor the original binding conformation for a large number of targets. For each interaction, we show the interface backbone RMSD of the lowest-energy model created for the derived peptide (dark blue bar), as well as for the best out of the 10 top scoring models (light blue bar). Red diamonds indicate for each target the fraction of binding energy that the derived peptide contributes (a dotted line is drawn at 50%). In 72% of the interactions (21 out of 29), a near-native conformation (below 2Å backbone-RMSD over peptide interface residues) is found among the top-10 models. In 35%, of the interactions, this is the top-scoring model. We note that we also calculated all-atom RMSD values for top-scoring models: these exceed backbone RMSD values on the average by 0.55±0.40Å. Thus, top-scoring models well below 2Å all-atom RMSD were created for a large fraction of the targets.
Figure 3B illustrates the energy landscape of a representative peptide derived from CAPRI target 20, describing a distinct funnel with a near-native energy well (additional energy funnels are shown in Supp. Figure S2). Overall, in most of the interactions (21 out of 29; 72%), a near-native conformation (below 2Å backbone-RMSD over peptide interface residues) was found among the 10 top-scoring models (Figure 3D). In 35% of the interactions, a near-native conformation was even obtained by the top-scoring model (see also Supp. Figure S3).
In an additional experiment, we performed docking simulations starting from the peptide conformation in the free monomer, for all complexes in docking Benchmark 3.0 (see Methods). In 76% of these protein interactions, we were able to identify a near-native peptide conformation (<2Å interface backbone-RMSD). This is not surprising, considering that 71% of Benchmark 3.0 is classified as “rigid-body docking”, indicating that no major backbone conformational changes occur in the protein monomers upon binding32 (Supp. Figure S4A). However, even in some difficult cases, where the structure of the derived peptide changes significantly between the unbound and the bound conformations, lowest-energy models in the docking simulation were near the native bound conformation (representative results are shown in Figure 3C and Supp. Figures S4B-C). Our docking experiments thus indicate that for many protein interactions, derived peptides have the potential to compete with the domain from which they originated by competing for the same binding mode.
Case study – design of a cyclic inhibitory peptide of the EphB4-EphrinB2 interaction
As a proof-of concept of our strategy, we chose to use our proposed strategy to design an inhibitor for the EphB4-EphrinB2 interaction. We first derived a linear peptide with predicted strong binding ability to the partner from the interface, and then proceeded towards the design of a high-affinity binding inhibitory peptide of potential therapeutic value using two different strategies, namely the design of a peptide sequence with improved binding affinity, and the cyclization of the peptide to improve its stability.
The interaction of the EphB4 receptor tyrosine kinase with its preferred ligand, EphrinB2, is implicated in pathological forms of angiogenesis, as well as in tumorigenesis30. Stimulation of EphrinB2 signaling by EphB4 was shown to promote tumor growth in vivo36, and EphB4 expression is up-regulated in several types of cancer37,38. Furthermore, disruption of this interaction (e.g. by the soluble EphB4 ligand binding domain) inhibits tumor growth in murine tumor xenograft models39. Accumulating data thus suggests that this interaction is a prime drug target candidate.
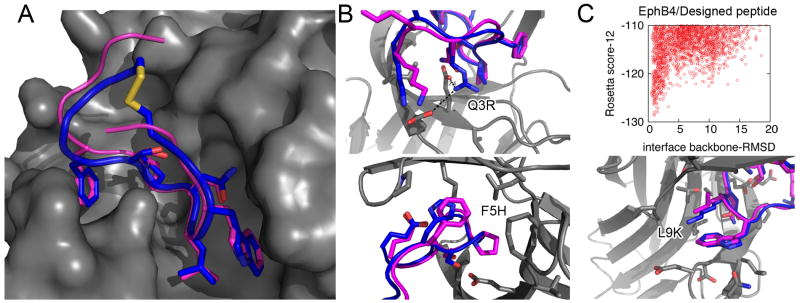
We analyzed the solved structure of the EphB4-EphrinB2 complex (pdb: 2HLE30) using our protocol to screen its interface for highest-affinity linear segments. We were indeed able to spot a dominant decamer in the G-H loop of EphrinB239 (Figure 1C). Manual inspection of the complex structure and the network of interactions at the interface (Supp. Figure S5) revealed that this identified decamer peptide in the G-H loop can be extended to a longer, 13-residue peptide that contributes 70% of the total interaction energy (residues 116-128 of EphrinB2; -20.9 REU; see Figure 4A). We next used our FlexPepDock peptide docking protocol6 to simulate the binding mode of the derived peptide to EphB4 and to sample the energy landscape around the binding pocket (along the lines described above; see Methods). Indeed, a steep energy funnel leads the derived peptide towards the original bound conformation of the G-H loop in its original context (Supp. Figure S6). Altogether, our protocol suggests the peptide derived from residues 116-128 of EphrinB2 as a good candidate for the inhibition of the EphB4-EphrinB2 interaction. We now show on this example case, how a derived peptides can be used as lead for designing more efficient inhibitors.
Figure 4. Design of an inhibitory peptide derived from the EphB4-EphrinB2 interaction.
A-B. The interaction of the EphrinB2 native G-H loop (residues 116-128; magenta) and of a proposed inhibitory designed peptide (blue) with EphB4 (gray). A. A docking model of the suggested S-S cyclized inhibitory peptide (blue) suggests that the stabilizing S-S bond can be introduced into the peptide without affecting the general binding mode or compromising the major binding determinants (0.75Å RMSD over all heavy atoms of residues 120-126). B. Models of the three proposed mutations (Q3R, F5H, L9K). C. The energy landscape of the designed EphrinB2 derived peptide shows a pronounced energy funnel leading towards the native binding mode. See also Supp. Figure S6.
a) Design of optimally binding sequence
Backbone flexibility has been demonstrated to improve the performance of computational sequence design40. We therefore used a set of backbone conformations produced by the peptide docking runs as starting points to design peptides with increased affinity towards EphB4 (see Methods). Three possible mutations were selected by combination of lowest scoring models and manual inspection: Q3R, F5H and L9K in the peptide ranging from 116-128 in EphrinB2 (see Figure 4B). According to our model, the Q3R mutation positions the new Arginine residue between the EphB4 residues Glu43 and Glu59. The F5H mutation might stabilize the interaction with the carboxy-terminal Glu13 residue of the peptide, while maintaining existing contacts with EphB4 Pro151. Finally, the L9K mutation keeps the hydrophobic interactions of the Leucine residue, but in addition adds a charge that points into the solvent, thereby improving the solvation energy of the peptide in its bound form, as well as possibly its cell penetration properties. As these mutations are spatially distinct, they can potentially be combined to achieve even higher binding affinity. Our docking experiments indicated that indeed a steep energy funnel leads the mutated peptide towards the native binding mode (Figure 4C).
b) Stabilization of peptide by S-S cyclization
Two hydrophobic residues 2 and 12 of the proposed inhibitory peptide face the interior of EphrinB2 within the context of the full protein, but face the solvent in the model created for the derived form (see Figure 4A). The spatial proximity of these residues provides optimal candidates for cyclization of the peptide. Peptide cyclization offers several advantages, such as improved stability17,41, protection from proteases41, and increased affinity due to reduced entropy of the unbound state42. We therefore mutated in-silico positions 2 and 12 to Cystine to introduce a disulfide bond. This cyclic peptide was then docked onto EphB4. Figure 4A displays the top-scoring model of this docking run. Strikingly, the main interactions that mediate the binding are fully accessible also to the cyclic peptide, and the key side-chain conformations at the interface are perfectly recovered (0.75Å RMSD over all heavy atoms of residues 120-126), in spite of the strong constraint imposed by cyclization. We should note here that since the binding mode is conserved, most sequence mutations proposed above are likely to also stabilize the interaction with the cyclic peptide (except for the F5H mutation, due to slightly different backbone orientations imposed by the cyclization, F5 is rather close to C2). Further cycles of sequence design and structure optimization can combine these two strategies together.
Discussion
Protein-protein interactions mediated by either globular domains or by short peptides are recognized to provide major components of the complex network of interactions within a cell, and therefore major efforts have been made to understand the basic principles that underlie these interactions. In the present study, we have investigated the possibility of deriving peptides from the interface of globular proteins to design inhibitors that would compete with their native interaction.
Hot linear binding segments
Our finding that short linear segments on the interface of globular proteins contribute in many interactions the main part of the overall binding affinity has profound implications not only on how these interactions could be inhibited in a targeted way by derived peptides, but also to the understanding of how protein-protein interactions are achieved in Nature. Previous studies that have investigated the structural nature of globular protein-protein interfaces and characterized the atomic details of the interactions in terms like size and shape (e.g. references 13 and 14), showed that a small number of so-called hotspot residues in this interface contribute the majority of the binding energy25. Linear binding motifs, on the other hand, are usually thought of within the context of recognition motif within disordered regions3. Here, we show that such “hot” linear segments play also a major role in the architecture of globular protein interfaces, including in non-trivial examples such as non-continuous antigen epitopes, where one would not necessarily expect to find a single dominant linear segment a-priori. It is also interesting to investigate how these high-affinity linear segments fit into the modular view of protein-protein interfaces 24,43 (see Supp. Figure S5 for two representative examples, namely the interfaces in TEM1-BLIP and EphB4-EphrinB2 interactions).
From high-affinity segments to inhibitory peptides
If linear segments often provide such a significant contribution to the protein-protein interaction, could they provide a basis for the design of short inhibitory peptides or molecules? It is assumed that free peptides do not in general adapt one preferred conformation44, and competitive binding to globular proteins requires that the correct conformation would be adopted upon binding. Our energy landscape analysis via docking experiments indicates that high-affinity segments indeed favor their original conformation within the context of the entire protein within some considerably large basin of attractions (Figure 3 and Supp. Figure S2). This makes them worthy candidates for inhibitory peptides, or at least promising leads for designing small inhibitory molecules and peptides with increased affinity, stability and bio-availability (see further discussion below). Notably, these candidates come from all categories of protein-protein interactions, and are not restricted to e.g., enzyme-inhibitor or antibody-antigen complexes (Supp. Tables S1 and S2).
Stabilization of lead peptides
In the Results section, we have provided an elaborate case-study example for deriving an inhibitory molecule from the EphB4/EphrinB2 interface. We subjected the lead peptide to subsequent computational sequence design and structural cyclization, in order to increase its stability and binding affinity towards its target. While it has been suggested that thanks to their smaller size, the effective binding ability of peptides compared to proteins is increased16, it is presumed that peptide binding affinity is decreased due to the loss of entropy upon binding. The stabilization of the peptide in its bound conformation suggested in this study by e.g., closing it to a cyclic peptide through a disulfide bond (thereby reducing the entropy of the unbound form42) may further shift the relative binding affinity between the protein and the derived peptide to the partner in favor of the peptide. It is worth noting that cyclic peptides also have many other advantages in terms of bioavailability and cell permeability17,41.
The road from inhibitory peptides to therapeutic drugs
The ability of peptides derived from a protein to bind independently to the target of the protein has been demonstrated for several cases26-28. This indicates that derived peptides can indeed be used as potential inhibitors of the original interactions, or as starting points for the design of more potent inhibitors41. Even though concerns have been raised regarding the binding affinity of such peptides (reviewed by Wells et al.16), and consequently their ability to inhibit the existing interaction, several recent progresses in drug design are based on the optimization of such peptides to bind with higher affinity to their target11, and their design for higher bio-availability and resistance to proteolysis 17,41,45. For instance, peptide-like molecules, such as peptoids46 may preserve the critical structural features of an inhibitory peptide, but are resistant to endogenous proteases. Another concern that might arise in the biological context is that in specific systems such as antibody-antigen complexes, the very binding of the peptide to its target receptor might elicit the unwanted e.g., immune response, instead of blocking it (although initial studies regarding synthetic vaccines indicate otherwise47). These issues should be resolved on a per system basis.
Underlying assumptions of this study and their implications
We note that this study is based on several assumptions to reach its conclusions. We assume that the binding energy of protein-protein, and peptide-protein interactions in particular, can be estimated by calculating the binding energy as the difference between the partners in their bound and isolated states, using the Rosetta Energy function. We further assume that the free backbone conformation is identical to the bound conformations, thereby ignoring the entropic effect of peptide folding upon binding48. For this reason, a more efficient inhibitory peptide might exist that was not detected due to entropic effects, and the same may apply for the sequence design procedure. Even though our assumptions might not provide accurate estimates, they can nevertheless provide initial guides and insights.
We recently showed that the Rosetta energy function is appropriate for creating high-resolution models of native peptide-mediated complexes6 from the peptiDB dataset4. Moreover, in the current study we found that a significant number of derived peptides are predicted to bind with similar estimated binding energies as the peptides from the PeptiDB dataset (Figure 1B). In many cases, they indeed correspond to peptides in regions suggested to contribute significantly to an interaction (e.g., the inhibitory loops of Trypsin and Subtilisin inhibitors, or the G-H loop in EphrinB2 that was previously identified as a critical component of the interaction with EphB439; see Results). Therefore, in spite of some gross simplifications made in this large-scale analysis, the suggested derived peptides are expected indeed to represent good candidates for competitive binding to the original protein partners, without their protein context.
Conclusions
In this study we have aimed to lay the ground for structured derivation of inhibitory peptides from interfaces of globular proteins. While it had been reported previously that linear peptides dominate the interaction of specific protein-protein interactions such as Trypsin – inhibitor complexes, this analysis represents to the best of our knowledge the first large-scale attempt to characterize the contribution of a single linear peptide in the interface to the affinity of binding between two proteins, the structural propensity of this peptide to adopt the bound conformation also without the context of the protein using docking experiments, and finally, the proceeding of such a peptide lead towards the computational design of inhibitory peptides of increased stability and affinity.
Methods
Characterization of interface peptides
We developed a simple protocol for the selection and evaluation of protein-protein interface-derived peptides. Given the complex structure of an interaction between proteins A and B, we first minimize shortly the structure using the Rosetta standard energy function (score12)29,49, to remove local clashes without changing the structure significantly. The protocol then slides a 10 amino acid window along the protein chain. We extract each fragment of length 10 from its protein context, add termini charges, and then estimate the interaction energy of this peptide with the other partner (see below). We select the peptide that contributes the best interaction energy to represent this interaction (the peptide can be located in either of the two protein partners). Here we analyze 10mers, but the same protocol can be applied to different peptide lengths.
Estimation of binding energy
In this study, we provide a coarse estimate of binding energy by evaluating the interface energy, defined as the energy of a peptide in complex with the protein partner compared to the energy of peptide and protein alone. The binding energy for a peptide derived from protein A to partner protein B is calculated as ΔΔGpepA-protB = ΔGpepA-protB - ΔGpepA - ΔGprotB. We select the peptide that contributes the best ΔΔGpepA-protB (or ΔΔGprotA-pepB) value to represent this interaction. The relative contribution of this peptide to the total binding energy is obtained by comparing its binding energy, ΔΔGpepA-protB, to the estimated binding energy of the full protein complex, ΔΔGprotA-protB = ΔGprotA-protB - ΔGprotA - ΔGprotB. This rough estimate is used for initial filtering of candidate inhibitory peptides.
Flexible peptide docking
Flexible peptide docking simulations were performed using FlexPepDock6. In short, this Rosetta protocol consists of iterative optimization of rigid body and peptide backbone Monte Carlo with Minimization (MCM) cycles, where side chain conformations are optimized on the fly, and the VdW attractive and repulsive terms are gradually decreased and increased, respectively, to their final weights in the Rosetta standard scoring function score1229,49. For a given starting conformation (e.g. a peptide derived from the free monomer conformation), 200 models were created using this protocol. For energy landscape mapping, half of the models were generated with the low-resolution pre-optimization mode of the protocol, to increase sampling range (see Raveh et al.6 for more details). We note that no information about the native side chain conformation was included in any of the docking simulations, to avoid bias towards the native structure.
Mapping the peptide binding energy landscape
For each derived peptide from the CAPRI dataset, a series of random perturbations was applied both in rigid body and in backbone torsion angle space, along the lines of the protocol described in Raveh et al.6. These perturbations create for each peptide-protein complex 45 random starting models of increasing RMSD (typically up to 15Å away from the native peptide) and backbone deviations (up to 100° per backbone torsion). Each of these starting models were subjected to a standard FlexPepDock simulation (see above). A plot of energy vs. RMSD of these 9000 models describes the near-native energy landscape of the peptide-protein interaction (see Figure 3B and Supp. Figure S2).
Supplementary Material
Acknowledgments
We thank the CAPRI organizers and the experimentalists that provide CAPRI targets for providing this stimulating platform to test and develop improved modeling protocols and new concepts. Protein structure figures were created by PyMOL (http://www.pymol.org).
Footnotes
We report here the analysis of peptides of length tenin order to be able to capture α-helices at the interface. Note however, that we repeated this analysis for peptides with varying lengths (5-10) with similar results (data not shown).
References
- 1.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300(5618):445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 2.Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr Opin Biotechnol. 2008;19(4):344–350. doi: 10.1016/j.copbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6(6):2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18(2):188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Vanhee P, Reumers J, Stricher F, Baeten L, Serrano L, Schymkowitz J, Rousseau F. PepX: a structural database of non-redundant protein-peptide complexes. Nucleic Acids Res. 2010;38(Database issue):D545–551. doi: 10.1093/nar/gkp893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raveh B, London N, Schueler-Furman O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins: Structure, Function, and Bioinformatics. 2010;78:2029–2040. doi: 10.1002/prot.22716. [DOI] [PubMed] [Google Scholar]
- 7.Antes I. DynaDock: A new molecular dynamics-based algorithm for protein-peptide docking including receptor flexibility. Proteins. 78(5):1084–1104. doi: 10.1002/prot.22629. [DOI] [PubMed] [Google Scholar]
- 8.Niv MY, Weinstein H. A flexible docking procedure for the exploration of peptide binding selectivity to known structures and homology models of PDZ domains. J Am Chem Soc. 2005;127(40):14072–14079. doi: 10.1021/ja054195s. [DOI] [PubMed] [Google Scholar]
- 9.Bordner AJ, Abagyan R. Ab initio prediction of peptide-MHC binding geometry for diverse class I MHC allotypes. Proteins. 2006;63(3):512–526. doi: 10.1002/prot.20831. [DOI] [PubMed] [Google Scholar]
- 10.Yaneva R, Schneeweiss C, Zacharias M, Springer S. Peptide binding to MHC class I and II proteins: new avenues from new methods. Mol Immunol. 47(4):649–657. doi: 10.1016/j.molimm.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Rubinstein M, Niv MY. Peptidic modulators of protein-protein interactions: progress and challenges in computational design. Biopolymers. 2009;91(7):505–513. doi: 10.1002/bip.21164. [DOI] [PubMed] [Google Scholar]
- 12.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13(3):284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 13.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285(5):2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 14.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93(1):13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fletcher S, Hamilton AD. Targeting protein-protein interactions by rational design: mimicry of protein surfaces. J R Soc Interface. 2006;3(7):215–233. doi: 10.1098/rsif.2006.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 17.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov Today. 2010;15(1-2):40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Murray JK, Gellman SH. Targeting protein-protein interactions: lessons from p53/MDM2. Biopolymers. 2007;88(5):657–686. doi: 10.1002/bip.20741. [DOI] [PubMed] [Google Scholar]
- 19.Gaestel M, Kracht M. Peptides as signaling inhibitors for mammalian MAP kinase cascades. Curr Pharm Des. 2009;15(21):2471–2480. doi: 10.2174/138161209788682299. [DOI] [PubMed] [Google Scholar]
- 20.Marchioni F, Zheng Y. Targeting rho GTPases by peptidic structures. Curr Pharm Des. 2009;15(21):2481–2487. doi: 10.2174/138161209788682334. [DOI] [PubMed] [Google Scholar]
- 21.Brunner HR, Gavras H, Laragh JH. Angiotensin II blockade in man by SAR1-ALA8 angiotensin II for understanding and treatment of high blood pressure. Lancet. 1973:1045–1049. doi: 10.1016/s0140-6736(73)92657-3. [DOI] [PubMed] [Google Scholar]
- 22.Hashemzadeh M, Furukawa M, Goldsberry S, Movahed MR. Chemical structures and mode of action of intravenous glycoprotein IIb/IIIa receptor blockers: A review. Exp Clin Cardiol. 2008;13(4):192–197. [PMC free article] [PubMed] [Google Scholar]
- 23.Pal A, Chakrabarti P, Bahadur R, Rodier F, Janin J. Peptide segments in protein-protein interfaces. J Biosci. 2007;32(1):101–111. doi: 10.1007/s12038-007-0010-7. [DOI] [PubMed] [Google Scholar]
- 24.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 26.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323(3):491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 27.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275(5302):983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 28.Slivka PF, Shridhar M, Lee GI, Sammond DW, Hutchinson MR, Martinko AJ, Buchanan MM, Sholar PW, Kearney JJ, Harrison JA, Watkins LR, Yin H. A peptide antagonist of the TLR4-MD2 interaction. Chembiochem. 2009;10(4):645–649. doi: 10.1002/cbic.200800769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das R, Baker D. Macromolecular modeling with Rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 30.Chrencik JE, Brooun A, Kraus ML, Recht MI, Kolatkar AR, Han GW, Seifert JM, Widmer H, Auer M, Kuhn P. Structural and biophysical characterization of the EphB4*ephrinB2 protein-protein interaction and receptor specificity. The Journal of biological chemistry. 2006;281:28185–28192. doi: 10.1074/jbc.M605766200. [DOI] [PubMed] [Google Scholar]
- 31.Janin J. Assessing predictions of protein-protein interaction: the CAPRI experiment. Protein Sci. 2005;14(2):278–283. doi: 10.1110/ps.041081905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santelli E, Bankston LA, Leppla SH, Liddington RC. Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- 34.Graille M, Heurgue-Hamand V, Champ S, Mora L, Scrima N, Ulryck N, Tilbeurgh Hv, Buckingham RH. Molecular basis for bacterial class I release factor methylation by PrmC. Molecular Cell. 2005;20:917–927. doi: 10.1016/j.molcel.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 35.Misaghi S, Galardy PJ, Meester WJN, Ovaa H, Ploegh HL, Gaudet R. Structure of the Ubiquitin Hydrolase UCH-L3 Complexed with a Suicide Substrate. Journal of Biological Chemistry. 2005;280:1512–1520. doi: 10.1074/jbc.M410770200. [DOI] [PubMed] [Google Scholar]
- 36.Noren NK, Lu M, Freeman AL, Koolpe M, Pasquale EB. Interplay between EphB4 on tumor cells and vascular ephrin-B2 regulates tumor growth. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(15):5583–5588. doi: 10.1073/pnas.0401381101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W, Ahmad SA, Jung YD, Reinmuth N, Fan F, Bucana CD, Ellis LM. Coexpression of ephrin-Bs and their receptors in colon carcinoma. Cancer. 2002;94:934–939. doi: 10.1002/cncr.10122. [DOI] [PubMed] [Google Scholar]
- 38.Berclaz G, Karamitopoulou E, Mazzucchelli L, Rohrbach V, Dreher E, Ziemiecki A, Andres AC. Activation of the receptor protein tyrosine kinase EphB4 in endometiral hyperplasia and endometrial carcinoma. Ann Oncol. 2003;14:220–226. doi: 10.1093/annonc/mdg072. [DOI] [PubMed] [Google Scholar]
- 39.Kertesz N, Krasnoperov V, Reddy R, Leshanski L, Kumar SR, Zozulya S, Gill PS. The soluble extracellular domain of EphB4 (sEphB4) antagonizes EphB4-EphrinB2 interaction, modulates angiogenesis, and inhibits tumor growth. Blood. 2006;107:2330–2338. doi: 10.1182/blood-2005-04-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandell DJ, Kortemme T. Backbone flexibility in computational protein design. Current Opinion in Chemical Biology. 2009;11:329–334. doi: 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Witt KA, Gillespie TJ, Huber JD, Egleton RD, Davis TP. Peptide drug modifications to enhance bioavailability and blood-brain barrier permeability. Peptides. 2001;22(12):2329–2343. doi: 10.1016/s0196-9781(01)00537-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhou HX. Loops, linkages, rings, catenanes, cages, and crowders: entropy-based strategies for stabilizing proteins. Acc Chem Res. 2004;37(2):123–130. doi: 10.1021/ar0302282. [DOI] [PubMed] [Google Scholar]
- 43.Reichmann D, Cohen M, Abramovich R, Dym O, Lim D, Strynadka NCJ, Schreiber G. Binding hot spots in the TEM1-BLIP interface in light of its modular architecture. Journal of Molecular Biology. 2007;365:663–679. doi: 10.1016/j.jmb.2006.09.076. [DOI] [PubMed] [Google Scholar]
- 44.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 45.Sillerud LO, Larson RS. Design and structure of peptide and peptidomimetic antagonists of protein-protein interaction. Current protein & peptide science. 2005;6:151–169. doi: 10.2174/1389203053545462. [DOI] [PubMed] [Google Scholar]
- 46.Simon RJ, Kania RS, Zuckermann RN, Huebner VD, Jewell DA, Banville S, Ng S, Wang L, Rosenberg S, Marlowe CK, et al. Peptoids: a modular approach to drug discovery. Proc Natl Acad Sci U S A. 1992;89(20):9367–9371. doi: 10.1073/pnas.89.20.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen SW, Van Regenmortel MH, Pellequer JL. Structure-activity relationships in peptide-antibody complexes: implications for epitope prediction and development of synthetic peptide vaccines. Curr Med Chem. 2009;16(8):953–964. doi: 10.2174/092986709787581914. [DOI] [PubMed] [Google Scholar]
- 48.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 49.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.