Abstract
Skeletal muscle differentiation relies on the coordinated activation and repression of specific subsets of genes. This reflects extensive changes in chromatin architecture, composition of chromatin-associated complexes and histone modifications at the promoter/enhancer elements of skeletal muscle genes. An early, key event in the activation of muscle-specific gene transcription is the disruption of the repressive conformation imposed by nucleosomes, which impede the access of pioneer transcription factors, such as the muscle-specific basic helix–loop–helix (bHLH) factors MyoD and Myf5, to their DNA-binding sites. This review focuses on our current understanding of the role of the SWI/SNF ATP-dependent chromatin-remodeling complex in the activation of the myogenic program, by inducing conformational changes permissive for muscle-gene expression. Recent findings suggest that specific combinations of individual SWI/SNF components can generate sub-complexes with specialized functions that are engaged at sequential stages of muscle-gene activation — e.g. initial displacement of the nucleosome followed by the loading of the complete myogenic transcriptosome that promotes gene transcription. SWI/SNF composition and function is regulated by the exchange of specific variants of structural sub-units. In turn, an exchange of histone variants and related epigenetic modifications might reflect the impact of distinct SWI/SNF complexes on the architecture and activity of target promoter/enhancer elements. Thus, the SWI/SNF complexes should be regarded not just as simple executors of the program imposed by transcription factors, but as multifaceted “readers” and “shapers” of the chromatin/DNA landscape within target muscle genes along the transition from myoblasts to myotubes.
Keywords: Skeletal myogenesis, Transcription, Chromatin, Epigenetics, SWI/SNF
Introduction
During cellular differentiation, the activation of tissue- and lineage-specific genes coincides with the disruption of the repressive conformation imposed by nucleosomes at previously silent loci. This task is achieved by the combinatorial activities of tissue-specific transcription factors and multi-sub-unit machineries that decompose the chromatin by disassembling and reassembling the nucleosomes [1]. For instance, during skeletal muscle differentiation the myogenic bHLH proteins MyoD and Myf5, which are expressed in myoblasts prior to the differentiation, face the challenge of gaining access to their binding sites (the so-called Eboxes-CANNTG) on muscle genes, by overcoming the unfavorable conformation of repressive chromatin [2]. Physical and functional interactions between muscle bHLH proteins and ATP-dependent SWI/SNF chromatin-remodeling complexes [3,4] promote the displacement of nucleosomes and generate the landscape conducive for the activation of transcription, by favoring the access of muscle bHLH proteins and MEF2 factors, and the assembly of the myogenic transcriptosome. The alternative presence of two catalytic sub-units, either Brg1 or Brm, confers on the SWI/SNF complex the ATPase activity necessary to remodel the chromatin [5]; moreover, different combinations of non-enzymatic, structural sub-units, such as the Brg1/Brm-associated factors (BAFs), can generate different sub-complexes, thereby providing the molecular variability by which SWI/SNF complexes select the interactions with different tissue-specific transcription factors and with other chromatin-modifying enzymes [6,7]. Thus, the specific composition of SWI/SNF sub-complexes determines their activity and specificity for particular subsets of genes. An exchange of SW/SNF variant sub-units at key transitions of the myogenic program might generate sub-complexes dedicated to specific tasks, such as facilitating the access of pioneer transcriptional activators and “reading” the combinations of histone marks generated by the activity of muscle-specific transcription factors and components of the epigenetic machinery.
Transcriptional control by chromatin remodelers during skeletal muscle differentiation
The muscle regulatory factors (MRFs) belonging to the bHLH family — MyoD, Myf5, MRF4 and myogenin — activates and perpetuates the skeletal myogenic program by promoting the gradual expression of muscle-specific genes in myoblasts [8]. This task is achieved with the contribution of other transcription factors, such as the MEF2 family members, which are expressed ubiquitously and cooperate with muscle bHLH proteins to activate muscle-gene transcription in myoblasts [9]. Recent evidence supports the existence of a network of functional interactions between different transcription factors induced by muscle bHLH proteins, MEF2 proteins and chromatin-modifying complexes that regulate the global and gene-specific chromatin conformation during the transition of myoblasts to myotubes [10–12].
The activation of muscle gene expression by muscle regulatory factors (MRFs)
The coordinated expression of muscle genes is ensured by the temporal regulation of MRF expression. Myf5 and MyoD are expressed in myoblasts prior to their differentiation, and are responsible for the myogenic determination of precursor cells and for the activation of downstream target genes, including the other muscle bHLH proteins, myogenin and MRF4 [13]. The cross-activation among muscle bHLH factors amplifies the repertoire of MRFs engaged to activate the expression of contractile, metabolic and structural muscle genes. For instance, at early gene promoters MyoD alone is sufficient for a full gene expression, whereas late muscle gene transcription requires the sequential binding of MyoD and myogenin, together with specific MEF2 isoforms [10,11,14]. This process culminates with the fusion of myoblasts into multinucleated myotubes, which are competent to perform contractile and metabolic functions.
Muscle bHLH proteins heterodimerize with ubiquitously expressed E-proteins (the products of the E2A gene) to productively bind the Ebox sequences in the regulatory region of many muscle-specific genes [15]. Activation of muscle gene expression by myogenic proteins requires the activity of a large variety of chromatin-modifying complexes, including the ATPase-dependent SWI/SNF chromatin-remodeling complexes on regulatory regions of target genes [12]. This process is regulated by the intracellular signaling pathways elicited by differentiation cues that control the composition of the complexes associated to muscle bHLH and MEF2 proteins on the chromatin of target genes [12].
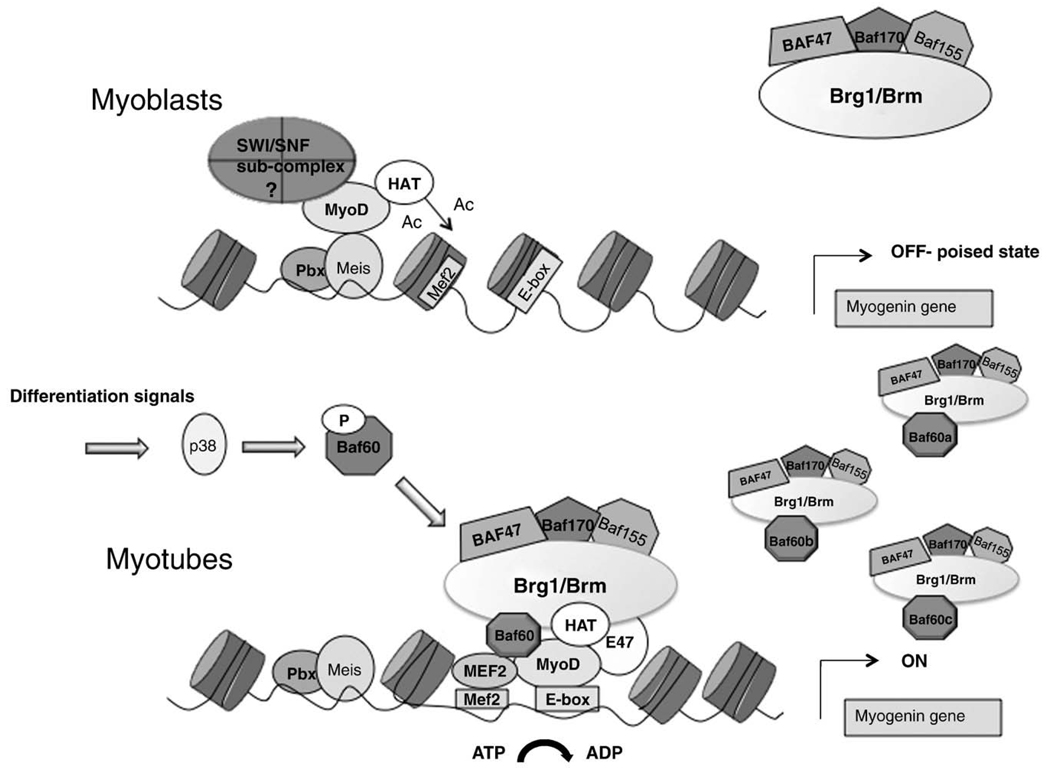
One intriguing and still unsolved issue relates to the mechanism by which pioneer MRFs recognize the binding sequences within the regulatory elements of their target genes within the repressive chromatin. In particular, MRFs face the challenge imposed by the nucleosomes that impede the access to cis-regulatory sequences in promoters of inactive genes, to preclude the inadverted expression of muscle differentiation genes in undifferentiated myoblasts. As such, the competition between nucleosomes and MRF muscle progenitors ensures the temporal coordination of gene expression at the transition from undifferentiated to terminally differentiated muscle cells. Nucleosomes typically cover the regions flanking the transcription start site (TSS) within the promoter of these regulated genes [1]. Thus, it is important to understand the mechanism by which the barrier imposed by nucleosomes is overcame by pioneer MRFs to initiate transcription at previously silent loci. Since at least one binding site in the promoter region of these genes might be partly exposed and thereby permits the access to pioneer transcription factors, an effort is currently being made to determine the identity and the mechanism of action of those transcription factors, which prime muscle loci for activation in skeletal myoblasts. While MyoD and Myf5 are the most obvious candidates, given their expression pattern and associated remodeling activities [16] that distinguish them from the other muscle bHLH factors, it is still unclear whether they can directly gain access to canonical Eboxes located on promoters covered by nucleosomes. Alternatively, pioneer MRFs can access their target sequences via binding to non-canonical Eboxes or through the association with other transcription factors that recognize DNA-binding sites that are exposed. An interesting model has been proposed in which the formation of a Pbx1/MyoD complex on a non-canonical Ebox in the distal region of myogenin promoter [17] permits the initial recruitment of chromatin remodelers that might contribute to nucleosome displacement and allow the subsequent recruitment of muscle bHLH and MEF2 proteins with the full complement of associated histone-modifying and chromatin-remodeling complexes. This model has been substantiated in MyoD-converted fibroblasts, in which Pbx1/MyoD complex might induce an early hyperacetylation of the distal region of the myogenic promoter, independent on Brg1 recruitment, but possibly via recruitment of a yet unidentified complex. These modifications displace the nucleosome and permit the subsequent binding of muscle bHLH and MEF2 proteins with an associated Brg1-containing SWI/SNF complex [18]. According to this model, MyoD (or other pioneer MRFs) initiates the chromatin modifications at target genes, prior to the recruitment of the epigenetic machinery necessary for activation of transcription (Fig. 1). Moreover, SWI/SNF activity is required to maintain the expression of muscle genes in myotubes [19]. This multistep mechanism of gene activation suggests that different chromatin-modifying complexes can perform specific tasks within the process of muscle-gene activation during skeletal myogenesis. In the following paragraphs, we will summarize the current evidence supporting the existence of different SWI/SNF sub-complexes that might be involved in the remodeling of the chromatin at different stages of the differentiation program.
Fig. 1.
The myogenin locus (which is representative of other muscle loci) is typically silent in myoblasts, due to the repressive conformation of the chromatin. A critical step in the activation of previously silent loci is the disruption of the nucleosome, which imposes a physical barrier to the promoter access of the pioneer muscle determination factors — MyoD or Myf5 — which possess the unique property of penetrating the repressive chromatin in muscle genes. This task is probably achieved by a sequence of events initiated by the indirect interaction of MyoD (or Myf5) with non-canonical Eboxes adjacent to the nucleosome. Pbx/Meis is one proposed mediator of this interaction, which results in the partial displacement of the nucleosome via engagement of histone acetyltransferases (HAT) and SWI/SNF sub-complexes not necessarily endowed with ATPase activity. The consequence of this initial event is the exposure of the canonical binding sites for muscle bHLH and MEF2 factors that allow the loading of the myogenic transcriptosome containing a full repertoire of chromatin remodelers. Different combinations of tissue-specific BAFs and ATPase variants might generate a number of different SWI/SNF sub-complexes with specialized functions and possibly gene-specific activity—see possible combinations of BAF60-based SWI/SNF complexes.
Networks in chromatin dynamics: histones and remodelers
How complicated is the task that MRFs face to change the architecture of target genes and initiate transcription?
In eukaryotic organisms, both the length and complexity of the genome impose a highly compacted and dynamic chromatin organization. The fundamental unit by which DNA is packaged is a 147 bp long stretch of DNA wrapped twice around an octamer of the core histones H2A, H2B, H3 and H4. These arrays of particles called nucleosomes are separated by a 20–50 bp of unwound linker DNA H1 [20]. This basic module can vary in the composition of histone variants and can be modulated by specific post-translational modifications of histone tail. The nucleosome represents the first level of compaction of DNA. The N-terminal tails of the four core histones protrude from the compact particle to contact DNA, other histones, and non-histone proteins, thus forming high order structures and specialized chromatin domains. Modifications of the chromatin architecture, such as DNA methylation, histone post-translational modifications, histone exchange and ATP-dependent chromatin remodeling allow a prompt response to environmental signals and control specific developmental programs. Chromatin modifiers can alter the histone–DNA contact by covalent modification or by ATP hydrolysis. In developmentally regulated genes (e.g. lineage- and tissue-specific genes), characterized by high nucleosome occupancy near the transcription start site [1], changes in gene expression require the action of ATP-dependent chromatin remodeling enzymes to promote the sliding or eviction of the nucleosomes and the exposure of binding sites for tissue-specific transcriptional activators. Among the ATP-dependent modifiers, the SWI/SNF family contributes to the activation of muscle genes during skeletal myogenesis, and the ATPase activity of Brg1 and Brm is essential for gene activation by myogenic transcription factors [3,4].
Moreover, SWI/SNF complexes might have an additional and more specific role. They may actively participate in the focal and global chromatin re-organization that occurs in myoblasts prior to their differentiation into myotubes. In this regard, increasing evidence indicates functional and physical interactions between SWI/SNF complex, myogenic bHLH factors and other histone-modifying complexes to control nucleosome position and accessibility at the transition from myoblasts to myotubes.
SWI/SNF interactions with other histoneodifying complexes and the chromatin reonfiguration at muscle loci during myoblast differentiation
Since SWI/SNF complexes can promote both gene activation and repression by nucleosome disruption and reconstruction [5], it is formally possible that they mediate both processes to coordinate gene expression and repression during skeletal myogenesis. While current data indicate a prominent role of SWI/SNF in the activation of muscle-gene expression, the physical and functional interactions reported in other cell types between SWI/SNF and other chromatin modifiers (see below) suggest a more complicated picture.
The ability of MyoD to promote myogenic conversion when ectopically introduced in non-muscle cells reflects the concerted activity of the associated epigenetic machinery that remodels the chromatin in the regulatory regions of target gene [16]. Of note, the composition of this machinery is extremely heterogeneous and dynamic, with a stage-specific association of distinct enzymatic complexes that execute different functions in myoblasts vs myotubes. In undifferentiated myoblasts, muscle-specific gene activation is precluded by the recruitment of transcriptional corepressors, such as histone deacetylases (HDACs) and histone methyltransferases, which contribute to generate the epigenetic marks associated with silent chromatin — H3–K9 dimethylation, H3K27 trimethylation and local hypoacetylation [12]. Upon differentiation cues, the chromatin at muscle promoters undergoes massive structural changes that reflect the elimination of HDACs and repressive methyltransferases, such as Suv39h1 and Polycomb complex [21–23], in favor of the recruitment and functional activation of positive regulators, such as histone acetyltransferases (HATs), arginine methyltransferases and SWI/SNF complexes. The result of these modifications is the deletion of pre-existing marks of transcriptional repression and the deposition of positive epigenetic modifications, such as H3–K4 trimethylation [12]. Whether these two processes occur simultaneously or sequentially is currently unknown. Likewise, the functional relationship between repressive and activatory chromatin-associated enzymatic complexes, which has been reported in other cell types, has not been yet investigated in muscle cells. In this regard, increasing interest is now directed toward the function of specific histone demethylases of the Jumonj family, as crucial effectors of the epigenetic switch during myoblast-to-myotube transition [24]. Moreover, other events, such as histone exchange, are likely involved in the switching of the epigenetic signature.
How do SWI/SNF complexes contribute to the epigenetic switch that occurs at muscle genes during the transition from myoblasts to myotubes?
A number of reported interactions between SWI/SNF and other chromatin-modifying complexes suggest that these interactions can be involved in some of the molecular events underlying the changes in chromatin architecture at muscle loci during myoblast differentiation.
An indirect link between SWI/SNF and other histone-modifying complexes was first established based on the increased affinity of the bromodomain-containing proteins for acetylated histones [25]. Bromodomains are present in the enzymatic sub-units of the SWI/SNF complex, Brg1 and Brm. As such, it has been proposed that interactions between Brg1 and Brm and acetylated histones contribute to stabilize SWI/SNF binding to hyperacetylated chromatin within the regulatory sequences of muscle genes. According to this model, HDAC-mediated histone deacetylation counters while HAT-dependent hyperacetylation promotes SWI/SNF recruitment to muscle genes. Other studies have illustrated a functional hierarchy in the recruitment of HATs and SWI/SNF complexes that is imposed by two intracellular cascades activated by regeneration cues — the MKK6>p38 and IGF1>Pi3K>AKT pathways. The IGF1 signaling promotes the recruitment of p300/CBP and PCAF HATs on the chromatin of muscle genes, thereby generating local hyperacetylation, which contribute to nucleosome disruption, possibly by cooperation with SWI/SNF activity [26]. Interestingly, the chromatin recruitment of SWI/SNF on muscle genes can be observed also in experimental conditions that prevent hyperacetylation — e.g. pharmacological blockade of the IGF1 signaling to HATs. Under these conditions, the p38 signaling appears sufficient to promote SWI/SNF recruitment on the chromatin of muscle genes, despite the local hypoacetylation; however, the chromatin-remodeling ability of SWI/SNF can only be detected once muscle-gene promoters are hyperacetylated in response to the IGF1 signaling to HATs. This evidence indicates a further level of functional relationship between SWI/SNF and HATs in response to external cues, whereby IGF1-mediated local hyperacetylation at muscle genes is required for the remodeling activity of the SWI/SNF complex, which is recruited in response to activation of the p38 signaling [26].
Independent studies have identified a multistep recruitment of the SWI/SNF complexes on muscle promoters. On myogenin promoter, the SWI/SNF recruitment is dependent on a prior association of MyoD with constitutively bound Pbx1 on a promoter region juxtaposed to, but not buried by, the nucleosome [17]. MyoD–Pbx1 complex mediates an initial recruitment of SWI/SNF complex possibly to displace the nucleosome, thereby allowing the full access of MyoD to the canonical Eboxes in the productive conformation for activation of gene expression — that is the heterodimer formed by MyoD and the E2A gene products, and the associated myogenic transcriptosome [12]. Moreover, SWI/SNF complexes can sequentially interact with two arginine methyltransferases, Prmt5 and CARM1, at early and late promoter respectively [27]. Interactions of the SWI/SNF complexes with other components of the muscle transcriptosome are regulated by chromatin-associated p38 alpha/beta kinases, which are activated in response to differentiation cues [28]. p38 signaling appears to promote multiple interactions within the muscle transcriptosome, including SWI/SNF recruitment, via BAF60 phosphorylation [4], formation of MyoD/E47 heterodimer, via E47 phosphorylation [29], and recruitment of the Ash2L-containing mixed-lineage leukemia (MLL) methyltransferases, via phosphorylation of MEF2D [30].
Other studies performed in different cell types reported on the functional interactions between SWI/SNF complexes and proteins implicated in repression of gene transcription, such as heterochromatin protein 1 (HP1) [31] and the Polycomb group proteins [32].
SWI/SNF sub-complexes and cellular differentiation: a tissue-specific regulation of sub-unit exchange
The multitude of interaction between SWI/SNF and other chromatin-associated proteins, as well as the temporal and gene-specific SWI/SNF distribution during skeletal myogenesis, suggest that distinct SWI/SNF sub-complexes can perform specific tasks and are subjected to different types of regulation. Indeed, increasing evidence is supporting the concept that SWI/SNF composition can vary depending on the cell type and in response to specific cues.
Tissue-and differentiation stage-pecific exchange of SWI/SNF BAF variants
SWI/SNF are highly conserved multi-protein complexes that during the evolution have lost, gained and replaced specific sub-units likely to respond to the increased complexity of the chromatin organization of multi-cellular organism. In vertebrates, these complexes are composed by a unique ATPase, either Brg1 or Brm, and 4–12 sub-units also referred as to Brg1 and Brm-associated factor (BAF). While the ATPase sub-unit provides the enzymatic activity necessary for chromatin remodeling, BAF sub-units regulate the ATPase activity and mediate interaction with specific transcription factors [7] thereby dictating cell type-specific and signal-regulated SWI/SNF distribution along the genome. Thus, varying the SWI/SNF compositionmay results in the formation of sub-complexes with specialized functions.
In mammals, the first variable is represented by the mutually exclusive presence of the ATPases Brmor Brg1. Brg1- and Brm-based SWI/SNF complexes typically coexist in the same cell types, but are likely to perform non-redundant functions. Indeed, Brg1−/− mice are embryonic lethal [33], whereas Brm−/− mice are viable and show increased body weight and alteration of cellular growth control [34]. In addition Brg1 is expressed constitutively, whereas the Brm protein concentration increases in G0-arrested cells and in cells induced to differentiate; and the expression of Brm, but not Brg1, is negatively regulated upon mitogenic stimulation oncogene-mediated transformation [35]. Finally, Brg1 binds to zinc finger-containing transcription factors through a unique N-terminal domain not present in Brm, while Brm interacts with two ankyrin repeat proteins that are crucial in the Notch signal transduction [36].
Another layer of variation is conferred on the combinatorial assembly of the SWI/SNF sub-units that are encoded by distinct families of genes and that can potentially give rise to hundreds of SWI/SNF sub-complexes with different function. The minimal “SWI/SNF remodeling core” is typically made by one ATPase — Brm or Brg1 — together with BAF155, BAF170 and INI1/BAF47 [37]. Studies in embryonic stem cells (ESCs) revealed the existence of a specific Brg1-based esBAF complex, which is devoid of Brm, BAF170 and the BAF60 variant, BAF60c [38]. This complex is essential for the maintenance of pluripotency and cannot be found in other cell types. As ESC differentiate into embryoid bodies (EBs) the esBAF complex also incorporates BRM and BAF60c, and this likely reflects the expanded repertoire of SWI/SNF functions in regulating lineage commitment and differentiation of different cell types [39]. A recent illuminating study has identified a specific BAF sub-unit exchange within the SWI/SNF complexes implicated in neural differentiation. This exchange occurs at the post-mitotic entry of differentiating neural progenitors and involves the replacement of BAF53a and BAF45a, which are present in neural stem cells, with neuron-specific BAF53b and BAF45b, to generate the SWI/SNF complex typical of post-mitotic neurons [40]. The sub-units expressed at distinct differentiation stages are not functionally interchangeable, and their exchange is regulated by neural-specific miRNA. In particular, the Baf53-sub-unit switching is achieved by the selective expression of specific miRNAs in post-mitotic neurons that target the Baf53a sub-unit, thereby ensuring the optimal composition expression of SWI/SNF complexes in the transition from neural proliferation progenitor to neurons [41].
Interestingly, during the evolution, vertebrate SWI/SNF complexes acquired three variants of the BAF60 sub-units — BAF60a, b and c — which are encoded by separate genes and show specific functions. Among them, BAF60c is preferentially expressed in developing heart and somites during embryogenesis, and Baf60c knockout mice show embryos with impaired cardiac and skeletal myogenesis [42]. Because BAF60 sub-units are specifically targeted by the p38 signaling, which promotes SWI/SNF recruitment on the chromatin of muscle genes [4], our lab is currently focusing on the identity of the BAF60 variant that mediate SWI/SNF responsiveness to intracellular signaling that promote myogenesis. Future studies should also define the specific role for each of the three BAF60 variants during skeletal myogenesis, regeneration and post-mitotic activity, as all three variants are expressed, albeit at different levels, in developing and adult skeletal muscles.
But how can the variant exchange impart the biological specificity to the SWI/SNF complexes? A logical consequence of the combinatorial assembly is the diversity of potential interacting partners. The acquisition of sub-unit equipped with bromodomain, chromodomain, PHD finger or DNA-binding domain can dictate differential association with transcriptional regulators and differential ability to “read” histone modifications. An example of such complexity is provided by the evidence that the double PHD finger-containing protein, DPF3, is associated with a SWI/SNF complex and mediates binding to methylated and acetylated lysine residues of histone 3 and 4 [43]. DPF3 is a downstream target of MEF2A, indicating a potential feed-forward loop by which myogenic transcription factors expand the number of SWI/SNF components to endow specific sub-complexes with the sub-unit that confers the ability to read chromatin modifications generated by previous events. Thus, the SWI/SNF complexes should be regarded beyond their conventional role of ATP-dependent chromatin remodelers, as readers of promoter-specific chromatin signatures that promote key transitions within the process of gene activation and repression.
When applied to skeletal myogenesis, this knowledge suggests that specific SWI/SNF sub-complexes are engaged to modify the chromatin of target genes along the functional transitions that permit first the promoter access to pioneer transcription factors, such as MyoD, and then to promote transcription in response to promyogenic cues (Fig. 1). Future studies should establish the identity, function and regulation of SWI/SNF sub-complexes involved in the activation of muscle gene expression during skeletal muscle differentiation. They should also determine the physical and functional relationship between these sub-complexes and the epigenetic marks generated by other chromatin-associated complexes on target genes.
SWI/SNF-mediated promoter remodeling by histone modifications and exchange
Chromatin architecture is deeply influenced by the composition of different histone variants, which facilitate or preclude gene transcription. It is therefore important to decipher the functional interactions between histone variants, post-translational modifications, chromatin remodelers and DNA-binding proteins in controlling higher order chromatin structure, nucleosome positioning and the dynamic nucleosome repositioning during developmental transitions [44].
Evidence indicates a functional link between chromatin remodelers and the incorporation of specific histone variants. For instance, certain histone variants, such as macroH2A, interfere with the remodeling activity of SWI/SNF [45], and the SWI/SNF-associated protein Atrx is required for localization of H3.3 at telomeres and for the repression of telomeric RNA [46].
The contribution of histone variants to the regulation of muscle gene expression has been poorly investigated. Increased synthesis of H2A.Z has been reported during the myoblast-to-myotube transition [47]. Of note, an exchange of histone variants has been implicated in the chromatin conformation, which establishes the epigenetic memory responsible for muscle-restricted expression of MyoD [48]. Likewise, linker histones play a role in the regulation of chromatin condensation in concert with chromatin remodelers. And the linker histone H1b has been shown to negatively affect the expression of MyoD [49].
Future effort should be devoted to understand the functional interactions between SWI/SNF chromatin remodelers and histone variants in the control of muscle gene expression.
Conclusions and perspectives
The fundamental contribution of chromatin remodelers to the reconfiguration of the chromatin architecture during skeletal myogenesis suggests the involvement of SWI/SNF complexes in many transcriptional events, including repression and activation of gene expression, but also regulation of control long-distance genome interactions and the non-coding (nc) epigenome. Indeed, the essential role of Brg1 in the activation of muscle-specific micro-RNAs activated by MyoD has recently been reported by the Imbalzano group [50]. Furthermore, the advance in deep-sequencing technologies (ChIP-seq) has recently revealed an unexpected broad-range of chromatin interaction of MyoD across the genome of muscle cells [51].
It will be of particular interest to understand whether distinct SWI/SNF sub-complexes are devoted to specific tasks, such as activation and repression of coding and non-coding epigenome, global chromatin modification and long-distance genome interactions that occur during skeletal muscle differentiation.
Acknowledgments
PLP is an Associate Scientist of Telethon Dulbecco Institute (DTI) and of the Sanford Children' Health Center at Sanford/Burnham Medical Institute, and was partially supported by AIRC and NIAMS (RO1AR052779).
REFERENCES
- 1.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461(7261):193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 2.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin. Cell. Dev. Biol. 2005;16(4–5):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 3.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 2001;27(2):187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 4.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004;36(7):738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 5.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006;7(6):461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 6.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136(2):200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi T. A BAF-centred view of the immune system. Nat. Rev. Immunol. 2004;4:965–977. doi: 10.1038/nri1501. [DOI] [PubMed] [Google Scholar]
- 8.Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell. Physiol. 2000;185(2):155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 9.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 10.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19(5):553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25(3):502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guasconi V, Puri PL. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19(6):286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75(7):1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 14.Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006;25(3):490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66(2):305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 16.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11(4):436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 17.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 18.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 2005;25(10):3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohkawa Y, Yoshimura S, Higashi C, Marfella CG, Dacwag CS, Tachibana T, Imbalzano AN. Myogenin and the SWI/SNF ATPase Brg1 maintain myogenic gene expression at different stages of skeletal myogenesis. J. Biol. Chem. 2007;282(9):6564–6570. doi: 10.1074/jbc.M608898200. [DOI] [PubMed] [Google Scholar]
- 20.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423(6936):145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 21.Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18(21):2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23(3):605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mal AK. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25(14):3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;15(22 (10)):1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111(3):369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 26.Serra C, Palacios D, Mozzetta C, Forcales SV, Morantte I, Ripani M, Jones DR, Du K, Jhala US, Simone C, Puri PL. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell. 2007;28(2):200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dacwag CS, Bedford MT, Sif S, Imbalzano AN. Distinct protein arginine methyltransferases promote ATP-dependent chromatin remodeling function at different stages of skeletal muscle differentiation. Mol. Cell. Biol. 2009;29(7):1909–1921. doi: 10.1128/MCB.00742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lluís F, Perdiguero E, Nebreda AR, Muñoz-Cánoves P. Regulation of skeletal muscle gene expression by p38 MAP kinases. Trends Cell Biol. 2006;16(1):36–44. doi: 10.1016/j.tcb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Lluís F, Ballestar E, Suelves M, Esteller M, Muñoz-Cánoves P. E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J. 2005;24(5):974–984. doi: 10.1038/sj.emboj.7600528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ, Dilworth FJ. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat. Struct. Mol. Biol. 2007;14(12):1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavigne M, Eskeland R, Azebi S, Saint-André V, Jang SM, Batsché E, Fan HY, Kingston RE, Imhof A, Muchardt C. Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PLoS Genet. 2009;5(12q) doi: 10.1371/journal.pgen.1000769. e1000769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306(5701):1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 33.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 34.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17(23):6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muchardt C, Bourachot B, Reyes JC, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI–SNF complex. EMBO J. 1998;17:223–231. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell. 2003;11(2):377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 37.Ryme J, Asp P, Böhm S, Cavellán E, Farrants AK. Variations in the composition of mammalian SWI/SNF chromatin remodelling complexes. J. Cell. Biochem. 2009;108(3):565–576. doi: 10.1002/jcb.22288. [DOI] [PubMed] [Google Scholar]
- 38.Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U. S. A. 2009;106(13):5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U. S. A. 2009;106(13):5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56(1):94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432(7013):107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 43.Lange M, Kaynak B, Forster UB, Tönjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, Mebus S, Lehrach H, Lurz R, Gobom J, Rottbauer W, Abdelilah-Seyfried S, Sperling S. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22(17):2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;8:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angelov D, Molla A, Perche PY, Hans F, Côté J, Khochbin S, Bouvet P, Dimitrov S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell. 2003;11(4):1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 46.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee YL, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140(5):678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wunsch AM, Reinhardt K, Lough J. Normal transitions in synthesis of replacement histones H2A.Z and H3.3 during differentiation of dystrophic myotube cells. A brief note, Mech. Ageing Dev. 1991;59(3):299–305. doi: 10.1016/0047-6374(91)90140-u. [DOI] [PubMed] [Google Scholar]
- 48.Ng RK, Gurdon JB. Epigenetic memory of an active gene state depends on histone H3.3 incorporation into chromatin in the absence of transcription. Nat. Cell Biol. 2008;10(1):102–109. doi: 10.1038/ncb1674. [DOI] [PubMed] [Google Scholar]
- 49.Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304(5677):1607–1609. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- 50.Mallappa C, Nasipak BT, Etheridge L, Androphy EJ, Jones SN, Sagerström CG, Ohkawa Y, Imbalzano AN. Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol Cell Biol. 2010 doi: 10.1128/MCB.00214-10. [Electronic publication ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18(4):662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]