Abstract
We describe a computationally efficient and robust fully-automatic method for large-scale electron microscopy image registration. The proposed method is able to construct large image mosaics from thousands of smaller, overlapping tiles with unknown or uncertain positions, and to align sections from a serial section capture into a common coordinate system. The method also accounts for nonlinear deformations both in constructing sections and in aligning sections to each other. The underlying algorithms are based on the Fourier shift property which allows for a computationally efficient and robust method. We demonstrate results on two electron microscopy datasets. We also quantify the accuracy of the algorithm through a simulated image capture experiment. The publicly available software tools include the algorithms and a Graphical User Interface for easy access to the algorithms.
Keywords: Image registration, electron microscopy, neural network reconstruction, connectome
1. Introduction
Transmission electron microscopy (TEM) has been an important imaging modality for studying three-dimensional ultrastructure in biology in general, and neuroscience in particular. Electron tomography based on computational assembly of tilt-series images (Hoppe, 1981; Sun et al., 2007) can provide high-resolution three-dimensional imagery but is restricted to very small volumes. Similarly, manual tracing of serial section TEM (ssTEM) imagery of synaptic relationships through small volumes has been a mainstay in network reconstruction. In such work, it is critical to acquire imagery at a resolution sufficient to unambiguously detect critical biological features, such as synapses and gap junctions. This sets the resolution at approximately 2 nanometers (nm) or better. This constraint has previously limited reconstruction volumes to sizes far smaller than the scale of canonical repeat units in the nervous system (Anderson et al., 2009).
Neural network reconstruction or connectomics (Sporns et al., 2005; Briggman and Denk, 2006; Mishchenko, 2008; Anderson et al., 2009), is the complete mapping of all individual neurons in a region, including their synaptic contacts, to create its canonical network map, also known as a connectome. Such complete mappings are long-standing problems in neuroscience. Progress has been hindered by impracticalities in acquisition, assembly and analysis of large scale TEM imagery. Complete connectome datasets have previously been attempted in very small invertebrate models, such as the roundworm C. elegans, which has just over 300 neurons and 6000 synapses (White et al., 1986; Hall and Russell, 1991; Chen et al., 2006). On the other hand, studying canonical samples of vertebrate neural systems (samples large enough to contain a statistically robust instances of the rarest elements in the network) require a new scale of imaging (Anderson et al., 2009).
Currently, there are several approaches to connectomics. Serial block-face scanning electron microscopy (SBFSEM) (Denk and Horstmann, 2004) uses electron backscattering from successively exposed block surfaces to capture a volume as a series of two-dimensional images. SBFSEM can provide sections as thin as 20 nm, but is currently limited by electron optics to 5–10 nm per pixel resolution in-section. An advantage of SBFSEM is the implicitly aligned nature of the images produced, nominally negating the need for elaborate registration schemes in software. Its disadvantages include sample destruction, limited in-section resolution, slow acquisition speed, incompatibility with molecular tagging methods, non-standard contrast generation, and the limited availability of SBFSEM platforms. While SBFSEM images often provide good contrast for cell membranes, much intracellular information is lost, rendering the efficient detection and classification of synapses from these images difficult. A very similar method is ion beam milling (Knott et al., 2008), which has the advantage of using a superior imaging platform (scanning TEM), though many of the deficits of destructive sampling remain, slow speed and limited platform access remain. A new, very exciting alternative is the Automatic Tape-collecting Lathe UltraMicrotome (ATLUM) (Hayworth et al., 2006) which sections a block and automatically collects the sections on a long Kapton® tape for imaging by scanning TEM.
We recently proposed ssTEM as a platform that provides the right combination of resolution, spatial coverage and speed for connectomics (Anderson et al., 2009). TEM microscopes and ultramicrotomes for serial sectioning are widely available. In TEM, sections are cut from a specimen and suspended in the electron beam, creating a projection image which may be captured on electron-sensitive film (and digitized later) or captured directly on electron-sensitive digital cameras. Slice thicknesses for ssTEM are typically in the 40–100 nm range, while in-section resolution is limited only by the resolution of TEM imaging (100–200 picometers). In practice, connectomics requires 2 nm resolution per pixel to resolve gap junctions. As noted above, acquisition and analysis of ssTEM data has been extremely time consuming, limiting neuronal mapping to projects involving small numbers of neurons (Cohen and Sterling, 1992; Harris et al., 2003; Dacheux et al., 2003). The complete C. elegans reconstruction (White et al., 1986; Hall and Russell, 1991; Chen et al., 2006) is reported to have taken more than a decade (Briggman and Denk, 2006). A major barrier in image acquisition has recently been overcome by implementing SerialEM automated ssTEM acquisition software (Mastronarde, 2005). But large volumes can also be acquired manually on film. Another major barrier has always been assembly of image mosaics and volumes from hundreds of thousands of ssTEM images. That computational barrier is the focus of this paper.
There are two image registration problems associated with assembling volumes from ssTEM imagery. First, the TEM field of view is insufficient to capture an entire section as a single image, and each section is imaged in overlapping tiles. For instance, a canonical area (Anderson et al., 2009) in the retina yields over 1000 tiles per section, where each tile is a 4096 × 4096 pixel, 16 bit image. If film is used, the pixel densities can be even higher, but positional metadata are lost requiring tile layout to be inferred as part of a software solution. In either case, warps due to the TEM aberrations and other distortions have to be corrected to generate seamless overlaps between tiles. We refer to the entire process of assembling a single section from multiple TEM tiles as section mosaicking. The second registration problem stems from the fact that each section is cut and imaged independently: mosaicked sections thus have to be aligned to each other. The coordinate transformation between sections includes unknown rotation and nonrigid deformations due to the section cutting and imaging processes. Typically, deformations in this section-to-section registration are larger than in the mosaicking stage. Once all sections are mosaicked and registered, a three-dimensional volume can be assembled.
1.1. Related Work
Image registration is a very active research topic in clinical imaging applications such as magnetic resonance imaging and computed tomography (Toga, 1999; Maintz and Viergever, 1999). In general, image registration methods can be classified according to a few criteria: types of features used for matching, coordinate transformation classes and targeted data modalities. Intensity-based methods compute transformations using image intensity information (Bajcsy and Kovacic, 1989; Toga, 1999). Landmark-based methods match a set of fiducial points between images (Evans et al., 1988; Thirion, 1994, 1996; Bookstein, 1997; Rohr et al., 1999, 2003). Fiducial points can be anatomical or geometrical in nature and are either automatically detected or input manually by a user. The range of allowed transformations include rigid, affine, polynomial, thin-plate splines or large deformations (Bookstein, 1989; Toga, 1999; Christensen et al., 1996; Davis et al., 1997; Rohr et al., 1999). A common theme among most clinical image registration methods is a variational formulation of the problem which can then be solved using the iterative optimization techniques such as gradient descent. Unfortunately, such optimization techniques are too slow and are too initialization dependent to be of practical use for large-scale ssTEM image registration. Solutions to ssTEM image registration problems must take into account the scale of the data. For instance, a ssTEM data set with sufficient resolution and size to reconstruct the connectivities of all ganglion cell types in retina is approximately 16 terabytes (Anderson et al., 2009).
Image mosaicking has been studied in many application areas. Irani et al. propose a method to compute direct mapping from video frames to a mosaic representation for mosaic based representation of video sequences (Irani et al., 1995). Panaromic image generation and virtual reality (Kanade et al., 1997; Davis, 1998; Peleg et al., 2000; Shum and Szeliski, 2002; Levin et al., 2004) other prominent applications areas for mosaicking. Vercauteren et al. propose a Lie group approach to finding globally consistent alignments for in vivo fibered microscopy (Vercauteren et al., 2006). Early work for ssTEM registration in the literature has been manual or semi-automatic (Carlbom et al., 1994; Fiala and Harris, 2001). For instance, Fiala and Harris proposed a method which estimates a polynomial transformation from fiducial points entered by a user (Fiala and Harris, 2001). Randall et al. propose an automatic method for registering electron microscopy images limited to rigid transformations (Randall et al., 1998). These and other earlier studies targeted TEM datasets three orders of magnitude smaller than those proposed in this paper: i.e. roughly 100 images instead of 100,000. We recently proposed a fast method for registering tiles within a section (mosaicking) which relies on the Fourier shift property (Tasdizen et al., 2006). In the same work a landmark based approach was used to register the adjacent sections to each other. Ultimately, we formulated the section-to-section registration in the Fourier shift framework as well (Koshevoy et al., 2007). A closely related study describes an automatic method for large-scale EM registration based on block matching using the normalized cross-correlation metric and the iterative closest point algorithm (Akselrod-Ballin et al., 2009). However, only rigid transformations are considered. Another related work uses phase correlation for EM image stitching (Preibisch et al., 2009). In (Anderson et al., 2009) we described a workflow for automatic, fast and robust solution to the mosaicking and section-to-section registration problems in ssTEM. That paper focussed on the acquisition technology, including methods for embedding molecular tags in the volume. In this paper, we discuss the technical details of the underlying algorithms and introduce a new, user-friendly graphical user interface (GUI) solution to allow laboratories without expertise in computer science to readily implement them. We also report the results of a quantitative experiment to assess the accuracy and reliability of the proposed approach. With the availability of these tools, the next hurdle in connectomics will be automation of image analysis, allowing reconstructions of very large numbers of neuronal connections and statistical analyses of network maps. Furthermore, the software tools described in this paper can be applied to data sets from other microscopy platforms for connectomics such as ATLUM (Hayworth et al., 2006) or even to other imaging modalities such as confocal microscopy. The software tools described in this paper are publicly available2.
2. Materials and Methods
2.1. Section mosaicking
Let and Wi : ℝ2 → ℝ2 denote the set of N two-dimensional image tiles constituting a mosaic image and the corresponding coordinate transformations mapping the image tiles into a common mosaic coordinate space. Also let Ωi,j denote the region in the mosaic space that is the overlap of image tiles gi and gj under their corresponding coordinate transforms. An energy function measuring the image intensity mismatch in overlap regions, given a set of coordinate transforms W1 … WN, can be computed as
| (1) |
where is the inverse transform from the mosaic space into the coordinate space of the i′th image tile and x denotes positions in the mosaic space. Figure 1 illustrates the relationship of the tiles with respect to each other and the mosaic space. The scalar function J is measures the smoothness of a given transformation and is known as a regularization function. We can then choose the transforms W1(x) … WN(x) that minimize this energy function. The parameter α in (1) controls the relative importance of the regularizer and therefore, the smoothness of the solution. Note that the coordinate transformations encode not only the positions of the tiles in the mosaic, but also the nonrigid warps that are needed to compensate for the deformation introduced in image acquisition. Also note that for pairs of nonadjacent images Ωi,j will be the empty set.
Figure 1.
Relationship of image tiles and the mosaic space.
A trivial minimizer for the first term in (1) that incurs no penalty is a set of translations that map the image tiles to entirely disjoint areas of the mosaic space with no overlap regions. In pairwise image registration applications, this problem can be circumvented in several ways. One possibility is to fix one image, defining a coordinate transform only for the moving image and to choose J to penalize the deviation of the moving image’s transform from the unity map (elastic deformation penalty). This solution is not applicable to our problem since all images need to be mapped into a common mosaic space which requires transformations with large translational components. Therefore, translational components of the transformations can not be penalized in J for our application. However, in the absence of such a penalty in J, the trivial solution mentioned above is not avoided. Another possibility for registering image pairs is to use the prior knowledge that the pair completely overlaps to define the integral in (1) over the entire domain of the fixed image rather than the overlap area. This solution is also not practical for our problem because pairs of tiles overlap only partially or not at all. A possible solution for the mosaicking problem when the average overlap area between adjacent tiles is known, is to minimize (1) under the additional constraint which prescribes the total area of the overlapping regions. Even then, a fundamental problem is the difficulty of finding the globally optimal solution for (1). Our approach is inspired by the variational formulation in (1), but it focuses on a fast, practical solution rather than formally finding an optimal solution to (1).
There are two alternative scenarios with differing workflows. In the first scenario, we assume no prior knowledge about the approximate locations of the tiles in the mosaic space, as in manual film acquisition. Our mosaicking workflow in this case is composed of several stages.
Find pairs of overlapping tiles and compute their pairwise displacement (Section 2.1.1).
Infer a layout of the mosaic using only the displacements found in first stage (Section 2.1.2).
Refine the mosaic using displacements (Section 2.1.3)
Refine the mosaic using nonrigid transformations (Section 2.1.4).
The first stage in the solution, finding overlapping pairs and computing their displacement, is a 𝒪(N2) operation where N is the number of tiles in the mosaic. In the second and simpler scenario (digital capture), an approximate displacement is known from positional metadata. This also implies that we know which pairs of tiles overlap; therefore, the first two stages of the above workflow are not necessary in this case. The refinement of the mosaic using rigid displacements (stage 3) is still necessary before nonrigid refinement (stage 4) because the positional metadata reported by the microscope has very low accuracy.
2.1.1. Closed-form estimate of pairwise tile displacement and overlapping tile pair detection
The first problem is to find pairs of overlapping tiles and their relative displacement. The main constraint at this stage of the algorithm is computational complexity because this procedure will applied to approximately N2 pairs where N is the total number of tiles in a section. If we restrict the class of allowed coordinate transformations between pairs of tiles to only translation, i.e. displacement, a fast closed-form solution based on phase correlation exists (Kuglin and Hines, 1975; Castro and Morandi, 1987; Girod and Kuo, 1989). While directly evaluating the cross correlation of two images via two-dimensional convolution and finding the displacement that produces the largest correlation is the most straightforward method for finding the unknown translation, this is computationally very expensive. The Fourier shift property and phase correlation present a much faster alternative. Let ℱ[g](u, υ) denote the two-dimensional Fourier transform of image g(x, y) where u and υ denote the variables in the frequency domain. For notational simplicity we will drop the frequency variables and use ℱ[g] to mean ℱ[g](u, υ). Given an image g of size Q × R, a (xo, yo) pixel circular shift of g is defined as
| (2) |
Then, the well known Fourier transform shift property (G. and Woods, 1992) provides a simple rule relating the Fourier transforms the image and its circularly shifted version
| (3) |
In other words, circularly shifting the image in the spatial domain corresponds to a multiplication with a complex exponential in the frequency domain. The complex exponential can be be isolated by computing the cross power spectrum. In general, the cross power spectrum of two images g and h is defined as
| (4) |
where ℱ* denotes the complex conjugate of the Fourier transform. For image g and its circularly shifted version gcirc(xo,yo) simplifies to
| (5) |
Since, in this case, the cross power spectrum isolates the complex exponential due to the circular shift, the displacement vector (xo, yo) can be recovered simply by taking the inverse Fourier transform of the cross power spectrum
| (6) |
where ℱ−1[·] is the inverse Fourier transform and δ(x−xo; y−yo) is the Dirac delta function located at (xo, yo). In other words, the inverse Fourier transform of S(g, gcirc(xo,yo) is an image which has a single non-zero entry at pixel location (xo, yo).
In practice, we have a linear shift rather than a circular shift. The method described in (Girod and Kuo, 1989) is for tracking an object over a flat background in a video sequence. In that case, the above derivation for circular shifts still holds for linear shifts. However, the phase correlation of overlapping but distinct EM images presents a more complex scenario. In this case, we can not expect to recover a single Dirac delta function at the correct shift location. Instead, we treat the inverse Fourier transform of the cross power spectrum between two EM tiles gi and gj as a displacement probability image
| (7) |
where for simplicity we use ℱi to denote ℱ[gi] and Real{·} is the real part of its complex argument. The addition of a small positive constant ε to the denominator avoids the problem of division by 0. Taking the real part of the inverse Fourier transform discards any imaginary component due to numerical accuracy limitations. Note that (7) also assumes that EM image tiles gi and gj are of the same size. If this is not the case, the EM image tiles should be padded to the size of the largest tile. While (7) does not strictly define an image of probabilities, it can be interpreted as such because larger values of Pi,j(x, y) correspond to displacements that are more probable. As discussed above, for partially overlapping images, the exact relationship Pi,j(x, y) = δ(x−xo, y−yo) does not hold. However, if the amount of overlap is sufficient, the maximum of Pi,j(x, y) should correspond to the true displacement between images gi and gj. In practice, finding this maximum is non-trivial because for most electron microscopy images the Pi,j(x, y) contains many spurious weak local maxima. This can be seen in Figure 2 (left) which shows Pi,j(x, y) computed for two tiles with approximately 10% overlap. Also, Pi,j for two non-overlapping images may contain several weak maxima, complicating the decision whether two image tiles overlap or not. These problems are not addressed in (Girod and Kuo, 1989) since the target application in that paper is object tracking from video. Our solution which addresses these practical difficulties is discussed next.
Figure 2.
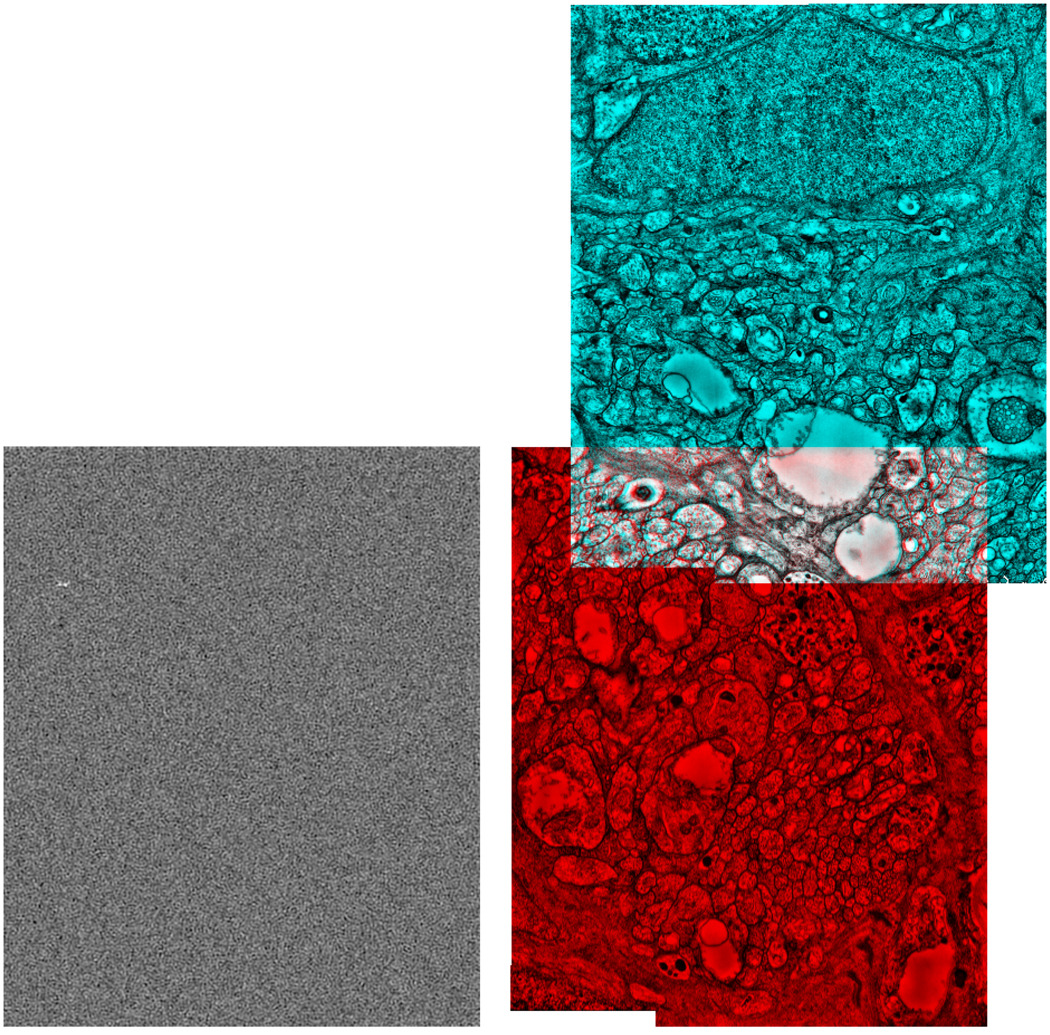
Left: Pij(x, y) for two image tiles with approximately 10% overlap. The location of the global maximum can be seen around the top left corner. Right: The two tiles shown in cyan and red displaced with the computed vector.
We have found that five steps are necessary in practice to identify the location of the correct maxima in Pi,j. These steps are performed by the executable ir-fft. The first step is to pre-smooth all image tiles gi to reduce the amount of noise and to smooth Pi,j(x, y) to reduce the number of spurious local maxima. The second step is to select and apply a threshold to Pi,j(x, y) to isolate the strongest peaks. We choose the threshold at the 99th quantile of the histogram of Pi,j(x, y). In other words, 1% of the total pixels in P are considered as possible displacement locations. Notice that if the image gi and gj do not overlap, some pixels in Pi,j(x, y) will still pass the threshold. In other words, at this stage the strongest peaks are identified only in a relative sense. In the third step, we look for a cluster of at least five 8-connected pixels that indicate a strong maximum. If the maxima pixels are scattered across Pi,j(x, y), it is likely there is no strong maximum. The coordinates of the maxima are calculated as the centers of mass of the corresponding clusters. Due to the periodicity of the Fourier transform, clusters that are broken across the image boundary are merged together. Notice that at this stage it is possible to have more than one displacement vector candidate per tile pair.
The fourth step is to verify which, if any, of the maxima found in the previous step is the true displacement between the image pair. Non-overlapping image pairs typically produce a Pi,j(x, y) with several maxima points at roughly the same value, while the Pi,j of two matching tiles produces one maximum significantly higher than the rest. If the strongest maxima is at least twice as large as the rest, it is marked as a good match; otherwise, we determine that the tiles do not overlap. We have found the proposed method works best for pairs of tiles that overlap at least 10% in area. In our application, adjacent image tiles have around 10 – 15% overlap. Therefore, displacement vectors resulting in less than 5% of overlap or greater than 30% overlap are discarded. Minimum and maximum allowable overlap percentages can be specified using the -ol flag in ir-fft.
Results of our image matching on two tiles with approximately 10% overlap is demonstrated in Figure 2. Notice that while Pi,j(x, y) has its global maximum at approximately at the correct displacement vector, there are many local maxima with strengths that are comparable to the global maxima. The ratio of the global maximum to the local maxima depends on the overlap area between the two tiles. For this reason, displacement vectors for tiles with very small overlaps can not be computed reliably.
Finally, due to the image periodicity assumption of the Fourier transform a global peak at (xo, yo) can correspond to any one of four possible displacement vectors (xo, yo), (I − xo, yo), (xo,R − yo) and (Q − xo,R − yo) where (Q,R) is the size of the image tiles. Therefore, once a valid global peak is identified at pixel location (xo, yo) in the fourth step, we have to generate all four possible displacements between the pair of images, compute the cross correlation of each and choose the displacement vector that results in the best match as the fifth step.
2.1.2. Mosaic layout
In Section 2.1.1 we described a method to compute the displacement vectors Ti,j between any overlapping image tile pair gi and gj. However, what is needed is a mapping Wi from each tile to a common mosaic space as described in (1) and illustrated in Figure 1. We will refer to the process of converting pairwise displacements Ti,j to displacement vectors Ui from gi to the mosaic space as the mosaic layout process. Vercauteren et al. provide a mathematically rigorous treatment of this problem using Lie groups (Vercauteren et al., 2006). In this paper, our focus is on the scalability of the solution to mosaics containing thousands of EM tiles as required in connectomics. The executable ir-fft implements the mosaic layout algorithm as well as the computation of the pairwise displacements described in Section 2.1.1.
Given a displacement vector Ti,j between any overlapping image tile pair gi and gj, the correlation coefficient between the tiles in the overlap area Ωi,j is defined as
| (8) |
where x denotes the (x, y) position of a pixel, and μi and μj are the average intensity of images gi and gj in the overlap region, respectively. Then, we define a cost function between gi and gj as the negative of the correlation coefficient: Ci,j = −ρi,j. Notice that Ci,j are fixed once the pairwise displacements Ti,j have been computed in Section 2.1.1. The mosaic layout process starts by placing an arbitrary tile as an anchor image in the mosaic space. Without loss of generality, let f0 be the anchor tile. Then, the tile with the lowest cost mapping to the anchor image is laid down into the mosaic. However, for a given pair of tiles, there can be two kinds of mappings. A direct mapping exists if the pair was determined to overlap in Section 2.1.1. We also consider indirect cascaded mappings via intermediate tiles. For example, there may exist a direct mapping f0 : f1 between tiles f0 and f1, and another mapping f1 : f4 between tiles f1 and f4. Then an indirect mapping f0 : f1 : f4 between tiles f0 and f4 can be created via the intermediate tile f1 by summing the displacement vectors for the mappings f0 to f1 and f1 to f4. This new cascaded displacement vector forms an alternative to the direct mapping f0 : f4. In fact, if f0 and f4 do not overlap, then the only possible mappings between those tiles will be indirect cascaded displacement vectors. For a mosaic with N tiles, we consider direct mappings and indirect mappings with 1 : N − 2 intermediate tiles.
We define the cost of a cascaded mapping to be the maximum of the pairwise costs Ci,j along the cascade. For instance, the cost of the cascaded mapping f0 : f1 : f4 is max(C0,1,C1,4). The mapping with the smallest cost is preferred even when it has greater cascade length. Typically, there are many redundant mappings between the anchor tile and any other tile in the mosaic. Using these redundant mappings between presents an opportunity to select the best mapping possible. This is important because we expect that a certain portion of the pairwise displacements found in Section 2.1.1 will be erroneous. Tiles are successively laid down in the same manner always choosing the best possible mapping to tiles already in the mosaic.
2.1.3. Displacement refinement
In the scenario where stage position information is available the pairwise tile displacement computations and the mosaic layout process becomes unnecessary. This reduces the overall computational cost from 𝒪(N2) to 𝒪(N). However, we have found that stage position information can be inaccurate, and attempting to perform nonrigid refinement starting from tile positions initialized directly from stage positions is prone to finding suboptimal solutions. Furthermore, for very large mosaics assembled without stage position information, the mosaic layout generated by the algorithms described in Sections 2.1.1 and 2.1.2 can also result in suboptimal solutions if directly followed by nonrigid refinement. Therefore, we first refine the displacement vectors in an iterative fashion before nonrigid refinement. Let 𝒩i denote the set of tiles that overlap tile i. We define the energy
| (9) |
where the weighting is the square of the correlation coefficient defined in (8). Notice that Ui − Uj denotes the preferred relative position of a pit of tiles where the tile-to-mosaic displacements. If microscope metadata provides approximate positions for each tile, we use this information only to determine adjacency relationships and compute Ui − Uj directly with the procedure described in Section 2.1.1 for adjacent tile pairs i, j. If no metadata is available, Ui are found with the procedures described in Sections 2.1.1 and 2.1.2 and Ui − Uj are then computed for adjacent tile pairs i, j. In equation (9), the tension vector (U′i −U′j)− (Ui −Uj) between gi and a neighboring tile gj which overlaps with it is defined to have a zero energy in the preferred relative position Ui − Uj. The weighting places more weight on pairs that have stronger correlation at their preferred displacement which makes the solution more robust. We minimize the energy defined in equation (9) with respect to the new displacement vectors U′i. For uniqueness of the solution, the first tile can be used as an anchor and U′1 treated as a constant vector equal to U1 rather than a free variable in the optimization. While equation (9) could be minimized in least squares form, we choose an iterative minimization strategy to impose an additional constraint on how far a tile is allowed to move away from its preferred position. We have found that 5–10 iterations are sufficient to find a good solution. Finally, since the transformations at this stage are one displacement vector per tile, scaling and rotation of tiles is not possible. These nonrigid transformations are addressed in the next section.
2.1.4. Nonrigid refinement
Each image tile undergoes an unknown warp due to electron optical aberrations and distortions induced by both the section process and intense electron beam exposure. Therefore, it is important to use a flexible, nonrigid transformation in image registration to generate seamless overlaps between tiles. The nonrigid image registration algorithm has two essential components: 1) a class of coordinate transformations, and 2) a method for finding the ”best” transform Wi for each image, from the class of transforms being considered. During the earlier stages of algorithm development, several continuous polynomial transforms were explored, in particular bivariate cubic Radial Distortion and Legendre polynomial transforms. These transforms suffer from a trade-off where the stability of the transform is related inversely to the degree of the polynomial. Our final approach uses a locally defined transform and is implemented by the executable program ir-refine-grid. A coarse rectilinear grid of control points are placed on to each tile. The number of control points (vertices) in the grid are defined by the -mesh flag of ir-refine-grid. Each vertex in this grid stores two sets of coordinates – the local tile coordinates and the mosaic space coordinates. The tile coordinates are fixed and the image is warped by changing the mosaic space coordinates directly. The mapping Wi(x) from any point x in the tile space into mosaic space is trivial due to the uniform structure of grid of control points. One has to find the mesh quad (solid rectangle in Figure 3) containing the tile space point and perform a bilinear interpolation between the mosaic space coordinates of the quad vertices.
Figure 3.
Tile-to-mosaic and mosaic-to-tile transformations.
For the class of transform described above, finding the ”best” transforms Wi is equivalent to finding the ”best” mosaic coordinates for the vertices in the grid. At each vertex, a small image neighborhood of the tile is sampled in the mosaic space (shown as dashed rectangle in Figure 3). A corresponding neighborhood is sampled also from all of the neighboring tiles in the mosaic. For sampling these neighborhoods in mosaic space, we need to be able to map any mosaic coordinate x′ back to tile coordinates as shown in Figure 3. For this purpose, the grid of control points is treated as a triangle mesh by breaking each quadrilateral element of the grid of control points into two triangles. To map a coordinate from the mosaic space into the tile space, the mesh is searched for the triangle containing the given mosaic space point (shown as triangle in Figure 3)). Then, the barycentric coordinates of the point are used to calculate the corresponding tile space point by interpolating the tile space vertex coordinates of the triangle. We use a triangle mesh due to ease and efficiency of implementation using OpenGL: The tile space coordinates correspond to the OpenGL texture coordinates, and the mosaic space coordinates correspond to the OpenGL triangle vertex coordinates.
Any two neighborhoods sampled as described above can be matched using the same Fourier shift method described in Section 2.1.1. When a tile overlaps with more than one neighboring tile, the resulting displacement vectors are averaged. The neighborhood has to be only as large as necessary to capture a meaningful amount of image texture for phase correlation to work. The size of the neighborhoods is chosen such that the control points are spaced at approximately 1/3 of the neighborhood size which creates overlapping neighborhoods for adjacent control points. For the retinal connectome volume (Anderson et al., 2009), we downscale tile images by a factor of 8 and use a 8 × 8 grid of control points. This translates into 96 × 96 pixel neighborhoods. Therefore, in this case, instead of pairs of tiles, we find the displacement between pairs of 96 × 96 pixel neighborhoods using the same method as in Section 2.1.1. The displacement vectors produced by this matching are used to correct the mosaic space coordinates of the vertex. Also, note that the -cell flag of ir-refine-grid can be used instead of the -mesh flag to specify the size of the neighborhoods to be used in matching rather than the number of control points in the grid. In this case the number of control points are again spaced at 1/3 the neighborhood size. The -mesh and -cell arguments of ir-refine-grid should not be used simultaneously.
The procedure described above can be used to compute mosaic positions of grid control points at the edges of the tiles that overlap with neighboring tiles. We still need to propagate this position information to the control points in the interior portions of the grid that do not overlap neighboring tiles. Furthermore, there can be errors in the mosaic position computation. To address these problems we take the following approach. The displacement vectors calculated at each vertex are median filtered to remove the outliers. The displacement vectors are then blurred with a Gaussian smoothing filter which propagates the information to interior control points.
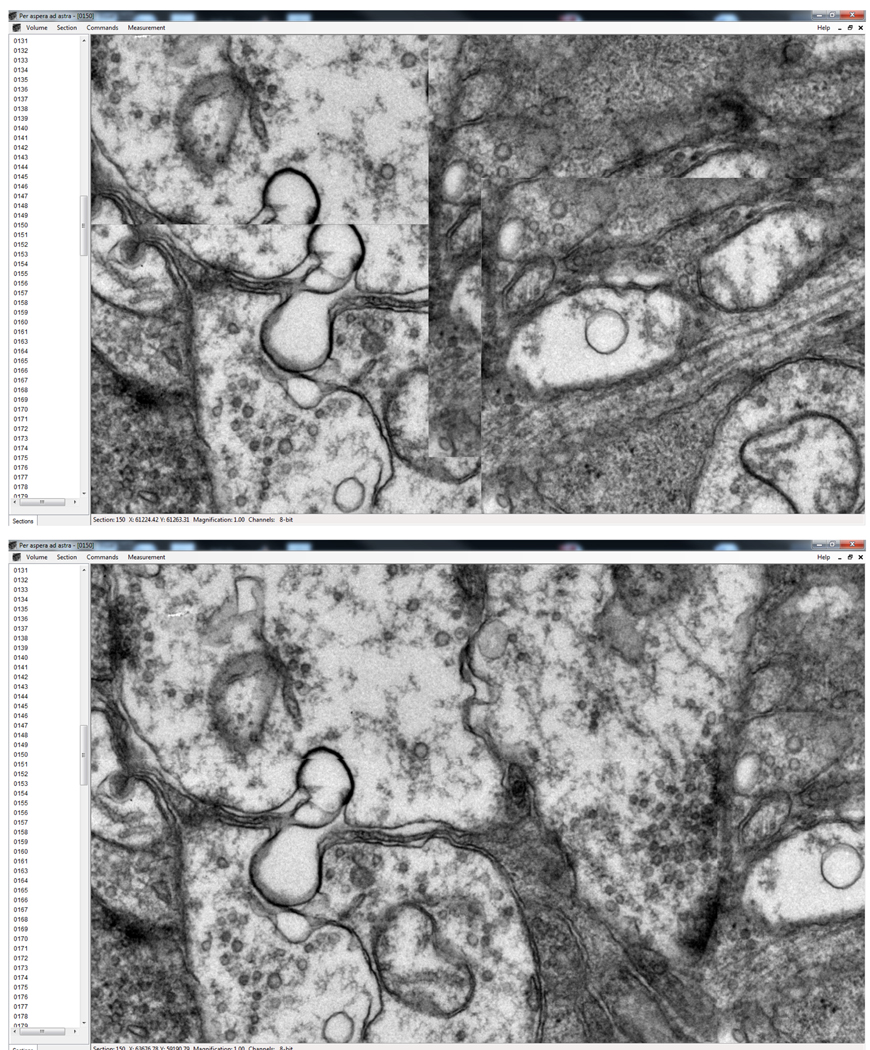
This algorithm requires several passes to ensure convergence. After each pass new neighborhoods are sampled using the newly updated mosaic coordinates of the control points and the position computation procedure is repeated. The number of passes can be set using the -it flag of ir-refine-grid. For the retinal connectome data we’ve found four passes to be sufficient. Figure 4 illustrates three different areas of the mosaic before and after the nonrigid refinement. Intensities in overlapping areas are averaged. Before nonrigid refinement this results in blurry images in overlapping areas due to the non-precise nature of the alignment (Figure 4 left). After nonrigid refinement averaging results in crisp images (Figure 4 right). Another way to visually assess the quality of the alignment is by assigning each pixel in the mosaic the intensity from the tile that it is closest to (Figure 5). Notice that tile boundaries are clearly visible if only the stage positions from the metadata are used (Figure 5 top). whereas tile boundaries are hard to detect with the eye after refinement (Figure 5 bottom).
Figure 4.
Several areas of the mosaic where multiple image tiles overlap before and after nonrigid refinement. Intensities are averaged from multiple overlapping tiles.
Figure 5.
An area where several tiles overlap aligned using stage positions from metadata only (top) and our mosaicking algorithms (bottom). Each pixel in the mosaic is assigned the intensity from the tile it is closest to. Note the clearly visible tile boundaries when only stage positions are used.
2.2. Section-to-section registration
Section-to-section registration is very similar to the distortion correction described above. As the orientation of the sections is arbitrary, however, we cannot use image correlation to directly estimate the section to section translation parameters. Thus we first perform a brute-force search for tile translation/rotation parameters on downscaled 128 × 128 pixel thumbnails of the mosaics. As downscaling eradicates nearly all image texture, the mosaics are pre-processed to enhance large blob-like features, e.g. cell bodies. The enhancement algorithm is implemented by the ir-blob executable and is defined as follows:
The image is partitioned into a regular square grid of roughly 17 × 17 pixels per square. The size of the squares can be controlled using the -r flag in ir-blob.
The intensity variance is calculated within each square.
The intensity variances for the squares computed in the previous step are sorted and the median variance (σmedian) is selected.
The algorithm iterates through all image pixels, and for each pixel calculates mean pixel variance within the local 17×17 pixel neighborhood centered at that pixel. Let σ(x, y) denote the variance computed in this manner at pixel location (x, y). Then, the output pixel value is proportional to .
As a result, areas with large variance are assigned low values while areas with small variance such as the interior of large blobs are assigned large values. The moving section is rotated in increments on 1 degree and matched against the fixed section by computing a translation between the fixed section and the rotated moving section. For each orientation, the translation is computed closed-form using phase correlation as described in Section 2.1.1. Then we choose the rotation angle which results in the largest correlation coefficient as defined by equation (8) for the displacement found by the phase correlation method for that rotation. This brute force matching algorithm is implemented by the ir-stos-brute executable. We have found that when preprocessed by ir-blob to enhance large blobby structures such as cell bodies, accurate rotation and displacement between image pairs at coarse resolution can be found reliably. Finally, the rotation and translation parameters corresponding to the best brute force match metric at coarse resolution are used to initialize the grid of control points for the non-rigid transform of the moving slice at a fine resolution. The transform is then computed as explained in Section 2.1.4, except the displacement vectors are applied to the moving slice only. The non-rigid transform refinement for section-to-section alignment is implemented by ir-stos-grid. Similar to ir-refine-grid, the user needs to specify the number of control points in the transform grid. This can be directly accomplished using the -grid flag in ir-stos-grid. Alternatively, the space in pixels between control points on the grid can be specified using the -grid_spacing flag. The size of the neighborhood associated with each control point can be specified using the -neighborhood flag. Finally, a volume can be built using the computed transformations using the ir-stom program.
2.3. Command line tools
In this section, we describe the command line tools which implement the algorithms discussed in Sections 2.1 and 2.2. All command line tools were implemented using the Insight Segmentation and Registration Toolkit (ITK) software framework (Ibanez et al., 2003). We also discuss the parameters used by the tools and the settings used in our workflow for building the retinal connectome (Anderson et al., 2009). These parameters can be customized by other users as needed for their applications.
2.3.1. ir-fft
The ir-fft program implements the pairwise tile displacement computation (Section 2.1.1) and mosaic layout (Section 2.1.2). The input to ir-fft is two or more image tiles. The input images are specified with the -data argument when running ir-fft from the command line. The input images do not have to be in any particular order since no prior adjacency information is assumed. The output of ir-fft is a text file with .mosaic extension containing the full tile transformation information Ui for the mosaic generated. We will refer to this file as the mosaic file in the rest of this paper. The name of the output mosaic file is specified with the -save argument. An actual output mosaic image is not created by ir-fft. There are two motivations for this choice. First, the transformations generated by ir-fft are rigid tile displacements only and typically computation of nonrigid transforms is also necessary before a final output is created. Second, as most TEM mosaics are very large, our Viking viewer3 interactively loads only those image tiles appearing on the screen and applies the transformations at run-time. For users wanting to create a single, fixed-resolution output image, we have also implemented a command line tool ir-assemble (discussed below) which reads in the input image tiles and the transforms computed either by ir-fft or by the nonrigid refinement tools and generates an actual image.
The following arguments are also supported by ir-fft :
-ol overlap_min overlap_max: Specify the minimum and maximum allowed overlap ratio (0: no overlap, 1: full overlap) between adjacent tiles. These arguments are useful for constraining the range of allowable displacement vectors if such prior knowledge from the image acquisition phase exists as described in Section 2.1.1. Default values are 0.05 and 1, respectively. For the retinal connectome we have used 0.05 and 0.3, respectively. This choice correspond to a minimum 5% allowed overlap between adjacent tiles.
-clahe slope: Specifies the contrast slope limit (greater than or equal to 1) for CLAHE (Zuiderveld, 1994) histogram equalization. The default is 1 which means no histogram equalization. Values larger than 1 results in the CLAHE algorithm being applied to 256 × 256 windows as a preprocessing step. This is useful for computing pairwise displacement vectors in image sets with poor contrast. For the retinal connectome we use a slope value of 6.
-sh downsample_factor: Downsample the image tiles by a specified factor for purposes of speeding up the transform computations. The default value is 1 (no downsampling). Note, images at full resolution can still be assembled later if a value greater than 1 is used; however, accuracy of the transformations may be reduced. In practice, we use a downsampling factor of 8 without noticing adverse effects.
Other arguments are supported which can be used to further customize the operation of ir-fft ; however, they are not listed here as their use are not necessary in a typical workflow. A list of available arguments can be obtained by executing any command line tool without any arguments.
2.3.2. ir-refine-translate
The command line tool ir-refine-translate implements the iterative displacement refinement discussed in Section 2.1.3. The input and output mosaic files are specified with the -load and -save arguments, respectively. The downscaling factor specified with -sh also applies to ir-refine-translate as described for ir-fft. A contrast enhancement may be specified using the -clahe argument as for ir-fft. Additionally, the following arguments are also supported:
-it iterations: Number of refinement iterations.
-max_offset dmax: Tiles are allowed to move a maximum distance dmax from their preferred positions.
-threads number_of_threads: The number of threads to be used can be set with the -threads argument. Default value is the number of hardware cores.
2.3.3. ir-refine-grid
The ir-refine-grid program implements the nonrigid warp computation discussed in Section 2.1.4. The input and output mosaic files are specified with the -load and -save arguments, respectively. The downscaling factor specified with -sh also applies to ir-refine-grid as described for ir-fft. A contrast enhancement may be specified using the -clahe argument as for ir-fft. The number of threads to be used can be set with the -threads argument. The following arguments are also supported:
-it iterations: Specifies the number of iterations described in Section 2.1.4. The default value is 10, in practice good results can be achieved with fewer iterations. Four iterations were used in building the retinal connectome.
-cell size: Specifies the neighborhood size associated with each control point in the grid transformation. When -cell is specified and -mesh is omitted, the number of control points is calculated automatically to produce a predetermined percentage of overlap between adjacent neighborhoods.
-mesh rows cols: Specifies the number of control points in the grid transformation. When -mesh is specified and -cell is omitted, the neighborhood size is calculated automatically to produce a predetermined percentage of overlap between adjacent neighborhoods. A 8 × 8 mesh was used for building the retinal connectome.
-displacement_threshold offset_in_pixels: The average displacement change threshold at which the tool should stop iterating. Defined in pixels. Default value is 1.
2.3.4. ir-assemble
Output images can be assembled from a mosaic file using ir-assemble. The input mosaic and output image files are specified with the -load and -save arguments, respectively. The down-scaling factor specified with -sh also applies to ir-assemble as described for ir-fft. The number of threads to be used can be set with the -threads argument. The following argument is also supported:
-feathering [none|blend|binary]: Selects the feathering mode used in the portions of the mosaic where multiple tiles overlap. The none mode simply averages pixels. The blend mode uses a weighted averaging where the weight of a tile’s contribution to a given mosaic pixels is inversely proportional to proximity. The binary mode uses the only the intensity value from the tile closest to the pixel of interest. This mode is especially useful for evaluating the quality of the image registration.
2.3.5. ir-blob
The ir-blob program implements the blob-like feature enhancement algorithm described in Section 2.2 which is used as a preprocessing step before the determination of unknown rotation and translation between a pair of sections. The input and output image files are specified with the -load and -save arguments, respectively. The downscaling factor specified with -sh also applies to ir-blob as described for ir-fft. The following argument is also supported:
-r radius: Variances are computed for (2 × r + 1) × (2 × r + 1) squares. The default values is 2. A value of r = 8 was used in building the retinal connectome.
2.3.6. ir-stos-brute
The ir-stos-brute program implements the brute force search for the unknown rotation and translation between a pair of sections. A mosaic file is specified using the -load argument. An output section-to-section transform file (.stos extension) name is specified using the -save argument. The downscaling factor specified with -sh also applies to ir-stos-brute as described for ir-fft. The following argument is also supported:
-refine: Specifies that the brute force registration results are refined in a multi-resolution fashion. Default value is not refine.
2.3.7. ir-stos-grid
The ir-stos-grid program refines a section-to-section registration initialized by ir-stos-brute by resampling the initial transform on to a mesh and using local neighborhood matching at the mesh vertices similar to ir-refine-grid. The input and output .stos files are specified with the -load and -save arguments, respectively. The downscaling factor specified with -sh also applies to ir-stos-grid as described for ir-fft. A contrast enhancement may be specified using the -clahe argument as for ir-fft. The following arguments are also supported:
-fft median_radius minimum_mask_overlap: The median radius specifies the radius of the median filter used to denoise the displacement vector image. The minimum mask overlap specifies the minimum area overlap ratio between neighbors. Default values are 1 and 0.5, respectively. For building the retinal connectome values of 2 and 0.25 were used.
-grid rows cols: Specifies the number of control points in the grid transformation.
-grid_spacing number_of_pixels: Specifies the grid spacing in pixels. A value of 192 was used for building the retinal connectome. Either -grid or grid_spacing should be used, but not both.
-neighborhood size: Specifies the size of the neighborhoods used for matching at each mesh vertex. A value of 128 was used in building the retinal connectome.
-it iterations: Specifies the number of iterations of mesh refinement. A value of 4 was used for building the retinal connectome.
-displacement_threshold offset_in_pixels: The average displacement change threshold at which the tool should stop iterating. Defined in pixels. Default value is 1.
2.3.8. ir-stom
The ir-stom program outputs a series of sections, specified by the -save argument, all registered to the first section using a series of .stos files as input. The .stos files specified with the -load argument are the section-to-section transforms between consecutive sections in a stack of sections. The n’th section is generated by cascading the first n-1 .stos files to get a transform mapping section n to the space of the first section.
2.4. Graphical user interface for mosaicking and section-to-section registration: Iris
We also developed a cross-platform graphical user interface called Iris that allows easy access to the ir-tools used in mosaicking and section-to-section registration. Iris provides a mosaic wizard and a volume wizard that automates these workflows with default parameters as well as a volume builder that allows detailed access to all the command line tools and associated parameters (Section 2.3) for advanced users. Iris also supports a batch mode, that allows running any multiple of tools as well as any selected number of sections. Each tool has a unique icon which is used to mark information about it. Figure 6 shows several screenshots illustrating the different modes of operation.
Figure 6.
Screenshots of Iris in action: (a) Mosaic wizard view, (b) Volume wizard view, (c) Volume builder access to ir-tools for advanced users, and (d) Customization of ir-tools parameters for advanced users.
The mosaic wizard facilitates building a mosaic from a user-specified directory of images. It loads the images, runs the tools (ir-fft, ir-refine-translate, ir-refine-grid, ir-assemble) with default parameters, and exports an assembled image. The volume wizard operates similarly. It loads the mosaics, runs the tools (ir-stos-brute, ir-stos-grid, ir-stom) with default parameters, and exports a stack of aligned images. A third wizard, the volume builder, allows users to customize tool parameters, change the pipeline flow by skipping tools or sections, reorganize sections, and finally export the data. This enables users to optimize volume assembly.
3. Results
3.1. Retinal connectome
The ir-tools described in this paper were used to build the first large-scale retinal connectome. The retinal connectome RC1 consists of 341 serial sections (nominally 70 nm thick), each captured and assembled as a mosaic of approximately 1000 tiles. Each tile is a 4096 × 4096 pixel imaged at 2.18 nm per pixel. This amounts to approximately 32 Gigabytes per section and 16 Terabytes (after processing) for the entire volume. The original image capture time for the volume was five months. Enhancements in the capture process now allow capture in 3 months (a rate of about 5000 images daily). The image assembly was performed on a single eight-core 3 Ghz computer. Mosaicking times for a single 1000 tile section was approximately 16 minutes for ir-refine-translate, 64 minutes for ir-refine-grid and 12 minutes for ir-assemble. Together with conversion from the MRC to TIF format approximately 8 sections can be mosaicked each day. The computational complexity of ir-refine-translate is 𝒪(NM logM) where N is the number of tiles in the section and M is the number of pixels per tile. The computational complexity of ir-refine-grid is 𝒪(PQlogQ) where P is the number of control points used and Q is the size of the neighborhood associated with each control point. Since approximate positions were known from metadata we did not need to use ir-fft in our workflow which has computational complexity 𝒪(N2M logM). Section-to-section alignment was less than one minute per pair for ir-stos-brute and approximately eight minutes per pair for ir-stos-grid. The computational complexity of ir-stos-grid is also 𝒪(PQlogQ). The total volume assembly time was three weeks.
Other datasets are also readily assembled. Figure 8 is an image of retina from rabbit expressing the rhodopsin P347L transgene. The image contains over 2200 TEM tiles captured and mosaicked in a completely automated fashion.
Figure 8.
[Color] A vertical section from a transgenic rabbit retina comprising over 2200 separate TEM tiles assembled in a completely automated fashion. Each tile is a 4096 × 4096 pixel image. Insets show several areas at varying levels of zoom to demonstrate the amount of information available in the mosaic.
3.2. Quantitative validation of accuracy
We also performed two experiments to quantify the accuracy of our image registration methods. First a simulation experiment was performed to assess the accuracy of ir-fft for various amounts of overlap between tiles. A single section from the RC1 dataset (see Figure 7) was chosen. We performed a simulated capture of a portion of this section by extracting 42 image tiles with 15% overlap, each 5000 × 5000 pixels. We then ran ir-fft with the default options to generate a new mosaic. When the positions of the tiles in the resulting mosaic were compared to ground truth, the maximum error in displacement was found to be 0.05 pixels, with a mean error of 0.01 pixels. This corresponds to a mean error of 0.1 nm in real image space. We performed the same experiment for several amounts of % overlap between tiles. The average errors in displacement are reported in Table 1.
Figure 7.
Four sections from the rabbit retinal connectome which comprises 341 sections. The sections shown are at approximately equal intervals in the volume progressing clockwise from top left. Each section is approximately 32 GB and comprises 1000 tiles.
Table 1.
Quantitative assessment of accuracy using a simulated image capture for various overlap amounts.
| Overlap (in % of tile size) | Average displacement (in pixels) |
|---|---|
| 15% | 0.013 |
| 10% | 0.028 |
| 8% | 0.066 |
For the second quantitative experiment, a mosaic consisting of 25 tiles (5 × 5 tile layout and each tile is 512×512 pixels) was manually and automatically registered using both rigid transforms only and non-rigid transforms. The mosaic is shown in Figure 9. Table 2 shows the root mean square difference in pixels between the control points in both cases. Notice that the manual and automatic registration are essentially identical for rigid transforms and are very close for the nonrigid case.
Figure 9.
5 × 5 tile mosaic used for comparison of manual vs. automatic registration.
Table 2.
Quantitative assessment of accuracy by comparison to manual registration.
| Average displacement (in pixels) | |
|---|---|
| Manual vs. automatic rigid transform % | 2.55 × 10−10 |
| Manual vs. automatic non-rigid transform | 1.45 |
It is not possible to directly quantify the accuracy of section-to-section alignment because there is no ground truth for the warping due to the section cutting, i.e. it is not known what changes are due to the cutting process and what changes are due to the neurons changing shape and position. Furthermore, an experiment performing alignment after a simulated warp would essentially be equivalent to the mosaicking experiment outlined above due to the similarity of the algorithms. However, we were able to track thousands of neurons across the volume which required correlating close to 200,000 profiles from one section to the next which would not be possible if the section-to-section alignment was low quality.
4. Discussion
We have developed a computationally efficient and robust fully-automatic method for large-scale image registration. The performance of the method was successful in both generating >10 terabyte-scale image volumes and extremely large single slices composed of over 2000 individual images. We have validated quantitative accuracy of the method for mosaicking these types of images using a simulated experiment. Though we envisioned the main application of our publicly available software tools would be biological neural network analyses, such as connectomics, the tools can easily be applied to other large image datasets and are not limited to studies of the nervous system. Thus large-scale volumetric ultrastructural analyses of other complex heterocellular tissues (histomics) are also feasible. Nor are these approaches limited to electron microscopy per se. Preliminary results indicate that the algorithms successfully mosaic and serial section data generation by high-resolution optical platforms. Future work will explore automated large-scale image volume construction in other imaging modalities such as confocal optical microscopy.
Research Highlights
A computationally efficient and robust, automatic solution for the registration of very large electron microscopy mosaics from thousands of image tiles.
A computationally efficient and robust, automatic solution for the section-to-section registration of a stack of electron microscopy image mosaics.
A user friendly graphical interface to the mosaicking and section-to-section registration algorithms.
Acknowledgements
This work was supported by NIH grants EB005832 (Tasdizen), EY02576 (Marc), EY015128 (Marc), EY0124800 Vision Core (Marc) and Research to Prevent Blindness Career Development Award (Jones). We thank the anonymous reviewers whose comments helped greatly improve the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The Neural Circuit Reconstruction (NCR) Toolset can be downloaded from http://www.sci.utah.edu/software.html
The Viking viewer is not discussed in this paper; however, a separate manuscript describing Viking is under preparation. Viking will be made publicly available
Contributor Information
Tolga Tasdizen, Electrical and Computer Engineering Department, University of Utah, Scientific Computing and Imaging Institute, University of Utah.
Pavel Koshevoy, Sorenson Media, Salt Lake City, Utah.
Bradley C. Grimm, Scientific Computing and Imaging Institute, University of Utah
James R. Anderson, Moran Eye Center, University of Utah
Bryan W. Jones, Moran Eye Center, University of Utah
Carl B. Watt, Moran Eye Center, University of Utah
Ross T. Whitaker, School of Computing, University of Utah, Scientific Computing and Imaging Institute, University of Utah
Robert E. Marc, Moran Eye Center, University of Utah
References
- Akselrod-Ballin A, Bock D, Reid RC, Warfield SK. Improved registration for large electron microscopy images. Proceedings of IEEE International Symposium on Biomedical Imaging; 2009. pp. 434–437. [Google Scholar]
- Anderson J, Jones B, Yang J-H, Shaw M, Watt C, Koshevoy P, Spaltenstein J, Jurrus E, U.V K, Whitaker R, Mastronarde D, Tasdizen T, Marc R. A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 2009;7(3):e74. doi: 10.1371/journal.pbio.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajcsy R, Kovacic S. Multiresolution elastic matching. Computer Vision Graphics and Image Processing. 1989;46(1):1–21. [Google Scholar]
- Bookstein F. Morphometric Tools for Landmark Data: Geometry and Biology. Cambridge University Press; 1997. [Google Scholar]
- Bookstein FL. Principal warps: Thin-plate splines and the decomposition of deformations. IEEE Trans Pattern Anal Mach Intell. 1989;11(6):567–585. [Google Scholar]
- Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006 October;16(5):562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Carlbom I, Terzopoulos D, Harris KM. Computer-assieted registration, segmentation, and 3d reconstruction from images of neuronal tissue sections. IEEE Trans Med Imaging. 1994 June;13(2) doi: 10.1109/42.293928. [DOI] [PubMed] [Google Scholar]
- Castro ED, Morandi C. Registration of translated and rotated images using finite fourier transforms. IEEE Trans Pattern Anal Mach Intell. 1987;9(5):700–703. doi: 10.1109/tpami.1987.4767966. [DOI] [PubMed] [Google Scholar]
- Chen BL, Hall DH, Chklovskii DB. Wiring optimization can relate neuronal structure and function. Proc Natl Acad Sci U S A. 2006;103(12):4723–4728. doi: 10.1073/pnas.0506806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen G, Rabbit R, Miller M. Deformable templates using large deformation kinematics. IEEE Trans Image Process. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- Cohen ED, Sterling P. Parallel circuits from cones to the on-beta ganglion cell. Eur J Neurosci. 1992;4:506–520. doi: 10.1111/j.1460-9568.1992.tb00901.x. [DOI] [PubMed] [Google Scholar]
- Dacheux RF, Chimento MF, Amthor FR. Synaptic input to the on-off directionally selective ganglion cell in the rabbit retina. J Comp Neurol. 2003;456(3):267–278. doi: 10.1002/cne.10521. [DOI] [PubMed] [Google Scholar]
- Davis J. Mosaics of scenes with moving objects. International Conference on Computer Vision and Pattern Recognition; 1998. pp. 354–360. [Google Scholar]
- Davis MH, Khotanzad A, Flamig DP, Harms SE. A physics-based coordinate transformation for 3-d image matching. IEEE Trans Med Imaging. 1997;16(3):317–328. doi: 10.1109/42.585766. [DOI] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2(11):e329. doi: 10.1371/journal.pbio.0020329. doi:10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AC, Beil C, Thompson SMCJ, Hakim A. Anatomical-functional correlation using an adjustable mri-based region of interest with positron emission tomography. J Cereb Blood Flow Metab. 1988;8:513–530. doi: 10.1038/jcbfm.1988.92. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Harris KM. Extending unbiased stereology of brain ultrastructure to three-dimensional volumes. J Am Med Inform Assoc. 2001;8(1):1–16. doi: 10.1136/jamia.2001.0080001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G RC, Woods RE. Digital Image Processing. Boston, MA, USA: Addison-Wesley Longman Publishing Co., Inc.; 1992. [Google Scholar]
- Girod B, Kuo D. Direct estimation of displacement histograms. Proceedings of the Optical Society of America Meeting on Understanding and Machine Vision; 1989. pp. 73–76. [Google Scholar]
- Hall DH, Russell RL. The posterior nervous system of the nematode caenorhaditis elegans: Serial reconstruction of identified neurons and complete pattern of synaptic interactions. J Neurosci. 1991;11(1):1–22. doi: 10.1523/JNEUROSCI.11-01-00001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Fiala JC, Ostroff L. Structural changes at dendritic spine synapses during long-term potentiation. Philos. Trans. R. Soc. Lond., Ser. B: Biol. Sci. 2003;358(1432):745–748. doi: 10.1098/rstb.2002.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayworth KJ, Kasthuri N, Schalek R, Lichtman JW. Automating the collection of ultrathin serial sections for large volume tem reconstructions. Microsc Microanal. 2006;12:86–87. [Google Scholar]
- Hoppe W. Three-dimensional electron-microscopy. Annu Rev Biophys Bioeng. 1981;10:563–592. doi: 10.1146/annurev.bb.10.060181.003023. [DOI] [PubMed] [Google Scholar]
- Ibanez L, Schroeder W, Ng L, Cates J. The ITK Software Guide: The Insight Segmentation and Registration. Toolkit: Kitware Inc.; 2003. [Google Scholar]
- Irani M, Anandan P, Hsu S. Mosaic based representations of video sequences and their applications. International Conf. on Computer Vision; 1995. pp. 605–611. [Google Scholar]
- Kanade T, Rander PW, Narayaman PJ. Virtualized reality: Constructing visrtual worlds from real scenes. IEEE Trans. on Multimedia. 1997;4(1):34–47. [Google Scholar]
- Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshevoy PA, Tasdizen T, Whitaker RT. Tech. Rep. UUSCI-2007-004. University of Utah: Scientific Computing and Imaging Institute; 2007. Automatic assembly of tem mosaics and mosaic stacks using phase correlation. [Google Scholar]
- Kuglin CD, Hines DC. The phase correlation image alignment method. Proc. Int. Conf. Cybernetics and Society; 1975. pp. 163–165. [Google Scholar]
- Levin A, Zomet A, Peleg S, Weiss Y. Seamless image stitching in the gradient domain. European Conf. on Computer Vision; 2004. pp. 377–389. [Google Scholar]
- Maintz JBA, Viergever MA. Navigated Brain Surgery. Oxford University Press; 1999. Ch. A Survey of medical image registration; pp. 117–136. [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mishchenko Y. Automation of 3d reconstruction of neural tissue from large volume of conventional serial section transmission electron micrographs. J Neurosci Methods. 2008 Sept; doi: 10.1016/j.jneumeth.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Rousso B, Rav-Acha A, Zomet A. Mosaicing on adaptive manifolds. IEEE Trans. on Pattern Analysis and Machine Intelligence. 2000;22(10):1144–1154. [Google Scholar]
- Preibisch S, Saafeld S, Tomancak P. Globally optimal stitching of tiled 3d microscopic image acquisitions. Bioinformatics. 2009;25(11):1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Fernandez A, Trujillo-Cenoz O, Apelbaum G, Bertalmio M, Vazquez L, Malmierca F, Morelli P. Neuro3d: an interactive 3d reconstruction system of serial sections using automatic registration. SPIE Proceedings Three-Dimensional and Multidimensional Microscopy: Image Acquisition and Processing V.1998. Jan, [Google Scholar]
- Rohr K, Fornefett M, Stiehl HS. Approximating thin-plate splines for elastic registration: Integration of landmark errors and orientation attributes. Lect Notes Comput Sci. 1999;1613:252–265. [Google Scholar]
- Rohr K, Fornefett M, Stiehl HS. Spline-based elastic image registration: integration of landmark errors and orientation attributes. Comput. Vision Image Understanding. 2003;90(2):153–168. [Google Scholar]
- Shum H-Y, Szeliski R. Construction of panaromic image mosaics with global and local alignment. Int. Journal of Computer Vision. 2002;48(2) [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput. Biol. 2005 Sep;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MG, Williams J, Munoz-Pinedo C, Perkins GA, Brown JM, Ellisman MH, Green DR, Frey TG. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat. Cell Biol. 2007;9:1057–1065. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- Tasdizen T, Koshevoy PA, Jones BW, Whitaker RT, Marc RE. Assembly of three-dimensional volumes from serial-section transmission electron microscopy. Proceedings of MICCAI Workshop on Microscopic Image Analysis with Applications in Biology. 2006:10–17. [Google Scholar]
- Thirion J. Extremal points: definition and application to 3d image registration. Proceedings of Computer Vision and Pattern Recognition. 1994:587–592. [Google Scholar]
- Thirion J. New feature points based on geometric invariants for 3d image registration. Int J Comput Vis. 1996;18(2):121–137. [Google Scholar]
- Toga A, editor. Brain Warping. New York: Wiley-Interscience; 1999. [Google Scholar]
- Vercauteren T, Perchant A, Malandain G, Pennec X, Ayache N. Robust mosaicing with correction of motion distortions and tissue deformations for in vivo fibered microscopy. Medical Image Analysis. 2006:673–692. doi: 10.1016/j.media.2006.06.006. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JNS, Brenner FRS. The structure of the nervous system of the nematode caenorhabditis elegans. Philos. Trans. R. Soc. Lond., Ser. B: Biol. Sci. 1986;314(1165):1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Zuiderveld K. Contrast limited adaptive histogram equalization. Graphics gems IV. 1994:474–485. [Google Scholar]