Abstract
A hierarchical approach has been developed for protein-protein docking. In the first step, a Fast Fourier Transform (FFT)-based docking algorithm is used to globally sample all putative binding modes, in which the protein is represented by a reduced model, that is, each side chain on the protein surface is represented by its center of mass. Compared to conventional FFT docking with all-atom models, the FFT docking method with a reduced model is expected to generate more hits because it allows larger side-chain flexibility. Next, the filtered binding modes (normally several thousands) are refined by an iteratively derived knowledge-based scoring function ITScorePP and by considering backbone/loop flexibility using an ensemble docking algorithm. The distance-dependent potentials of ITScorePP were extracted by a physics-based iterative method, which circumvents the long-standing reference state problem in the knowledge-based approaches. With this hierarchical protocol, we have participated in the CAPRI experiments for Rounds 15–19 of 11 targets (T32-T42). In the predictor experiments, we achieved correct binding modes for six targets: three are with high accuracy (T40 for both distinct binding modes, T41, and T42), two are with medium accuracy (T34 and T37), and one is acceptable (T32). In the scorer experiments, of the seven target complexes that contain at least one acceptable mode submitted by the CAPRI predictor groups, we obtained correct binding modes for four targets: three are with high accuracy (T37, T40, and T41) and one is with medium accuracy (T34), suggesting good accuracy and robustness of ITScorePP.
Keywords: protein-protein interaction, CAPRI experiments, scoring function, reduced model, molecular docking
1 Introduction
Protein-protein docking is a valuable computational tool for studying interactions between proteins that play important roles in many biological processes.1–5 Based on the structures of individual proteins, the docking process attempts to predict the structure of the complex by sampling putative binding modes of one protein around the other protein and by scoring/ranking the constructed complex structures with an energy function. Many protein-protein docking algorithms have been developed using different search/sampling methods,4 including local shape matching methods such as DOCK and PatchDock,6–11 direct global search method such as SOFTDOCK, BiGGER, GAPDOCK, ICM, RosettaDock, ATTRACT, and HADDOCK,12–18 and FFT-based seach algorithms19 such as 3D-DOCK, GRAMM, DOT, ZDOCK, MolFit, and PIPER.19–25 Among these methods, FFT-based algorithms have been widely used in protein-protein docking in the past decade and obtained considerable success because of its high computational efficiency. It can reduce the conventional search time O(N6) in the six-dimenstion (three translational plus three rotational) space to O(N3 log(N3)) by implementing a fast Fourier transform for the translation search.
Despite significant progresses, there remain several challenges in the field of protein-protein docking, One of them is how to account for protein flexibility.26 It is desirable but challenging to explicitly consider protein flexibility and particularly backbone flexibility because of the large number of atoms in a protein and thereby high degrees of freedom. Many docking algorithms such as FFT-based docking algorithms treat proteins as rigid bodies and consider the flexibility implicitly by allowing some favorable overlap between receptor and ligand protein surface layers.19 This kind of implicit treatment is capable of considering only small atomic movements on the protein surface and is not effective for the incorporation of conformational changes of side chains. Some direct search docking algorithms such as ICM, RosettaDock, and ATTRACT can account for side-chain flexibility and even larger loop and/or backbone flexibility by using Monte Carlo methods. However, these direct search methods are computationally expensive and thus are usually used for local protein-protein docking with the information of binding site(s) available. Therefore, for a truly global exhaustive search, FFT-based docking is still one of the commonly-used protein-protein docking algorithms or the first step in many post-docking methods.
To account for moderate protein flexibility and to retain the merit of FFT-based methods on highly efficient global search, we have developed a hierarchical approach for protein-protein docking. In this approach, putative binding poses are first generated by an FFT-type docking method using a reduced protein model, in which each side chain on the protein surface is represented by a single point located at its center of mass. Compared to conventional all-atom FFT-type docking methods, the reduced-model docking method can generate more hits by allowing larger side-chain flexibility. Similar reduced protein representations have also been proposed previously for direct global search algorithms.16;17 The initial binding poses by the FFT method are filtered by a shape complementarity criterion, leaving usually several thousands poses to be further refined by a recent iteratively-derived knowledge-based scoring function (ITScorePP).27 The backbone/loop flexibility are also considered by using an ensemble docking algorithm.28;29 To validate the hierarchical protein docking algorithm (MDockPP) and the iterative knowledge-based scoring function (ITScorePP), we participated in the CAPRI (Critical Assessment of Prediction of Interactions) experiments — a community-wide blind test for protein-protein interactions,30–32 from round 15 to round 20 (a total of 11 targets).
2 Materials and Methods
2.1 FFT-based, reduced-model docking
The putative binding modes between proteins were generated by a reduced-model, FFT-based docking algorithm for better incorporation of side-chain flexibility. The details of the algorithm will be described else where (manuscript in preparation). Briefly, each surface residue is represented by its center of mass for both the receptor and the ligand. Here, the receptor is defined as the larger protein in a complex, and the ligand is the smaller protein. The surface residues are the residues of which the solvent accessible surface area is more than 10% of the standard surface area, using a water probe radius of 1.4 Å.33 Next, the receptor protein is fixed and the ligand protein is rotated by an interval of 15° Euler angles in the rotational space. Shape complementarity is evaluated using fast Fourier transform (FFT).19;23 Compared to all-atom models, the reduced representation for the surface residues enables a more efficient consideration of side chain flexibility while retaining the reasonable description of the protein atomic coordinates.
2.2 The scoring function ITScorePP
The iterative knowledge-based scoring function ITScorePP that we recently developed was used to re-fine the filtered binding modes generated by the reduced-model FFT docking algorithm. The details of ITScorePP have been described in our previous work.27 Briefly, to derive ITScorePP, we used an iterative method to circumvent the reference state problem in traditional knowledge-based potentials. The basic idea is to improve the inter-atomic pair potentials {uij(r)} through iterations by comparing the experimentally observed structures and the predicted binding modes.34;35 The specific iterative equation is expressed as
| (1) |
Here, k stands for the iterative step. i and j represent the types of a pair of atoms in the receptor and the ligand. is the pair distribution function for atom pair ij observed in the experimentally determined (i.e., native) protein-protein complex structures. is the pair distribution function calculated from the ensemble of possible interaction modes including the native structure and 1000 decoys (i.e., incorrect models) for each complex, using the trial potentials { } at the k-th step. { } are the improved potentials after { }. At the end of the iterations, the final pair potentials are able to distinguish native structures from decoys. Details are described in ref 27.
ITScorePP was derived based on a large training set of 851 biological protein-protein complex structures, and has been extensively validated for binding mode prediction using three unbound docking test sets, including a combined test set from the three benchmarks (Benchmark 0.0, 1.0 and 2.0) prepared by Weng and colleagues,36;37 the ZDOCK decoy set,23;38 and the RosettaDock unbound perturbation decoy set.16
2.3 A hierarchical protocol for docking and scoring
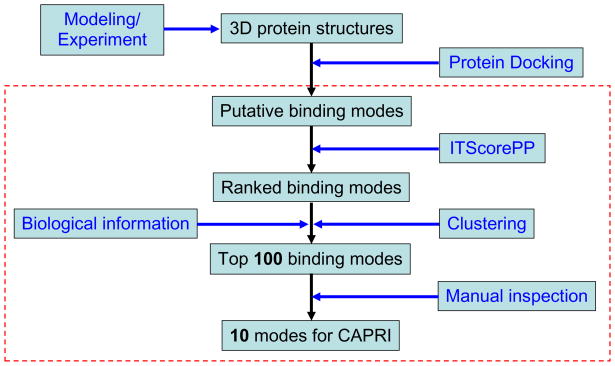
Figure 1 shows an illustration of the hierarchical protocol we used for the CAPRI predictor and scorer experiments. The only difference between these two types of experiments is the source of initial putative binding modes. For the predictor experiments, we generated our own putative binding modes by using available experimental structures or our modeled protein structures. For the scorer experiments, the initial putative binding modes were provided by CAPRI from different predictor participants.
Figure 1.
A flowchart for the hierarchical scheme that we used in docking and scoring for the CAPRI experiments. The three-dimensional protein structures were either obtained from the experimentally-determined structures provided by the CAPRI organizers or modeled from the given protein sequences. The procedures shown in the dashed box were used for the CAPRI scoring experiments, in which the putative binding modes were downloaded from the CAPRI site. Note: We used ZDOCK 2.1 for protein docking to generate the putative binding modes in Round 15 of Targets 32–36 before our reduced-model FFT docking program was developed.
Specifically, for the CAPRI predictor experiments, we first generated putative binding modes based on shape complementarity using the aforementioned reduced-model FFT docking algorithm. For the docking calculations, the grid spacing was set to 1.2 Å, and the interval of the Euler angles was set to 15° which resulted in 4416 rotations of the ligand in the Euler space. For each rotation, according to the FFT calculation, one relative translation of the ligand with the best shape complementarity to the receptor was kept for further refinement, yielding a total of 4416 putative binding modes for a docking run. Then, these 4416 filtered binding modes were scoring/optimized at atomic level using ITScorePP. If available, the biological information about the binding site was also applied at this refinement step to sort out the binding modes that satisfy the biological information. The ranked binding modes were then clustered. For two binding modes with rmsd < Rclu, only the ligand orientation with lower ITScore-PP score was kept. Here, Rclu is calculated based on the backbone atoms of the ligand, which was set to 8 Å unless otherwise specified. The top 100 binding modes after clustering were kept for manual inspection to assure that the biological information was properly applied. Ten binding modes were selected and submitted to CAPRI.
For the CAPRI scorer experiments, the protocol is similar except that the putative binding modes were directly downloaded from the CAPRI site that were kindly provided by the CAPRI predictors. Ten binding modes were finally submitted to CAPRI.
3 Results and Discussion
3.1 Overall Performance
Since the 3rd CAPRI Evaluation Meeting in 2007 (Toronto, Canada), there have been seven rounds (i.e. Rounds 13–19) of CAPRI experiments including 14 targets in which Target 31 has not been assessed yet. We joined CAPRI at round 15 and participated in the rounds 15–19 of 11 targets.
Table I summarized our CAPRI results. For the predictor experiments, we have predicted at least one acceptable binding modes for six targets, including three high-accuracy predictions for Targets 40, 41, and 42, two medium-accuracy predictions for Targets 34 (a protein-RNA complex) and 37, and one acceptable prediction for Target 32. The complexes with correct predictions cover different types of unbound docking tests, including unbound/unbound, unbound/bound, and unbound/homology tests.
Table I.
Performance of our docking/scoring method in CAPRI rounds 15–19.
| Targeta | Complexb | Typec | Bio.d Info. | Predicting |
Scoring |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| fnat (%) | Lrmsd (Å) | Irmsd (Å) | accuracye | fnat (%) | Lrmsd (Å) | Irmsd (Å) | accuracyf | ||||
| 32 | Savinase/BASI | U/U | Y | 43.5 | 11.55 | 3.40 | 1* | 0.5 | 13.55 | 4.87 | 0 |
| 33 | RNA/Enzyme | H/H | Y | 12.2 | 34.41 | 18.44 | 0 | 0.9 | 29.63 | 20.27 | 0 |
| 34 | RNA/Enzyme | B/H | Y | 48.7 | 3.19 | 1.67 | 3/2** | 49.6 | 3.06 | 1.71 | 7/2** |
| 35 | CBM22/GH10 | H/H | – | 0.0 | 28.80 | 11.89 | 0 | 0.05 | 33.58 | 8.72 | 0 |
| 36 | CBM22/GH10 | B/H | – | 0.0 | 27.04 | 7.81 | 0 | 0.08 | 24.18 | 6.17 | 0 |
| 37 | ARF6/LZ2 | U/H | Y | 79.6 | 3.28 | 1.08 | 3/1** | 93.9 | 1.15 | 0.78 | 4/2*** |
| 38 | Centaurin-α1/FHA | U/H | ? | 0.0 | 29.76 | 14.30 | 0 | 0.0 | 29.59 | 14.30 | 0 |
| 39 | Centaurin-α1/FHA | U/B | ? | 0.0 | 55.52 | 25.70 | 0 | 0.02 | 35.85 | 16.37 | 0 |
| 40 | Trypsin/API-A | U/B | Y | 88.2 | 0.78 | 0.40 | 9/3*** | 89.5 | 0.76 | 0.66 | 5/1*** |
| 41 | Colicin E9/Im2 | U/U | Y | 93.2 | 0.87 | 0.55 | 9/6*** | 94.9 | 1.50 | 0.61 | 10/2*** |
| 42 | TPR oligomer | U/U | – | 87.9 | 1.66 | 0.51 | 1*** | – | – | – | – |
We did not participate in the first two rounds (i.e., Rounds 13 and 14 of three targets: T29, T30 and T31).
The first one is assigned as the receptor, and the second one is the ligand.
The symbol “B” stands for the bound experimental structure, “U” for the unbound experimental structure, and “H” for the homology-modeled structure.
“Y” represents that valid biological information is available for the binding site, “–” represents that no or little biological information is available, and the question mark “?” means that the available experimental information about the binding site is not consistent with the crystal structure.
The accuracy is categorized by three parameters following the CAPRI criteria:30;31 The percentage of the native residue-residue contacts fnat, the ligand rmsd Lrmsd, and the interface rmsd Irmsd. “***” stands for high-accuracy, “**” for medium-accuracy, “*” for acceptable accuracy, and “0” for no correct prediction, respectively. Example: “9/3***” means that among 10 submitted binding modes for CAPRI, nine modes have an at least acceptable accuracy of which three are with high accuracy (***).
There were no scorer experiments for Target 42.
For the scorer experiments, our results are similar to those of the corresponding predictor experiments except for Target 32 and Target 37. With Target 32, we achieved one acceptable model in the predictor experiment (i.e., docking) but failed in the scorer experiment (i.e., scoring). With Target 37, our scorer experiment achieved high accuracy whereas our predictor experiment achieved medium accuracy, suggesting the relative accuracy and robustness of the ITScorePP scoring function.
3.2 Target 32 (Savinase/BASI)
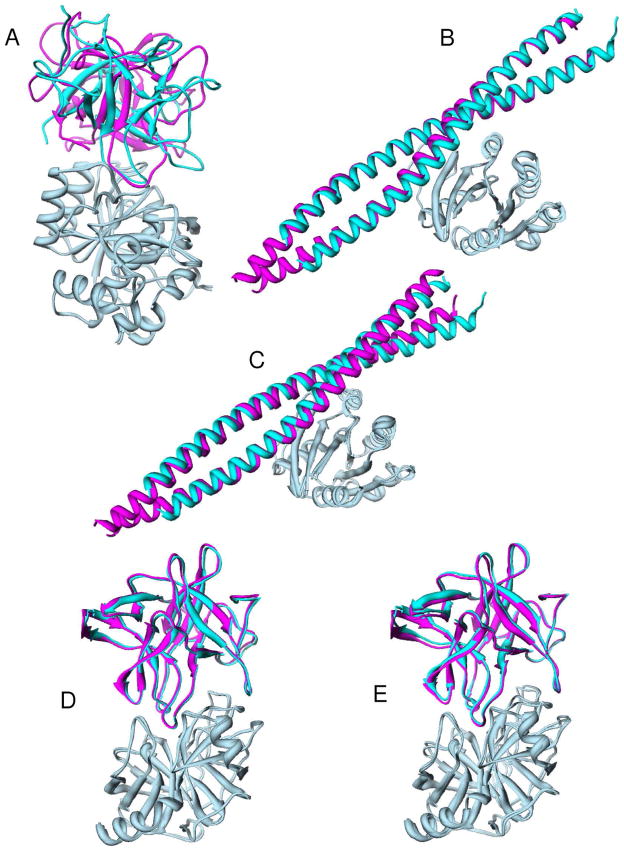
Target 32 is a complex formed by the protease Savinase and its inhibitor protein BASI.39 There exist unbound structures for both Savinase (1SVN)40 and BASI (1AVA)41 in the Protein Data Bank (PDB)42, as provided by the CAPRI site. Literature search shows that the active and substrate-binding site of Savinase consist of four subsites: S1 is formed by residues 155–166 and residue 191, S2 is a narrow “cleft” formed by residues Leu96, His64 and Gly100, S3 is the protein surface surrounded by S1, S2, and S4, and S4 is formed by the hydrophobic residues Val104, Ile107, and Leu135 and the hydrophilic side chains of Ser128, Pro129, and Ser130.43 Therefore, we assumed that the bi-functional inhibitor BASI binds to the same site on Savinase as its substrates. Mutagenesis experiments also suggest that residue Y87 on BASI could be involved in the Savinase-binding site.44 For this target, the putative binding modes were generated using ZDOCK 2.1 with the default parameters and then scored/minimized by using ITScorePP. Loop flexibility of BASI in the region near Y87 was also considered by generating multiple loop copies with the program LOOPY45;46 for multiple docking runs. The results from different docking runs were merged and ranked according to their binding scores, followed by a clustering procedure using a rmsd cutoff of 8 Å. The top 100 binding modes in which Y87 of BASI is within 6 Å of the S1–S4 sites of Savinase were kept for manual inspection. For the predictor experiment, our best model gave an acceptable prediction with an interface rmsd of 3.4 Å for this target [Fig. 2(A)]. However, our scorer experiment did not have any success. This is not as surprising as it sounds, because being first-time participants we did not upload our 10 predictor models to CAPRI for scoring and therefore the acceptable model identified by ITScorePP in our predictor experiment was not in the initial CAPRI predictor set for the scorer experiment.
Figure 2.
Comparison between the predicted binding mode (magenta) and experimentally determined crystal structure (cyan) where the complexes are aligned according to the receptor proteins (light blue). (A) Target 32: Savinase/BASI, predictor; (B) Target 37: ARF6/LZ2, predictor; (C) Target 37: ARF6/LZ2, scorer; (D) Target 40: Trypsin/API-A, predictor; (E) Target 40: Trypsin/API-A, scorer.
3.3 Targets 33/34 (RNA/Enzyme)
Targets 33 and 34 are the same complex between an RNA and a methyl transferase (unpublished results, Louis Renault, LEBS, Gif-sur-Yvette, France). In Target 33, the RNA structure required to be modeled based on homology, whereas in Target 34 the bound structure of the RNA was provided by CAPRI. For both targets, the 3D structure of the enzyme was modeled from its homologous protein RlmAI. The crystal structure of RlmAI implied that the enzyme may bind the RNA next to the S-adenosyl-L-methionine (SAM) binding site47. For docking or the predictor experiment, we modeled the 3D structure of the enzyme from the crystal structure of RlmAI (PDB code: 1P91) using Modeller.48 For Target 33, we used the RNA models from the Bonvin group through the CAPRI site. ZDOCK 2.1 was used to generate the putative binding modes between the RNA and the enzyme. The backbone flexibility of the enzyme was considered by docking multiple protein models generated with Modeller.48 To incorporate the binding site information, we constrained the docked/scored binding modes with a criterion that the RNA is within 5 Å of the SAM binding site of the enzyme. Similarly, the top 100 binding modes after clustering with an rmsd cutoff of 8.0 Å were kept for final manual inspection. For Target 34, our algorithms achieved medium-accuracy predictions for both the predictor (Irmsd = 1:67 Å) and scorer (Irmsd = 1:71Å) experiments. However, for Target 33, due to the large conformational change between the modeled and bound RNA structures, no acceptable solutions were predicted for docking or scoring by all the CAPRI groups.
3.4 Targets 35/36 (CBM22/GH10)
Targets 35 and 36 represent the same domain-domain interaction between the polysaccharide binding module CBM22 and the catalytic module GH10 of the xylanase Xyn10B from C. thermocellum (PDB code: 2W5F)49. In Target 35, the structures of CBM22 and GH10 both required homology modeling. However, in Target 36, CBM22 was replaced with a bound crystal structure. Although it is known that the two domains are covalently linked, the linked region is missing in the structures and there is a lack of clear information about the interaction interface. We modeled the 3D structures of CBM22 based on the crystal structure of its homologous protein (PDB code: 1YDO) 50 and modeled GH10 based on 1N82 using Modeller.48 We did not predict any acceptable or higher accuracy mode for both Target 35 and 36. For Target 35, our best models gave an interface rmsd of 11.89 Å and 8.72 Å for the predictor and scorer experiments, respectively, though the results for Target 36 are better with an interface rmsd of 7.81 Å and 6.17 Å respectively. One possible reason for the failure is that these two targets represent domain-domain interactions which could be different from conventional interactions between individual proteins.
3.5 Target 37 (ARF6/LZII)
Target 37 is a protein-protein complex between the G-protein ARF6 and LZII, the second leucine zipper domain of JIP4 (JNK-interacting protein 4)51. ARF6 has an unbound crystal structure (2A5D)52 and LZII was modeled using GCN4 leucine zipper (2ZTA)53. The complex between bacterial CTA1 and human ARF6-GTP revealed that the switch I, interswitch, and switch II regions (residues T40-T79) on ARF6 are the important protein binding site.52 Starting from Target 37 (round 16), to implement ITScorePP and to incorporate side-chain flexibility in docking, we have developed a reduce-model FFT docking program for hierarchical docking. Namely, since Target 37, the putative binding modes were generated/scored by our ITScorePP-implemented reduce-model FFT docking program. To utilize the biological information, we constrained the binding modes to those solutions in which any atom of LZII is within 5.0 Å from the binding site of ARF6. For this target, our best predicted models from the predictor experiment reached medium accuracy with the fraction of native residue contacts of 79.6% and interface rmsd of 1.08 Å [Fig. 2(B)], and best models from the scorer experiment reached high accuracy with the fraction of native residue contacts of 93.9% and interface rmsd of 0.78 Å [Fig. 2(C)], respectively.
3.6 Targets 38/39 (Centaurin-α1/FHA)
Targets 38 and 39 are the same protein-protein complex formed between centaurin-alpha 1 and the FHA domain of KIF13B (PDB code: 3FM8)54. An unbound crystal structure was provided for centaurin-alpha 1 by CAPRI. In Target 38, the structure of the FHA domain was modeled based on 2G1L using Modeller. In Target 39, the FHA domain was provided with a bound crystal structure. Due to being misled by the inappropriate experimental information that the FHA domain will bind to residues 1–126 of centaurin-alpha 1,55 no acceptable binding modes were found by all the CAPRI groups for Target 38, and only two groups submitted correct models in the predictor experiments for Target 39. Interestingly, without applying the biological information, our algorithm would be able to predict a high-accuracy binding mode in the top three solutions for Target 39, suggesting one has to be careful about using biological information in protein docking.
3.7 Target 40 (Trypsin/API-A)
Target 40 is a complex between the bovine trypsin and the double-headed arrowhead protease inhibitor API-A.56 This is an unbound/bound docking experiment where the bovine trypsin was provided with an unbound crystal structure (1BTY)57 and API-A was provided with a bound crystal structure.56 Previous studies about the complexes between trypsin and other proteins revealed that the region near SER195 in the active site is the putative protein binding site.58;59 The information provided by the authors (R. Bao, USTC, Hefei, China) showed that Leu87 and Lys145 of API-A are involved in potential protein binding. Since there are potentially two different binding modes, we submitted six models that are near Leu87 of API-A and four models that are near Lys145. For this target, we predicted the high-accuracy binding modes in both the predictor and scorer experiments for the two distinctive modes. Our best models achieved a high accuracy of fnat = 88.2% and Irmsd = 0.40 Å for the predictor experiment [Fig. 2(D)] and fnat = 89.5% and Irmsd = 0.66 Å for the scorer experiment [Fig. 2(E)], respectively.
3.8 Target 41 (Colicin E9/Im2)
Target 41 is a protein-protein complex between Colicin E9 DNase domain and Im2 immunity protein (PDB code: 2WPT).60 The task of Target 41 is to predict the binding mode between the DNase domain of colicin E9 and the IM2 immunity protein where colicin E9 is an unbound structure (1FSJ) and IM2 was taken from the NMR ensemble (2NO8). The interactions between colicin E9 and other proteins revealed that the segment of residues 75–90 is the possible binding site on colicin E9 and the region of residues 48–58 could be involved in binding for IM2 based on its homologous protein IMP9.61 To consider the backbone flexibility of IM2, we also modeled five more structures of IM2 for ensemble docking28 based on its homologous protein IM9 (1BXI, 1EMV, 1E0H, 1IMQ, and 1FR2) in addition to the NMR structures 2NO8. We predicted the high-accuracy models for both the predictor (fnat = 93.2% and Irmsd = 0.55 Å) and scorer (fnat = 94.9% and Irmsd = 0.61 Å) experiments for this target.
3.9 Target 42 (TPR/TPR)
Target 42 is to predict the oligomeric form of a designed tetratricopeptide repeat (TPR) (PDB code: 2WQH)62 that was modeled on Lynn Regan’s idealized TPR structure (1NA3).63 The oligomer (A:B:C) contains two different interfaces of A:B (symmetrical) and B:C (non-symmetrical). No information about the binding site was found in the literature. Therefore, the generated binding modes were purely ranked by our scoring function ITScorePP, followed by a clustering with an rmsd cutoff of 8.0 Å. Our best model achieved a high-accuracy classification with a native residue contact of 87.9% and an interface rmsd of 0.51 for the predictor experiment.
Although we achieved a high-accuracy classification for this target, we also predicted a binding mode that is distinctive from the experimentally determined oligomeric structure. In this second mode which has an even better energy score (i.e., more energetically stable), the oligomeric structure has identical binding interfaces; namely, the three TPRs bind to one another in a head-to-tail mode with helical symmetry. This finding is consistent with the polymeric characteristic of this target. Another CAPRI team (the Vajda group) also reported a similar finding. However, in the experimental structure it is an interesting symmetrical oligomer.62 The possibility of the presence of this second binding mode with helical symmetry might be worth for future experimental investigation at a different crystallographic condition.
4 Conclusion
We have developed an efficient hierarchical approach for protein-protein docking (MDockPP) in which the putative binding modes were generated by a reduced-model FFT-based protein docking and then scored/optimized by the iterative knowledge-based scoring function ITScorePP. The method was applied to the CAPRI experiments — a community-wide blind test for protein-protein interactions. For the predictor experiments, we predicted correct models for six targets out of 11 targets, including three high-accuracy, two medium-accuracy, and one acceptable predictions. For the scorer experiments, our scoring function achieved correct modes for four targets out of the seven target complexes that contain at least one acceptable mode submitted by the CAPRI predictors, including three high-accuracy and one medium-accuracy models. In spite of the success, lessons have also been learned through the CAPRI experiments of 11 targets. Biological information about the binding site is valuable for improving the selection of correct modes (Targets 32, 33, 34, 37, 40, and 41), despite that some experimental information may be misleading (Targets 38 and 39). Protein flexibility remains a major challenge for protein-protein docking (e.g. Target 33). Considering that many modeled structures are used in practical protein docking, inclusion of an intramolecular interaction component in ITScorePP may improve its scoring performance.
Acknowledgments
Support to XZ from OpenEye Scientific Software Inc. (Santa Fe, NM) and Tripos, Inc. (St. Louis, MO) is gratefully acknowledged. XZ is supported by NIH grant GM088517, Cystic Fibrosis Foundation grant ZOU07I0, Research Board Award RB-07-32 and Research Council Grant URC 09-004 of the University of Missouri. The work is also supported by Federal Earmark NASA Funds for Bioinformatics Consortium Equipment and additional financial support from Dell, SGI, Sun Microsystems, TimeLogic, and Intel.
References
- 1.Wodak SJ, Janin J. Computer analysis of protein-protein interaction. J Mol Biol. 1978;124:323–342. doi: 10.1016/0022-2836(78)90302-9. [DOI] [PubMed] [Google Scholar]
- 2.Smith GR, Sternberg MJ. Prediction of protein-protein interactions by docking methods. Curr Opin Struct Biol. 2002;12:28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 3.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–43. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 4.Schneidman-Duhovny D, Nussinov R, Wolfson HJ. Predicting molecular interactions in silico: II. Protein-protein and protein-drug docking. Curr Med Chem. 2004;11:91–107. doi: 10.2174/0929867043456223. [DOI] [PubMed] [Google Scholar]
- 5.Gray JJ. High-resolution protein-protein docking. Curr Opin Struc Biol. 2006;16:183–193. doi: 10.1016/j.sbi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromoleculeligand interactions. J Mol Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 7.Shoichet BK, Kuntz ID. Protein docking and complementarity. J Mol Biol. 1991;221:327–346. doi: 10.1016/0022-2836(91)80222-g. [DOI] [PubMed] [Google Scholar]
- 8.Lorber DM, Udo MK, Shoichet BK. Protein-protein docking with multiple residue conformations and residue substitutions. Protein Sci. 2002;11:1393–1408. doi: 10.1110/ps.2830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolfson HJ, Lamdan Y. Geometric hashing: A general and efficient model-based recognition scheme. Proceedings of the IEEE Int Conf on Computer Vision; Tampa, FL. 1988. pp. 238–249. [Google Scholar]
- 10.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nuclei Acid Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bordner AJ, Gorin AA. Protein docking using surface matching and supervised machine learning. Proteins. 2007;68:488–502. doi: 10.1002/prot.21406. [DOI] [PubMed] [Google Scholar]
- 12.Jiang F, Kim SH. Soft docking: matching of molecular surface cubes. J Mol Biol. 1991;219:79–102. doi: 10.1016/0022-2836(91)90859-5. [DOI] [PubMed] [Google Scholar]
- 13.Palma PN, Krippahl L, Wampler JE, Moura JJ. BiGGER: a new (soft) docking algorithm for predicting protein interactions. Proteins. 2000;39:372–384. [PubMed] [Google Scholar]
- 14.Gardiner EJ, Willett P, Artymiuk PJ. Protein docking using a genetic algorithm. Proteins. 2001;44:44–56. doi: 10.1002/prot.1070. [DOI] [PubMed] [Google Scholar]
- 15.Abagyan R, Totrov M, Kuznetsov D. ICM – A new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15:488–506. [Google Scholar]
- 16.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 17.Zacharias M. Protein-protein docking with a reduced protein model accounting for side-chain flexibility. Protein Sci. 2003;12:1271–1282. doi: 10.1110/ps.0239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 19.Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–9. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J Mol Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 21.Vakser IA. Evaluation of GRAMM low-resolution docking methodology on the hemagglutininantibody complex. Proteins. 1997;(Suppl 1):226–230. [PubMed] [Google Scholar]
- 22.Mandell JG, Roberts VA, Pique ME, Kotlovyi V, Mitchell JC, Nelson E, Tsigelny I, Ten Eyck LF. Protein docking using continuum electrostatics and geometric fit. Protein Eng. 2001;14:105–113. doi: 10.1093/protein/14.2.105. [DOI] [PubMed] [Google Scholar]
- 23.Chen R, Weng ZP. A novel shape complementarity scoring function for protein-protein docking. Proteins. 2003;51:397–408. doi: 10.1002/prot.10334. [DOI] [PubMed] [Google Scholar]
- 24.Heifetz A, Katchalski-Katzir E, Eisenstein M. Electrostatics in protein-protein docking. Protein Sci. 2002;11:571–587. doi: 10.1110/ps.26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 26.Bonvin AM. Flexible protein-protein docking. Curr Opin Struct Biol. 2006;16:194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Huang S-Y, Zou X. An iterative knowledge-based scoring function for protein-protein recognition. Proteins. 2008;72:557–579. doi: 10.1002/prot.21949. [DOI] [PubMed] [Google Scholar]
- 28.Huang S-Y, Zou X. Ensemble docking of multiple protein structures: Considering protein structural variations in molecular docking. Proteins. 2007;66:399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 29.Huang S-Y, Zou X. Efficient molecular docking of NMR structures: Application to HIV-1 protease. Protein Sci. 2007;16:43–51. doi: 10.1110/ps.062501507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janin J, Henrick K, Moult J, Ten Eyck L, Sternberg MJE, Vajda S, Vasker I, Wodak SJ. CAPRI: a critical assessment of predicted interactions. Proteins: Struct Funct Genet. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 31.Méndez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 32.Lensink MF, Wodak SJ, Méndez R. Docking and scoring protein complexes: CAPRI 3rd edition. Proteins. 2007;69:704–718. doi: 10.1002/prot.21804. [DOI] [PubMed] [Google Scholar]
- 33.Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- 34.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: I. Derivation of interaction potentials. J Comput Chem. 2006;27:1865–1875. doi: 10.1002/jcc.20504. [DOI] [PubMed] [Google Scholar]
- 35.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: II. Validation of the scoring function. J Comput Chem. 2006;27:1876–1882. doi: 10.1002/jcc.20505. [DOI] [PubMed] [Google Scholar]
- 36.Chen R, Mintseris J, Janin J, Weng Z. A protein-protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- 37.Mintseris J, Wiehe K, Pierce B, Anderson R, Chen R, Janin J, Weng Z. Protein-protein docking benchmark 2.0: an update. Proteins. 2005;60:214–216. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- 38.Chen R, Li L, Weng ZP. ZDOCK: An initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 39.Micheelsen PO, Vévodová J, De Maria L, Ostergaard PR, Friis EP, Wilson K, Skjøt M. Structural and mutational analyses of the interaction between the barley alpha-amylase/subtilisin inhibitor and the subtilisin savinase reveal a novel mode of inhibition. J Mol Biol. 2008;380:681–690. doi: 10.1016/j.jmb.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 40.Betzel C, Klupsch S, Papendorf G, Hastrup S, Branner S, Wilson KS. Crystal structure of the alkaline pro-teinase Savinase from Bacillus lentus at 1.4 Å resolution. J Mol Biol. 1992;223:427–445. doi: 10.1016/0022-2836(92)90662-4. [DOI] [PubMed] [Google Scholar]
- 41.Vallée F, Kadziola A, Bourne Y, Juy M, Rodenburg KW, Svensson B, Haser R. Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution. Structure. 1998;6:649–659. doi: 10.1016/s0969-2126(98)00066-5. [DOI] [PubMed] [Google Scholar]
- 42.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georgieva DN, Stoeva S, Voelter W, Genov N, Betzel C. Differences in the specificities of the highly alka-lophilic proteinases Savinase and Esperase imposed by changes in the rigidity and geometry of the substrate binding sites. Arch Biochem Biophys. 2001;387:197–201. doi: 10.1006/abbi.2000.2249. [DOI] [PubMed] [Google Scholar]
- 44.Bøsager BC, Nielsen PK, Abou Hachem M, Fukuda K, Præorius-Ibba M, Svensson B. Mutational analysis of target enzyme recognition of the b-trefoil fold barley aamylase/subtilisin inhibitor. J Biol Chem. 2005;280:14855–14864. doi: 10.1074/jbc.M412222200. [DOI] [PubMed] [Google Scholar]
- 45.Xiang Z, Soto CS, Honig B. Evaluating conformational free energies: The colony energy and its application to the problem of loop prediction. Proc Natl Acad Sci USA. 2002;99:7432–7437. doi: 10.1073/pnas.102179699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soto CS, Fasnacht M, Zhu J, Forrest L, Honig B. Loop modeling: sampling, filtering and scoring. Proteins. 2008;70:834–843. doi: 10.1002/prot.21612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das K, Acton T, Chiang Y, Shih L, Arnold E, Montelione GT. Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc Natl Acad Sci USA. 2004;101:4041–4046. doi: 10.1073/pnas.0400189101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marti-Renom MA, Stuart A, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 49.Najmudin S, Pinheiro BA, Prates JAM, Romao MJ, Fontes CMGA. Putting an N-terminal end to the Clostrid-ium thermocellum xylanase Xyn10b story: Crystallographic structure of the CBM22–1-GH10 modules complexed with xylohexaose. 2010 doi: 10.1016/j.jsb.2010.07.009. submitted. [DOI] [PubMed] [Google Scholar]
- 50.Forouhar F, Hussain M, Farid R, Benach J, Abashidze M, Edstrom WC, Vorobiev SM, Xiao R, Acton TB, Fu Z, Kim JJ, Miziorko HM, Montelione GT, Hunt JF. Crystal structures of two bacterial 3-hydroxy-3-methylglutaryl-CoA lyases suggest a common catalytic mechanism among a family of TIM barrel metal-loenzymes cleaving carbon-carbon bonds. J Biol Chem. 2006;281:7533–7545. doi: 10.1074/jbc.M507996200. [DOI] [PubMed] [Google Scholar]
- 51.Isabet T, Montagnac G, Regazzoni K, Raynal B, El Khadali F, England P, Franco M, Chavrier P, Houdusse A, Ménétrey J. The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4. EMBO J. 2009;28:2835–2845. doi: 10.1038/emboj.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 53.O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 54.Shen L, Tong Y, Tempel W, MacKenzie F, Arrowsmith CH, Edwards AM, Bountra C, Weigelt J, Bochkarev A, Park H. Crystal structure of full length centaurin alpha-1 bound with the FHA domain of KIF13B. 2010 to be published. [Google Scholar]
- 55.Venkateswarlu K, Hanada T, Chishti AH. Centaurin-alpha1 interacts directly with kinesin motor protein KIF13B. J Cell Sci. 2005;118:2471–2484. doi: 10.1242/jcs.02369. [DOI] [PubMed] [Google Scholar]
- 56.Bao R, Zhou ZC, Jiang C, Lin SX, Chi CW, Chen Y. The ternary structure of double-headed arrowhead protease inhibitor API-A complexed with two trypsins reveals a novel reactive site conformation. J Biol Chem. 2009;284:26676–26684. doi: 10.1074/jbc.M109.022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katz BA, Finer-Moore J, Mortezaei R, Rich DH, Stroud RM. Episelection: novel Ki approximately nanomolar inhibitors of serine proteases selected by binding or chemistry on an enzyme surface. Biochemistry. 1995;34:8264–8280. doi: 10.1021/bi00026a008. [DOI] [PubMed] [Google Scholar]
- 58.Blow DM, Janin J, Sweet RM. Mode of action of soybean trypsin inhibitor (Kunitz) as a model for specific protein-protein interactions. Nature. 1974;249:54–57. doi: 10.1038/249054a0. [DOI] [PubMed] [Google Scholar]
- 59.Huber R, Kukla D, Bode W, Schwager P, Bartels K, Deisenhofer J, Steigemann W. Structure of the complex formed by bovine trypsin and bovine pancreatic trypsin inhibitor. II. Crystallographic refinement at 1.9 A resolution. J Mol Biol. 1974;89:73–101. doi: 10.1016/0022-2836(74)90163-6. [DOI] [PubMed] [Google Scholar]
- 60.Meenan NAG, Sharma A, Fleishman SJ, MacDonald C, Morel B, Boetzel R, Moore GR, Baker D, Kleanthous C. The structural and energetic basis for high selectivity in a high affinity protein-protein interaction. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.0910756107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kühlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C. Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J Mol Biol. 2000;301:1163–1178. doi: 10.1006/jmbi.2000.3945. [DOI] [PubMed] [Google Scholar]
- 62.Krachler AM, Sharma A, Kleanthous C. Self-association of TPR domains: Lessons learned from a designed, consensus-based TPR oligomer. Proteins. 2010;78:2131–2143. doi: 10.1002/prot.22726. [DOI] [PubMed] [Google Scholar]
- 63.Main ER, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11:497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]