Abstract
In CAPRI rounds 13–19, the most native-like structure predicted by RosettaDock resulted in two high, one medium and one acceptable accuracy model out of 13 targets. The current rounds of CAPRI were especially challenging with many unbound and homology modeled starting structures. Novel docking methods, including EnsembleDock and SnugDock, allowed backbone conformational sampling during docking and enabled the creation of more accurate models. For Target 32, α-amylase/subtilisin inhibitor-subtilisin savinase, we sampled different backbone conformations at an interfacial loop to produce five high-quality models including the most accurate structure submitted in the challenge (2.1 Å ligand rmsd, 0.52 Å interface rmsd). For Target 41, colicin-immunity protein, we used EnsembleDock to sample the ensemble of nuclear magnetic resonance (NMR) models of the immunity protein to generate a medium accuracy structure. Experimental data identifying the catalytic residues at the binding interface for Target 40 (trypsin-inhibitor) were used to filter RosettaDock global rigid body docking decoys to determine high accuracy predictions for the two distinct binding sites in which the inhibitor interacts with trypsin. We discuss our generalized approach to selecting appropriate methods for different types of docking problems. The current toolset provides some robustness to errors in homology models, but significant challenges remain in accommodating larger backbone uncertainties and in sampling adequately for global searches.
Keywords: SnugDock, EnsembleDock, Flexible Backbone, Protein-Protein Docking, Flexible Loop Docking, Docking NMR Models
INTRODUCTION
Proteins are one of the most important classes of molecules in biology, and protein-protein and protein-nucleic acid interactions are responsible for important cellular functions. Advances in high-throughput proteomics allow identification of protein-protein complexes with high binding affinities. Rational engineering of proteins to improve binding affinity or alter binding specificities requires structural insights, but structure determining experimental tools like x-ray crystallography and NMR are laborious, time consuming and expensive. In the absence of experimentally obtained structures, the development of computational techniques for prediction of protein-protein interactions allows generation of structural models, and steady advances in computational power enables increasingly more thorough sampling of the available conformational space to generate physically realistic high resolution structures. The Critical Assessment of PRotein Interactions (CAPRI),1 a blind, community-wide challenge to computationally predict new experimentally solved structures of protein complexes, serves as a testing platform for the effectiveness of docking protocols. Our docking software, RosettaDock,2 has continually evolved by incorporating novel scoring and sampling strategies, and it has been successful in all rounds of CAPRI.3–5
CAPRI has become more challenging, evolving from initial rounds where most targets involved docking starting with bound protein partners, via intermediate rounds where the starting monomers were unbound structures, to one of the most challenging docking problems in the current rounds that require homology modeling of the starting monomers for most targets. It is becoming increasingly clear that backbone flexibility during docking is the logical next step for successful docking predictions.6,7 In CAPRI rounds 13–19, 7 of 13 targets required homology modeling, compared to 3 of 8 targets in the previous sets of rounds.8 Homology models are imperfect, especially when the sequence identity of the query sequence to the template sequence is poor. Correct docking solutions are precluded by homology models that exhibit significant deviation of the binding patch from that in the bound orientation. RosettaDock is meeting the increasingly complex docking challenge by incorporating backbone flexibility during docking to sample conformations that bridge the gap between the unbound/homology modeled structures and the bound structure.
Our early attempts at incorporating backbone flexibility in docking in the previous rounds of CAPRI underscored the inherent challenges involved in both sampling realistic backbone conformations and in energetically discriminating near-native structures with varying backbone conformations.5 In Target 20, HemK plus eRF1, we pre-generated multiple loop conformations along a flexible interface loop prior to docking but did not sample a near-native loop conformation. In Target 24, Arf1-GTP plus ARHGAP10, we modeled a 15-residue loop and sampled various backbone conformations of a 33-residue C-terminal tail during docking, but found that the docking simulations resulted in non-compact and unrealistic backbone conformations. In both cases all of our predictions were incorrect.
Since then we have developed two new techniques to more realistically capture backbone conformational change. First, our recently developed EnsembleDock9 protocol follows the conformer-selection model of binding by using a partition function-based selection of candidate backbone conformations from an ensemble of NMR models or a set of refined unbound structures. Second, SnugDock10 is a flexible docking protocol for docking of antibody-antigen complexes that structurally optimizes the paratope during docking to simulate an induced-fit. That is, SnugDock samples the relative orientation of the antibody light and heavy chains and the backbone conformations of the complementarity determining region loops while docking to the antigen. In local docking tests,9,10 recovery of models created by SnugDock and EnsembleDock outperform rigid-backbone RosettaDock, and the combination of EnsembleDock and SnugDock for docking homology modeled starting structures approaches that as with crystal structures using standard RosettaDock. We were eager to test the approaches in CAPRI. While there were no antibody targets in the rounds, we were able to adjust the flexible loop building methods for Target 32, and EnsembleDock was directly applied to Targets 29, 35–37 and 41.
TARGETS AND PREDICTIONS
In CAPRI rounds 13–19, RosettaDock with and without flexible docking generalizations (EnsembleDock and SnugDock) predicted two high, one medium and one acceptable quality most native-like model according to the standard CAPRI criteria. All decoys were evaluated using the ligand root mean square deviation (Lrmsd), interface rmsd (Irmsd) and fraction of native contacts (fnat).11 Table I shows a summary of the docking techniques and the results for all targets. We employed different strategies for each prediction, depending on the available data. When no experimental information was available for the binding interface, we performed a global docking simulation. For cases where experimental biochemical information or sequence conservation bioinformatics data were available to identify the binding patch, a local docking perturbation sufficed. When multiple structures were available for either of the docking partners (e.g. a set of NMR structures), we used EnsembleDock to dock the ensemble of structures (Target 41). For cases (Target 32) where a loop at the binding surface was known to be important for binding, we applied SnugDock methods for simultaneously optimizing the loop and docking. When an unbound structure was provided, and evidence suggested that the protein does not change its conformation upon binding, we applied standard RosettaDock involving rigid-body moves with side-chain flexibility. Additionally, for some cases (Target 40) available biochemical information was used as post-processing filters to eliminate low-scoring incorrect predictions (false-positives).
Table I.
Summary of docking techniques and results for all targets. Complex shows the names of the two proteins. Type indicates the origin of the starting monomer coordinates: bound (B), unbound (U) or homologous (H) structures. Percent sequence identity, SeqID for homology targets is shown by the number (##) following ‘H’, denoted as ‘H##’. If a set of NMR structures is provided for a monomer, it is indicated by NMR in parentheses. Search Scale indicates if a global or local docking perturbation. Method indicates the docking strategy used. Best Model is the submitted structure with the lowest ligand root mean square deviation (Lrmsd). The three metrics that decide the CAPRI quality rating are provided for the best model: fraction of native contacts (fnat), Lrmsd and interface root mean square deviation (Irmsd). The CAPRI rating of the Best Model with the respective submission rank is also provided. High, medium, acceptable and incorrect quality predictions are indicated by ***, **, * and - respectively. The last column summarizes the quality of the ten submitted structures for each target.
| Target | Complex | Type/SeqID (%) | Search Scale | Method | Metrics of Best Model |
Model Quality (High/Medium/Acceptable) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Model Rank | fnat | Lrmsd (Å) | Irmsd (Å) | Quality | ||||||
| 29 | Trm8-Trm82 | U-B | Global | EnsembleDock | 8 | 0.13 | 25.3 | 13.3 | - | |
| 30 | Rac1-Plexin B1 | U-U(NMR) | Local | Standard | 5 | 0.27 | 15.5 | 4.3 | - | |
| 32 | Subtilisin-Inhibitor | U-U | Local | SnugDock | 2 | 0.75 | 2.1 | 0.5 | *** | 5***/2**/1* |
| 32.AD | Subtilisin-Inhibitor | U-U | Local | SnugDock | 6 | 0.00 | 38.0 | 9.3 | - | |
| 33 | Methyltransferase-RNA | H31-U | Local | Protein-RNA | 8 | 0.11 | 27.9 | 5.1 | - | |
| 34 | Methyltransferase-RNA | H31-B | Local | Protein-RNA | 3 | 0.21 | 4.6 | 2.6 | * | 0***/0**/3* |
| 35 | Xylanase-Xylan BD | H33-H23 | Global | DomainInsertion | 9 | 0.00 | 34.3 | 11.5 | - | |
| 36 | Xylanase-Xylan BD | H33-B | Global | DomainInsertion | 3 | 0.00 | 33.4 | 13.9 | - | |
| 37.1 | Arf6GTP-LZ2 JIF4 | U-H20 | Global | EnsembleDock | 1 | 0.00 | 29.0 | 10.8 | - | |
| 37.2 | Arf6GTP-LZ2 JIF4 | U-H20 | Global | EnsembleDock | 1 | 0.00 | 28.7 | 10.6 | - | |
| 38 | Centaurin a1-KIF13B | U-H38 | Global | EnsembleDock | 4 | 0.00 | 53.4 | 18.0 | - | |
| 39 | Centaurin a1-KIF13B | U-B | Global | Standard | 9 | 0.08 | 24.9 | 12.7 | - | |
| 40.CA | B-Trypsin-API | U-B | Global | Standard | 4 | 0.83 | 2.3 | 0.6 | *** | 6***/0**/4* |
| 40.CB | B-Trypsin-API | U-B | Global | Standard with biochemical filter | 2 | 0.76 | 1.8 | 0.6 | *** | 3***/0**/7* |
| 41 | E9DNase-Im2 | U-U(NMR) | Local | EnsembleDock | 9 | 0.59 | 5.0 | 1.8 | ** | 0***/1**/7* |
| 42.AB | TPR oligomer | H95.2-H95.2 | Global | Symmetric | 7 | 0.00 | 15.6 | 4.6 | - | |
| 42.BC | TPR oligomer | H95.2-H95.2 | Global | Symmetric | 5 | 0.00 | 31.6 | 5.1 | - | |
Flexible Backbone Docking
Target 32: Barley α-Amylase/Subtilisin Inhibitor (BASI)-Subtilisin Savinase
Target 32 required prediction of the BASI-subtilisin complex starting with the unbound crystal structures of BASI (Protein Data Bank12 (PDB) ID: 1AVA:C)13 and subtilisin (1SVN).14 A previous paper describes the complex structure of proteinase K inhibitor PK13 and proteinase K, which are similar to BASI and bacterial subtilisin respectively, suggesting that the interaction complex may adopt a similar conformation.15 While a crystal structure was not deposited in the PDB for the complex described previously, the paper identifies a catalytic loop in the inhibitor (residues 84–94), and describes atomic interactions between proteinase K and its inhibitor. The atomic interactions were used to align the starting structures for a local docking perturbation. A modified version of the SnugDock protocol was applied to optimize the backbone conformation of the inhibitor loop responsible for the β-sheet formation and the C-terminus. The traditional SnugDock protocol, applicable to antibody-antigen complexes, was modified to make the catalytic loop flexible instead of the antibody complementarity determining region loops. Loop conformation was perturbed by a combination of small and shear moves16 loop closure using cyclic coordinate descent,17 and quasi-Newton minimization. As a post-processing step, decoys were scanned for the hydrogen bonding ladder reported in the homologous complex structure.
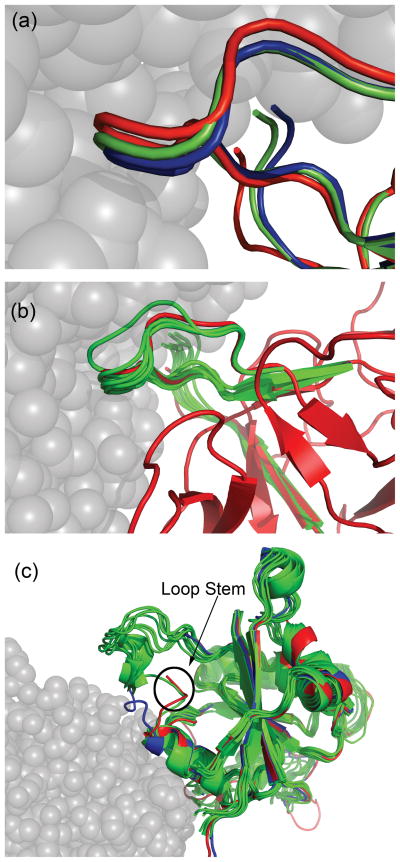
The local docking perturbation incorporating backbone flexibility and biochemical information resulted in five high quality, two medium quality and one acceptable model. The top-ranked model was of high quality. The best high quality model is our second prediction with 2.1 Å Lrmsd, 0.52 Å Irmsd and 0.75 fnat; this model was the best structure submitted among all CAPRI participants. Figure 1(a) shows that SnugDock’s backbone flexibility enabled the catalytic loop to move slightly closer to the bound conformation (3BX1)18 by 0.1 Å Cα global rmsd to the bound structure solution. In the bound structure, threonine 88 (in the catalytic loop), arginine 81, and aspartatic acid 99 have multiple conformations suggesting a flexible loop. Such flexibility can be captured by loop conformation diversity generated by SnugDock’s backbone conformational sampling as shown in Figure 1(b). Both Figures 1(a) and 1(b) also show that the narrow C-terminus conformational space sampled by the conservative perturbations of SnugDock does not span the large deviation of the bound C-terminus conformation from the starting conformation. More accurate docking predictions are precluded by the inability of the C-terminus to move significantly away, because the initial BASI C-terminus conformation clashes with serine 132 of the bound subtilisin savinase conformation.
Figure 1.
Backbone conformational sampling during docking. (a) Target 32: Protein loop representations showing that the best SnugDock-generated model of BASI (green) is closer to the bound conformation (red) than the starting unbound conformation (blue). The partner subtilisin savinase is in the bound conformation (gray spheres). The SnugDock generated model and the starting unbound structure of BASI has been superimposed on the BASI of the co-crystal (3BX1). (b) Target 32: Protein loop conformations of the ten lowest-scoring SnugDock-generated BASI molecules (green) show that the conformational space sampled by SnugDock spans the bound conformation. The BASI models are superimposed on the bound conformation of BASI in the co-crystal (red). Subtilisin savinase is shown in the bound conformation (gray). (c) Target 29: Superposition of unbound Trm8 structure (blue) and ensemble of unbound relaxed structures with the modeled loop (green) on bound Trm8 (red) in the co-crystal complex (2VDU) shows that the stems of the predicted loop are similar to that in the bound conformation. Only one representative loop stem from the ensemble of relaxed structures is shown for clarity. The docking partner Trm82 is in the bound conformation (translucent gray spheres).
Target 41: Colicin-Immunity Protein
In Target 41, we were provided unbound coordinates of the DNase domain of colicin E9 (1FSJ)19 and an NMR ensemble for the IM2 immunity protein (2NO8).20 We found an x-ray crystal structure of the homologous complex between the DNase domain of colicin E7 and immunity protein IM7 (7CEI)21 in the PDB. This structure was used as a template for coarse structural alignment of the given proteins. A local ensemble docking run was then carried out keeping the backbone of the DNase domain of colicin E9 rigid while including all alternate backbone structures from the NMR ensemble for the IM2 immunity protein. Because EnsembleDock samples backbone conformations in the low-resolution stage of docking, the computational time is increased 2.2 fold over standard RosettaDock, which is very efficient sampling of the sixty NMR models of the IM2 immunity protein monomer. Each of the ten lowest-scoring models obtained by ensemble docking were subjected to local refinement involving high-resolution rigid body docking to generate the final structures. This exercise resulted in seven acceptable predictions and one medium prediction (1.7 Å Irmsd and 0.59 fnat). Structural comparison cannot be performed because the coordinates of the bound structure (2WPT22) have not yet been released.
Target 29: Trm8-Trm82
We were given the bound structure of Trm82 (2VDU:B)23 and the unbound structure of Trm8 (2VDV).23 Inspection of a sequence alignment of Trm8 revealed a surface loop (residues 183–198) that was highly conserved across eukaryotes that could potentially play a role in forming a complex with Trm82.24 This loop is largely disordered in the unbound Trm8 structure, and we used a fragment-based Rosetta loop building protocol25 to create a complete loop. The structure with the loop was then used as an input for RosettaRelax,26 an optimization of the side-chain and backbone atomic co-ordinates in full atomistic detail, as described in the original EnsembleDock paper. We used an ensemble of ten relaxed structures and docked them with the bound conformation of Trm82.
We selected the ten lowest-energy clusters of solutions from the global docking run to serve as our final CAPRI predictions. Unfortunately, all models were incorrect. However, retrospective analysis showed that although the loop apex is still unresolved in the bound crystal structure,23 the loop-building and refinement methods were successful in recovering the disorder-to-order transition of the stem regions of the highly conserved surface loop with remarkable structural similarity to the bound conformation (Figure 1(c)). Although ultimately unsuccessful, these results demonstrate the potential of combining loop modeling and refinement with ensemble docking to overcome the challenges of flexible and disordered regions in unbound structures. EnsembleDock simulations starting with multiple loop conformations instead of multiple refined states of one loop may have helped recover more native-like models.
Target 35–36: Xylanase-Xylan BD
In Target 35, we were given the amino-acid sequences for xylanase and xylan BD and asked to find the quaternary structure of an end-to-end fusion protein. We used the crystal structures of a related xylan binding domain (1DYO,27 sequence identity 23%) to generate a homology model of xylan BD, and the crystal structure of a related xylanase (1N82,28 sequence identity 33%), to generate a model for xylanase. We used Robetta29 to generate five homology models for each partner and modified Rosetta’s DomainInsertion protocol30 to incorporate ensemble docking in its low-resolution phase. Our combination protocol alternated between rigid-body moves, selection of xylanase and xylan BD conformers, and loop building steps to explore conformations of the inter-domain linker. The algorithm generated structures of the covalently linked fusion protein of xylanase-xylan BD complex. In Target 36, we carried out the same procedure with the bound structure of xylan BD. Unfortunately, in both cases the protocol failed to achieve even an acceptable quality prediction.
Target 37: Arf6GTP-LZ2 JIF4
In Target 37 we were given the unbound crystal structure of Arf6-GTP (2A5D)31 and the amino acid sequence of the leucine zipper 2 motif of JIP. We received an ensemble of 16 homology models of LZ2 JIP from Alexandre Bonvin32 created from the two symmetrical homodimer (coiled coil) templates (2ZTA,33 sequence identity 19%; 1GK6,34 sequence identity 20%). We carried out global ensemble docking using these structures, clustered the low energy decoys, and submitted the largest cluster centers as our predictions, but were unable to achieve an acceptable or better quality prediction. Retrospectively, local docking using the unbound structures superimposed on the released crystal structure of the complex (2W83)35 showed a relatively pronounced energy funnel, however this was not found in global docking, perhaps due to the unusually extended shape of the LZ2 motif of JIP.
Use of Biochemical Information
Target 40: Trypsin-Protease Inhibitor
In Target 40, we were given the unbound structure of bovine trypsin (1BTY)36 and the bound coordinates for the double-headed arrowhead protease inhibitor API-A (3E8L).37 We were informed that each molecule of API-A binds two molecules of trypsin simultaneously with reactive sites at leucine 87 and lysine 145. We carried out a global docking run using RosettaDock and subjected the resulting lowest-scoring structure to a local docking perturbation leading to the prediction of one of the binding modes (reactive site lysine 145) of the inhibitor.
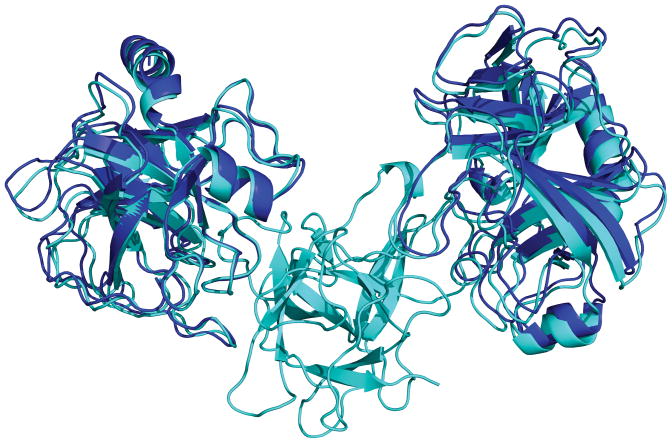
To arrive at the second possible complex, we screened the interface of all the decoys generated during the global docking run and filtered for decoys with leucine 87 at the interface. The lowest-energy structure from the filtered list was then subjected to local docking perturbations and the resulting structures were submitted along with the structures generated for the first binding mode. This exercise resulted in nine high-quality predictions (five for the first binding site and four for the second binding site). Figure 2 shows the remarkable native-like successful high-quality predicted ternary complex structure by superimposing the inhibitor molecule of the models with the lowest Lrmsd for the first binding mode (Target 40.CA) with 2.3 Å Lrmsd, 0.6 Å Irmsd and 0.83 fnat, and the second binding mode (Target 40.CB) with 1.8 Å Lrmsd, 0.6 Å Irmsd and 0.76 fnat.
Figure 2.
Target 40: High quality prediction of two molecules of bovine trypsin in complex with the double-headed arrowhead protease inhibitor API-A. The inhibitor of the predicted complex with the lowest Lrmsd (deep blue) of targets 40.CA and 40.CB is superimposed on the respective inhibitor as found in the crystal structure of the complex (cyan).
Standard RosettaDock
T30: Rnd1-Plexin B1
We were given the unbound structure of Rnd1 (2CLS:A)38 and one unbound NMR model of the Ras binding domain of plexin B1 (2R2O).39 Since we were only given a single model of the NMR structure of plexin B1, we were unable to utilize our ensemble docking method. Instead we used standard RosettaDock and carried out global docking on the target and submitted eight of the largest low-energy clusters as our predictions. Additionally, based on an alternate hypothesis that plexinB1 forms an extended β-sheet structure with Rnd1 as in the related Cdc42/f-ACK complex,40 we used a modified peptide docking algorithm and submitted the two lowest energy structures as our final predictions. Our closest prediction came from global docking and was just short of “acceptable” quality with 4.3 Å Irmsd and 0.27 fnat. The released complex structure of Target 30 (2REX:AB)41 revealed a substantial loop conformation change along residues 1808–1814 of plexinB1 at the interface of the complex which likely precluded correct prediction of the target.
Targets33–34: Methyltransferase-RNA
In Target 33 we were given the unbound structure of the 23S RNA and the sequence for Rlma2 methyltransferase. We used Robetta to generate a homology model for Rlma2 based on the crystal structure of Rlma1 (1P91, sequence identity 31%).42 We visually aligned the 23S RNA structure so that the substrate nucleotide, guanine 748 of hairpin 35, was placed near the active site of Rlma2. We then modified the RosettaDock algorithm to allow docking of RNA molecules using parameters from RosettaLigand43 for the nucleotides and phosphate backbone. RosettaDock sampled extensively using high-resolution local docking around this structure, but it failed to achieve an acceptable prediction. In Target 34, we carried out the same procedure with the bound structure of 23S RNA and achieved three predictions of acceptable quality. The closest prediction had 4.6 Å Lrmsd, 2.3 Å Irmsd, and 0.21 fnat.
Target 38–39: Centaurin A1-KIF13B
In Target 38 we were given the unbound structure of centaurin a1 (3FEH)44 and the amino acid sequence of the FHA domain of KIF13β. We used SWISS-MODEL45 to generate a homology model of KIF13β using the crystal structure of a related FHA domain protein (2G1L, sequence identity 38%).46 Previous experimental data47 that truncated various sections of centaurin a1 and observed its binding with KIF13B suggested the minimal construct of centaurin a1 needed to interact with KIF13B was residues 1–133, containing the GAP domain and N-PH domain, but not the C-PH domain. Consequently, we included only the first two domains of centaurin a1 in global docking. For Target 39, we were given the bound structure of KIF13B and carried out the same procedure. In both cases RosettaDock failed to achieve an acceptable quality prediction. The released crystal structure of the complex (3FM8)48 shows that, contrary to the experimental data, the interaction occurs almost entirely through the C-PH domain that we truncated.
Target 42: Designed Oligomer
Target 42 involved prediction of the structure of an oligomeric TPR protein given the sequence of the protein and structure of a homologous monomer (1NA3).49 For this challenge, we threaded the given sequence over a homologous TPR protein x-ray crystal structure (2FO7).50 The resulting structural model was then used in a global symmetry docking run wherein two monomers were simultaneously docked maintaining an overall C2 symmetry for the complex.51 The resulting structures were then sorted based on their total energies and the complexes with minimum free energy (calculated by the Rosetta energy function) were submitted. Although our predicted conformation of the TPR motif monomer was very close to the bound structure (2WQH52), RosettaDock scored the non-native docked models with larger interfaces better (lower) than the native co-crystal structure with its small binding interface.
DISCUSSION
Since CAPRI rounds 6–125 we have developed a number of new flexible docking methods, including EnsembleDock, which allows simultaneous docking of multiple backbone conformations, and SnugDock, which optimizes interface loops and backbone conformations during docking. In CAPRI rounds 13–19 we have implemented these methods over a wide range of docking targets, in both local and global docking, for both bound and unbound crystal and NMR structures as well as homology models. Here we outline a general approach to flexible backbone docking in RosettaDock that best utilizes these new docking tools, based on our experiences in the present CAPRI rounds.
There are primarily four types of starting structures used in docking for CAPRI: bound structures, unbound crystal structures, unbound NMR structures, and homology models. Ultimately, the flexible docking strategy selected is dependent on the expected deviation of the bound conformations of both partners from their given starting structures. Unsurprisingly, for a number of targets with bound starting structures, rigid-body docking techniques are often sufficient for accurate structure prediction. Examples include Target 34 where we had a number of acceptable-quality predictions, and Target 40, where we had a number of high-quality predictions, both using rigid-body docking. However, it is important to consider the expected deviation from the bound conformation for both partners. In Target 40, we were confident that unbound structure of β-trypsin underwent minimal binding-induced conformation changes based on existing structures of the enzyme in the PDB. By contrast, in Target 29, we used flexible docking because even though we had the bound structure of Trm82, the unbound structure of Trm8 contained a disordered loop that was thought to be important for interaction.
Unbound crystal structures are generally the most accurate starting structures for truly blind predictive docking, but they have two challenges which sometimes require the use of flexible docking: binding induced conformation changes and disordered regions. For modest binding-induced conformation changes, both EnsembleDock and SnugDock can be used. In EnsembleDock, an ensemble of structures can be generated starting from the unbound crystal structure9 and used for docking. If a particular interface loop is thought to change conformation slightly upon binding, SnugDock can be used. In Target 32, the catalytic loop of the enzyme subtilisin-savinase which was known to interact with the inhibitor BASI was made flexible through SnugDock, leading to the most accurate predictions among all CAPRI predictors. In many unbound crystal structures, there are disordered regions among surface loops that may play a role in binding. In such cases it is advisable to rebuild a complete loop using loop modeling in Rosetta25 prior to docking. In Target 29, we used loop-building to rebuild a disordered loop in Trm8 thought to interact with partner Trm82. In this case, we chose to use EnsembleDock for docking, but SnugDock could have been used as well. Although we were unsuccessful in predicting a structure near the native, the structure of the disorder-to-order transition of the loop in Trm8 was recovered with a remarkable degree of accuracy, demonstrating the utility of this approach in accommodating disordered regions in protein docking.
NMR structures constitute approximately 15% of the PDB, and are the given starting structures for a number of CAPRI targets. Since NMR structures typically consist of a set of structural models that best fit the NMR data, EnsembleDock is the natural choice of docking method. We have previously demonstrated that using EnsembleDock with an NMR ensemble outperforms rigid-body docking using a single NMR conformer.9 In Target 41, we used the entire 60-conformer ensemble from the NMR structure of IM2 to dock with colicin e9 using EnsembleDock and achieved a number of medium-quality predictions. In CAPRI targets where the NMR starting structure has not yet been released into the PDB, we were given only a single conformer, precluding the use of using EnsembleDock with an NMR ensemble (e.g. Target 30); we hope that in the future, we will be able to access the entire NMR ensemble or the primary NMR data from which an ensemble can be generated, to more accurately reflect blind docking from available structure information.
Homology models have seen steadily increasing use in CAPRI targets; in the current rounds more than half of the targets had a least one partner represented by a homology model. Homology models often differ significantly from the bound conformation, making them the single greatest challenge in flexible docking. Our general approach is to use EnsembleDock with a set of multiple homology models, instead of rigid-body docking with a single homology model, with the idea that an ensemble of models may overcome the structural inaccuracies inherent in using a single model. Although in the present rounds our approach did not achieve much success in homology model docking, we have recently conducted a study systematically docking antibody homology models with antigen crystal structures using a combination of EnsembleDock and SnugDock with promising results.10
Despite the recent advances, significant challenges in blind predictive docking remain. There are a number of possible reasons that could account for the inability to make successful predictions. Often the homology models used are not accurate enough to provide an accurate assessment of the interface between the two proteins, especially if there are inaccuracies in the residues at the interface. When EnsembleDock is used, the ability of the algorithm to predict the correct structure depends on both the accuracy of the structures in the ensemble and the ability of the algorithm to assign lower scores to the complex structures with the most bound-like monomer(s) in the ensemble.
The single most important determinant to successful docking is whether there is prior experimental or homology information localizing the docking search. Among the five targets in rounds 13–19 where information was available localizing the binding site of at least one partner, acceptable-quality or better predictions were made in four cases. Among the remaining nine targets where no such information was available, we failed to make a single successful prediction. The failures suggest that work is needed to address sampling issues in global searches. Eventual high-throughput interactome-scale protein-protein docking will rely heavily on both homology model docking and global docking, making these twin challenges central in future CAPRI development efforts.
Acknowledgments
We thankfully acknowledge the efforts of the CAPRI organizers in finding and evaluating targets and facilitating communication between protein docking groups. We are grateful to the crystallographers for offering their complexes as CAPRI targets. This work was supported by National Institute of Health (NIH) grant R01-GM078221 and U.C.B S.A. We also thank Alexandre Bonvin for providing the homology models for Target 37, Ingemar Andre for helping us with symmetric docking for Target 42 and Brian Weitzner for feedback on the manuscript. The Rosetta protein structure modeling suite is developed collaboratively, and all code is available from the RosettaCommons (www.rosettacommons.org).
References
- 1.Vajda S, Vakser IA, Sternberg MJ, Janin J. Modeling of protein interactions in genomes. Proteins. 2002;47(4):444–446. doi: 10.1002/prot.10112. [DOI] [PubMed] [Google Scholar]
- 2.Gray JJ, Moughon SE, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid body displacement and side-chain conformations. J Mol Biol. 2003;331(1):281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 3.Gray JJ, Moughon SE, Kortemme T, Schueler-Furman O, Misura KM, Morozov AV, Baker D. Protein-protein docking predictions for the CAPRI experiment. Proteins. 2003;52(1):118–122. doi: 10.1002/prot.10384. [DOI] [PubMed] [Google Scholar]
- 4.Daily MD, Masica D, Sivasubramanian A, Somarouthu S, Gray JJ. CAPRI rounds 3–5 reveal promising successes and future challenges for RosettaDock. Proteins. 2005;60(2):181–186. doi: 10.1002/prot.20555. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhury S, Sircar A, Sivasubramanian A, Berrondo M, Gray JJ. Incorporating biochemical information and backbone flexibility in RosettaDock for CAPRI rounds 6–12. Proteins. 2007;69(4):793–800. doi: 10.1002/prot.21731. [DOI] [PubMed] [Google Scholar]
- 6.Bonvin AM. Flexible protein-protein docking. Curr Opin Struct Biol. 2006;16(2):194–200. doi: 10.1016/j.sbi.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Zacharias M. Accounting for conformational changes during protein-protein docking. Curr Opin Struct Biol. doi: 10.1016/j.sbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Janin J. The targets of CAPRI rounds 6–12. Proteins. 2007;69(4):699–703. doi: 10.1002/prot.21689. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhury S, Gray JJ. Conformer selection and induced fit in flexible backbone protein-protein docking using computational and NMR ensembles. J Mol Biol. 2008;381(4):1068–1087. doi: 10.1016/j.jmb.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sircar A, Gray JJ. SnugDock: paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Comput Biol. 2010;6(1):e1000644. doi: 10.1371/journal.pcbi.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60(2):150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- 12.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vallee F, Kadziola A, Bourne Y, Juy M, Rodenburg KW, Svensson B, Haser R. Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution. Structure. 1998;6(5):649–659. doi: 10.1016/s0969-2126(98)00066-5. [DOI] [PubMed] [Google Scholar]
- 14.Betzel C, Klupsch S, Papendorf G, Hastrup S, Branner S, Wilson KS. Crystal structure of the alkaline proteinase Savinase from Bacillus lentus at 1.4 A resolution. J Mol Biol. 1992;223(2):427–445. doi: 10.1016/0022-2836(92)90662-4. [DOI] [PubMed] [Google Scholar]
- 15.Pal GP, Kavounis CA, Jany KD, Tsernoglou D. The three-dimensional structure of the complex of proteinase K with its naturally occurring protein inhibitor, PKI3. FEBS Lett. 1994;341(2–3):167–170. doi: 10.1016/0014-5793(94)80450-8. [DOI] [PubMed] [Google Scholar]
- 16.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 17.Canutescu AA, Dunbrack RL., Jr Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 2003;12(5):963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Micheelsen PO, Vevodova J, De Maria L, Ostergaard PR, Friis EP, Wilson K, Skjot M. Structural and mutational analyses of the interaction between the barley alpha-amylase/subtilisin inhibitor and the subtilisin savinase reveal a novel mode of inhibition. J Mol Biol. 2008;380(4):681–690. doi: 10.1016/j.jmb.2008.05.034. [DOI] [PubMed] [Google Scholar]
- 19.Kuhlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C, Hemmings AM. Structure of the E9 DNase domain in comparison with the inhibited structure of the E9 DNase/Im9 complex. http://dx.doi.org/10.2210/pdb1fsj/pdb.
- 20.Macdonald CJ. NMR Studies of Order and Disorder In Protein-Protein Interactions. University of East Anglia; 2009. [Google Scholar]
- 21.Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999;7(1):91–102. doi: 10.1016/s0969-2126(99)80012-4. [DOI] [PubMed] [Google Scholar]
- 22.Meenan NAG, Sharma A, Fleishman SJ, MacDonald C, Morel B, Boetzel R, Moore GR, Baker D, Kleanthous C. The structural and energetic basis for high selectivity in a high affinity protein-protein interaction. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0910756107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure. 2008;16(1):52–61. doi: 10.1016/j.str.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 24.Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8(10):1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373(2):503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 26.Misura KM, Baker D. Progress and challenges in high-resolution refinement of protein structure models. Proteins. 2005;59(1):15–29. doi: 10.1002/prot.20376. [DOI] [PubMed] [Google Scholar]
- 27.Charnock SJ, Bolam DN, Turkenburg JP, Gilbert HJ, Ferreira LM, Davies GJ, Fontes CM. The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry. 2000;39(17):5013–5021. doi: 10.1021/bi992821q. [DOI] [PubMed] [Google Scholar]
- 28.Solomon V, Teplitsky A, Shulami S, Zolotnitsky G, Shoham Y, Shoham G. Structure-specificity relationships of an intracellular xylanase from Geobacillus stearothermophilus. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 8):845–859. doi: 10.1107/S0907444907024845. [DOI] [PubMed] [Google Scholar]
- 29.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32(Web Server issue):W526–531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berrondo M, Ostermeier M, Gray JJ. Structure prediction of domain insertion proteins from structures of individual domains. Structure. 2008;16(4):513–527. doi: 10.1016/j.str.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309(5737):1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 32.De Vries SJ, Melquiond ASJ, Kastritis PL, Karaca E, Bordogna A, Rodrigues J, Bonvin AMJJ. Strengths and weaknesses of data-driven docking in CAPRI. Proteins. doi: 10.1002/prot.22814. Submitted. [DOI] [PubMed] [Google Scholar]
- 33.O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 34.Strelkov SV, Herrmann H, Geisler N, Wedig T, Zimbelmann R, Aebi U, Burkhard P. Conserved segments 1A and 2B of the intermediate filament dimer: their atomic structures and role in filament assembly. EMBO J. 2002;21(6):1255–1266. doi: 10.1093/emboj/21.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isabet T, Montagnac G, Regazzoni K, Raynal B, El Khadali F, England P, Franco M, Chavrier P, Houdusse A, Menetrey J. The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4. EMBO J. 2009;28(18):2835–2845. doi: 10.1038/emboj.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz BA, Finer-Moore J, Mortezaei R, Rich DH, Stroud RM. Episelection: novel Ki approximately nanomolar inhibitors of serine proteases selected by binding or chemistry on an enzyme surface. Biochemistry. 1995;34(26):8264–8280. doi: 10.1021/bi00026a008. [DOI] [PubMed] [Google Scholar]
- 37.Bao R, Zhou CZ, Jiang C, Lin SX, Chi CW, Chen Y. The ternary structure of the double-headed arrowhead protease inhibitor API-A complexed with two trypsins reveals a novel reactive site conformation. J Biol Chem. 2009;284(39):26676–26684. doi: 10.1074/jbc.M109.022095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pike ACW, Yang X, Colebrook S, Gileadi O, Sobott F, Bray J, Wen Hwa L, Marsden B, Zhao Y, Schoch G, Elkins J, Debreczeni JE, Turnbull AP, Von Delft F, Arrowsmith C, Edwards A, Weigelt J, Sundstrom M, Doyle D. The Crystal Structure of the Human Rnd1 Gtpase in the Active GTP Bound State. http://dx.doi.org/10.2210/pdb2cls/pdb.
- 39.Tong Y, Chugha P, Hota PK, Alviani RS, Li M, Tempel W, Shen L, Park HW, Buck M. Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain. J Biol Chem. 2007;282(51):37215–37224. doi: 10.1074/jbc.M703800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mott HR, Owen D, Nietlispach D, Lowe PN, Manser E, Lim L, Laue ED. Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK. Nature. 1999;399(6734):384–388. doi: 10.1038/20732. [DOI] [PubMed] [Google Scholar]
- 41.Tong Y, Tempel W, Shen L, Arrowsmith CH, Edwards AM, Sundstrom M, Weigelt J, Bochkarev A, Park H. Crystal structure of the effector domain of PLXNB1 bound with Rnd1 GTPase. http://dx.doi.org/10.2210/pdb2rex/pdb.
- 42.Das K, Acton T, Chiang Y, Shih L, Arnold E, Montelione GT. Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc Natl Acad Sci U S A. 2004;101(12):4041–4046. doi: 10.1073/pnas.0400189101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65(3):538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 44.Shen L, Tong Y, Tempel W, MacKenzie F, Arrowsmith CH, Edwards AM, Bountra C, Weigelt J, Bochkarev A, Park H. Crystal structure of full length centaurin alpha-1. http://dx.doi.org/10.2210/pdb3feh/pdb.
- 45.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Tempel W, Shen Y, Shen L, Arrowsmith C, Edwards A, Sundstrom M, Weigelt J, Bochkarev A, Park H. Crystal structure of the FHA domain of human kinesin family member C. http://dx.doi.org/10.2210/pdb2gil/pdb.
- 47.Venkateswarlu K, Brandom KG, Yun H. PI-3-kinase-dependent membrane recruitment of centaurin-alpha2 is essential for its effect on ARF6-mediated actin cytoskeleton reorganisation. J Cell Sci. 2007;120(Pt 5):792–801. doi: 10.1242/jcs.03373. [DOI] [PubMed] [Google Scholar]
- 48.Shen L, Tong Y, Tempel W, MacKenzie F, Arrowsmith CH, Edwards AM, Bountra C, Weigelt J, Bochkarev A, Park H. Crystal structure of full length centaurin alpha-1 bound with the FHA domain of KIF13B. http://dx.doi.org/10.2210/pdb3fm8/pdb.
- 49.Main ER, Xiong Y, Cocco MJ, D’Andrea L, Regan L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure. 2003;11(5):497–508. doi: 10.1016/s0969-2126(03)00076-5. [DOI] [PubMed] [Google Scholar]
- 50.Kajander T, Cortajarena AL, Mochrie S, Regan L. Structure and stability of designed TPR protein superhelices: unusual crystal packing and implications for natural TPR proteins. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 7):800–811. doi: 10.1107/S0907444907024353. [DOI] [PubMed] [Google Scholar]
- 51.Andre I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc Natl Acad Sci U S A. 2007;104(45):17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krachler AM, Sharma A, Kleanthous C. Self-association of TPR-domains: natural and engineered. Proteins. 2010 doi: 10.1002/prot.22726. accepted. [DOI] [PubMed] [Google Scholar]