Summary
It is currently thought that antennal target genes are activated in Drosophila by the combined action of Distal-less, homothorax, and extradenticle, and that the Hox gene Antennapedia prevents activation of antennal genes in the leg by repressing homothorax. To test these ideas, we analyze a 62 bp enhancer from the antennal gene spineless that is specific for the third antennal segment. This enhancer is activated by a tripartite complex of Distal-less, Homothorax, and Extradenticle. Surprisingly, Antennapedia represses the enhancer directly, at least in part by competing with Distal-less for binding. We show that Antennapedia is required in the leg only within a proximal ring that coexpresses Distal-less, Homothorax and Extradenticle. We conclude that the function of Antennapedia in the leg is not to repress homothorax, as has been suggested, but to directly repress spineless and other antennal genes that would otherwise be activated within this ring.
Introduction
Mutations of several genes in Drosophila cause transformations of antenna toward second leg. The best known of these mutations are dominant gain-of-function alleles of the Hox gene Antennapedia (Antp), which can cause the antenna to develop as a complete leg. Struhl (1981; 1982a) showed that loss-of-function alleles of Antp have the opposite effect, causing transformation of leg structures to antenna, but have no effect on development of the antenna itself. He proposed that Antp is normally expressed in the legs but not the antenna, and that its function is to repress the activation of antenna-specific genes in the leg. The gain-of-function alleles were suggested to cause ectopic expression of Antp in the antenna. Molecular studies confirmed that Antp is expressed as inferred by Struhl (Frischer et al. 1986). However, until recently, the identities of the “antennal genes” controlled by Antp remained uncertain, as it was not known how antennal identity is specified.
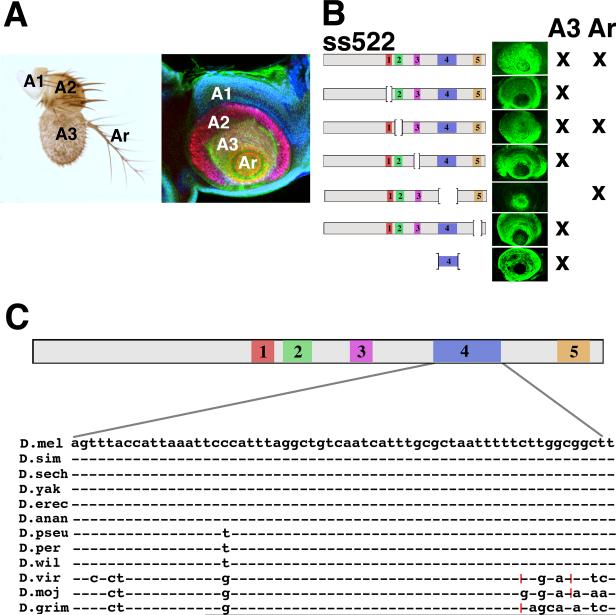
We now know that the identity of most of the antenna is specified by the combined action of homeodomain transcription factors encoded by the homothorax (hth) and Distal-less (Dll) genes (Casares and Mann 1998; Dong et al. 2000). These genes are coexpressed extensively in the antenna, whereas in the leg they are coexpressed in only a narrow proximal ring of cells. Several antennal genes have been shown to be activated independently by combined Hth and Dll expression (Dong et al. 2002). One of the most important of these targets is spineless (ss), which encodes a bHLH transcription factor homologous to the mammalian dioxin receptor (Duncan et al. 1998). The expression patterns of Dll, hth and ss in the antennal imaginal disc, and an adult antenna are shown in Fig. 1A.
Figure 1.
(A) Left: A wild-type adult antenna. The first (A1), second (A2), and third (A3) antennal segments and the arista (Ar) are indicated. Right: A mature antennal disc stained for Hth (blue), Dll (red), and the ss reporter B6.9/lacZ (Emmons et al. 2007) (green). Hth is expressed in the primordia of A1, A2, and A3; Dll is expressed in A2, A3, and the arista; and ss is expressed in A3 and the arista. (B) Five conserved domains within ss522 and their deletion derivatives are indicated. The antennal expression each drives in vivo is shown to the right. (C) Conservation of the sequence of domain 4 in 12 Drosophila species; dashes indicate identity, red hatch marks indicate 3 bp insertions relative to the D. melanogaster sequence.
Hth is required for normal identity of the entire antenna, and is expressed throughout the antennal disc in the first and second larval instars. hth- mitotic recombination clones induced at these times transform the entire antenna to a leg-like appendage (Casares and Mann 1998). Subsequently, Hth expression is lost in the most distal portion of the disc, the primordium of the arista, whose development becomes independent of hth (Emmons et al. 2007). Hth is also expressed in the most proximal segments of the leg, where it is required for normal growth and proper formation of segment boundaries (Abu-Shaar and Mann 1998; Wu and Cohen 1999; Casares and Mann 2001). Hth functions as a heterodimer with the homeodomain protein Extradenticle (Exd) (Rieckhof et al. 1997; Pai et al. 1998; Kurant et al. 1998), which is also required for antennal specification and proximal leg development (González-Crespo and Morata 1995). In addition to these roles, Hth and Exd serve as important cofactors that increase the binding specificity of the Hox proteins (for review see Mann et al. 2009).
Dll is required for the development of distal structures in all of the ventral appendages (Cohen et al. 1989). In the antenna, Dll is expressed in the primordia of the second (A2), and third (A3) antennal segments and the arista, and this entire expression domain is deleted in Dll- mutants (Cohen and Jürgens 1989). However, weak alleles of Dll cause transformations of antenna toward leg (Sunkel and Whittle 1987; Dong et al. 2000), suggesting that Dll has a role in specifying antennal identity that is distinct from its general role of specifying distal limb structures. Dong et al. (2000) proposed that Dll acts in concert with Hth (and presumably also Exd) to define antennal identity. This proposal is supported by the effects of hth- and Dll- alleles on the expression of antenna-specific target genes and by the effects of combined ectopic expression of Hth and Dll (Duncan et al. 1998; Dong et al. 2000; 2002; Emmons et al. 2007).
Many of the identity functions of Hth and Dll in the distal antenna are executed by the target gene ss (Dong et al. 2002; Emmons et al. 2007), which is expressed in the primordia of A3 and the arista. In ss- mutants, A3 lacks all olfactory sensilla, and the arista is transformed to distal leg (Struhl 1982b; Duncan et al. 1998). In previous work (Emmons et al. 2007), we identified the antennal enhancer from ss and showed that its expression depends upon Dll and Hth, and that it is repressed by ectopically expressed Antp. The enhancer is also repressed in A2 by the homeodomain protein Cut (Blochlinger et al. 1988).
In this report, we address two major unresolved questions. First, how are inputs from Dll, Hth, and Exd integrated at antennal target genes? To date, no antennal enhancers have been characterized at the molecular level, so the mechanism of action of these factors has remained uncertain. Second, how does Antp repress antennal identity in the leg? Based on the finding that Antp- clones in the leg sometimes show ectopic distal expression of Hth, Casares and Mann (1998) proposed that the primary function of Antp is to repress hth in the distal leg, which then prevents activation of antennal target genes. Although this view is widely accepted, it has not been subject to direct test.
To address these questions, we focused our attention on the antennal enhancer of ss. We identify a 62 bp subregion of this enhancer that drives expression specifically in A3. Like the full antennal enhancer, the A3 enhancer requires Dll, Hth, and Exd for expression. All three of these factors interact directly with the enhancer. The binding of Dll shows strong cooperativity with Hth and Exd, indicating that these proteins bind as a complex. This Dll/Hth/Exd tripartite binding suggests that Dll behaves much like a Hox protein in specifying antennal identity. Surprisingly, we find that Antp also interacts directly with the A3 enhancer. Antp binds cooperatively with Hth and Exd, and represses the enhancer at least in part by competing with Dll for binding.
Our finding that Antp interacts directly with the A3 enhancer led us to reexamine the role of Antp in leg development. We find that the A3 enhancer is sometimes activated within Antp- clones in the leg, consistent with the transformation to antenna that such clones can cause. However, this activation occurs only within a narrow ring of cells in the proximal leg that coexpresses Dll, Hth, and Exd (Wu and Cohen 1999). Subsequently, some of the Antp- cells in which the A3 enhancer has been activated begin to express Ss, Cut, and other antennal markers, indicating a transformation to antenna. Importantly, we find that expression of Hth and Dll in the proximal ring is unaffected in Antp- clones, indicating that Antp does not block antennal development in the leg by repressing hth, as has been thought. Rather, we conclude that the main, and perhaps sole, function of Antp in the leg imaginal disc is the direct repression of antennal genes that would otherwise be activated by the combined expression of Dll, Hth, and Exd in the proximal ring.
Results
Dissection of the ss522 antennal enhancer
In a previous report (Emmons et al. 2007), we showed that the antennal expression pattern of ss is reproduced by lacZ reporters containing a 522 bp fragment from the ss 5' region. This fragment contains five conserved (41-90% identity) domains (Stark et al. 2007), each of which was deleted and tested for effect on expression in vivo. Expression in the arista and the third antennal segment (A3) prove to be under separate control; expression in the arista requires domains 1, 3 and 5, whereas expression in A3 is lost only when domain 4 is deleted (Fig. 1B). Moreover, reporters containing domain 4 alone show expression in A3 and nowhere else in imaginal discs. Thus, domain 4 is both necessary and sufficient for A3-specific expression. Domain 4 (D4) is 62 bp in length and is highly conserved, being invariant at 50/62 base pairs in the 12 Drosophila species sequenced (Fig. 1C).
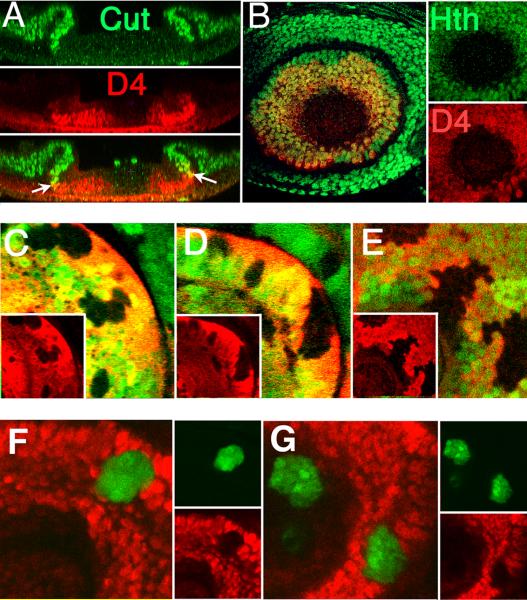
We first established the boundaries of D4/lacZ reporter expression relative to Homothorax (expressed in A3 and more proximally) and Cut (expressed in A2 and more proximally) in mature third instar antennal discs. As shown in Fig. 2A-B, the distal boundary of D4/lacZ expression coincides with the distal limit of Hth expression, and the proximal boundary largely coincides with the distal boundary of Cut expression. D4/lacZ is therefore expressed throughout A3. D4/lacZ expression often overlaps Cut expression slightly, indicating that the reporter may also be expressed in a few cells in distal A2.
Figure 2.
(A) Cross section of an antennal disc showing expression of Cut and D4/lacZ. The distal boundary of Cut expression and the proximal boundary of D4/lacZ expression closely match, although a few cells at the interface often express both (arrows). (B) Antennal disc stained for expression of Hth and D4/lacZ. The distal boundaries of expression coincide precisely. (C - E) Expression of D4/lacZ (red) is lost within clones mutant for Dll (C), hth (D), or exd (E). All clones are marked by the loss of GFP (green). (F,G) Clones expressing either Antp alone (F) or both Antp and Hth (G) (green) fully repress D4/lacZ (red). Expression of Hth was confirmed by antibody staining (not shown).
Trans regulation of D4
D4/lacZ expression is lost in clones homozygous for null alleles of Dll, hth, and exd (Fig. 2C-E), indicating that Dll, Hth, and Exd are all required for expression. We also examined clones expressing either or both Hth and Dll proteins ectopically (data not shown). Clones expressing Hth show activation of D4/lacZ in the aristal region of the antenna and the distal part of the leg, regions where Dll is expressed. Similarly, clones expressing Dll activate the reporter in the proximal antenna and wing, regions where Hth is expressed. Clones expressing both Hth and Dll activate D4/lacZ expression in most locations. A notable exception is the proximal region of the leg discs (see below).
We also find that D4/lacZ is repressed within antennal clones ectopically expressing Antp (Fig. 2F). Since ectopic Antp is known to repress hth in the antenna (Casares and Mann 1998), we tested whether repression of hth accounts for the loss of D4/lacZ expression within Antp-expressing clones by examining antennal clones that express both Antp and Hth ectopically. Surprisingly, D4/lacZ is fully repressed within such clones (Fig. 2G), just as in clones that express Antp alone, indicating that repression of D4/lacZ by ectopic Antp is not due to the loss of Hth.
Dll, Hth, Exd, and Antp all interact directly with D4
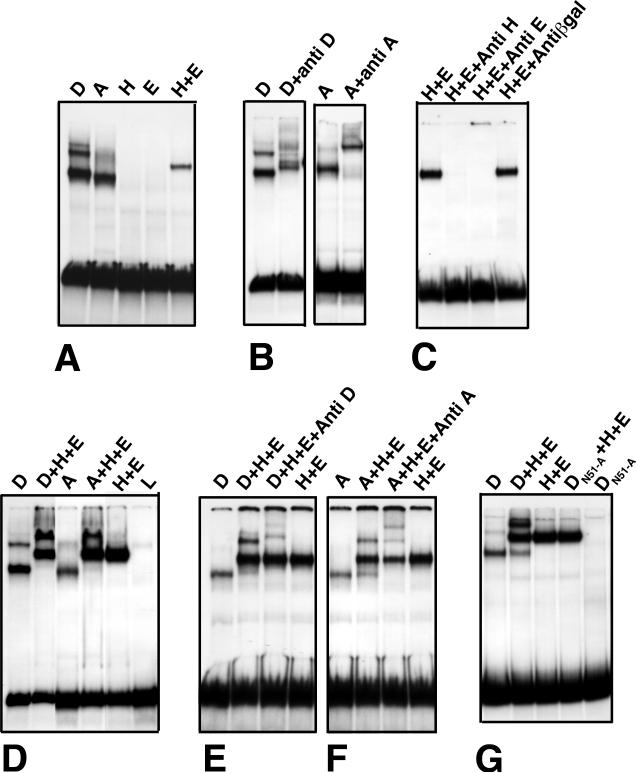
Gel-shift and footprinting studies demonstrate that all four regulators defined above bind D4 directly. In vitro translated Dll and Antp both produce prominent gel retardation bands in gel-shift assays (Fig. 3A). These retardation bands are supershifted by anti-Dll or anti-Antp antibodies, indicating that both Dll and Antp are present in their respective retardation complexes (Fig. 3B). Although Hth and Exd do not produce retardation bands on their own in our assays, when mixed they bind cooperatively to produce a prominent retardation complex (Fig. 3A). Anti-Hth and anti-Exd do not supershift this retardation band, but instead dramatically reduce its intensity, suggesting that these antibodies interfere with the heterodimerization or DNA binding of these proteins (Fig. 3C).
Figure 3.
(A) Binding of Dll (D), Antp (A), Hth (H), and Exd (E) to D4. Dll produces three retardation bands, whereas Antp produces a single major retardation band. Hth and Exd produce no shift on their own, but generate a prominent retardation band when mixed. (B) Anti-Dll and anti-Antp supershift the respective retardation complexes. (C) Anti-Hth and anti-Exd antibodies block production of the Hth+Exd retardation band. (D) When combined with Hth and Exd, both Dll and Antp produce slowly migrating bands, but show very little of the singly bound species produced by Dll or Antp on their own. L = lysate control. (E-F) Antibodies to Dll (E) and Antp (F) supershift the slow moving bands formed when these proteins are mixed with Hth and Exd. (G) Dll protein in which asn51 of the homeodomain has been changed to ala does not bind D4 on its own or when mixed with Hth and Exd. In vitro translation of the mutant protein was confirmed by 35S-methionine labeling (not shown).
When Dll is mixed with Hth and Exd, strong cooperative binding to D4 is seen (Fig. 3D); the band corresponding to binding of a single molecule of Dll is replaced by an intense band located higher in the gel than the Hth + Exd band. This upper band is supershifted by anti Dll (Fig. 3E), indicating that it contains Dll. These observations indicate that when all three proteins are present, almost all Dll is bound to probe that is also bound by Hth and Exd. This striking cooperativity implies protein-protein interactions between Dll and Hth and/or Exd. Dll carrying a change of asn51 to ala in the homeodomain, which eliminates DNA binding in other homeodomain proteins (Ades and Sauer 1995), fails to bind D4 on its own or in combination with Hth and Exd (Fig. 3G). Thus, DNA binding of Dll appears to be required for its interaction with Hth and Exd on D4. Antp also binds cooperatively with Hth and Exd (Fig. 3D,F)), although we have not tested whether the ability of Antp to bind DNA is essential for this interaction.
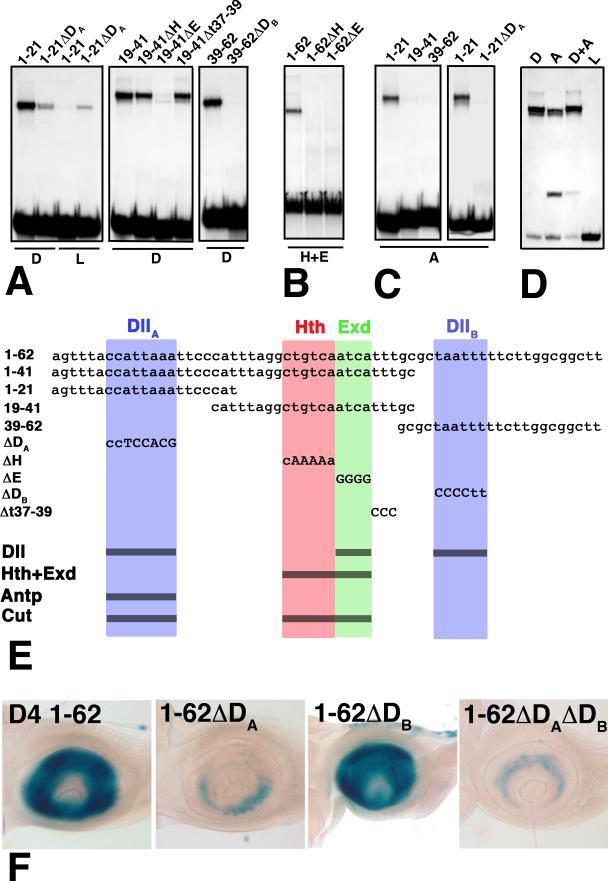
Binding sites for Hth/Exd, Dll, and Antp were defined by footprinting (data not shown) and testing mutant oligonucleotides in gel-shift assays. The sites defined are summarized in Fig. 4. Hth and Exd bind to directly adjacent consensus binding sites (Chang et al. 1997), and mutations in these sites block cooperative binding of these factors (Fig. 4B). Dll binds three sites in D4. To characterize these sites, D4 was subdivided into three oligonucleotides (bp 1-21, 19-41, and 39-62), each containing a single Dll binding site. The Dll binding sites present in oligonucleotides 1-21 and 39-62 are designated Dlla and Dllb, respectively (Fig. 4E). Mutations in these sites almost completely eliminate binding by Dll (Fig. 4A). The central 19-41 oligonucleotide, which contains the Hth/Exd site, also binds Dll. Mutation of the Exd site blocks this binding, indicating that Dll and Exd bind overlapping or identical sites (Fig. 4A). Dll produces three distinct retardation bands when bound to full-length D4; we interpret these bands as having one, two, or all three binding sites occupied by Dll. Antp binds only one site in D4, which overlaps or coincides with the Dlla site (Fig. 4C). Mutation of this site blocks all binding of Antp.
Figure 4.
Gel-shift assays of mutant and wild-type derivatives of D4. The sequences of full length D4, four fragments, and five clustered site mutations are shown in (E). Abbreviations as in Fig. 3. (A) Dll binds three sites in D4, one in each of the subfragments 1-21, 19-41, and 39-62. Binding to these fragments is almost completely eliminated by the ΔDA, ΔE, and ΔDB mutants, respectively. (B) Cooperative binding of Hth and Exd is eliminated in both the ΔH and ΔE mutants, indicating that Hth and Exd bind adjacent consensus sites. (C) Antp binds only the 1-21 fragment, and this binding is lost in the ΔDA mutant. Lysate (L) control lanes were blank (not shown) for all but the 1-21ΔDA probe. (D) Purified Dll (D) and Antp (A) compete for binding to the 1-21 probe. The faster migrating band in the Antp lanes is likely due to the binding of a breakdown product generated during purification. (E) Summary of the DNA sequences tested in (A-D). (F) Effects of the ΔDA and ΔDB mutations on antennal expression in vivo.
The finding that Antp binds Dlla raises the possibility that it represses D4 by competing with Dll for binding. To test this possibility, the ability of combined Dll and Antp to gel-shift the 1-21 oligonucleotide was examined. To achieve robust binding, both Dll and Antp were purified from in vitro translation reactions by oligonucleotide selection (Ozyhar et al. 1992). As shown in Fig. 4D, under conditions in which the majority of the 1-21 probe is shifted by either Dll or Antp alone, no additional slower mobility band is seen when these proteins are mixed. This result indicates that Antp and Dll do compete for binding to Dlla.
Finally, to assess the importance of the Hth/Exd, Dlla, and Dllb binding sites in vivo, D4/lacZ reporters carrying mutations in each site were reintroduced into flies. Mutation of the Hth or Exd half sites eliminated enhancer activity in all P-element transformants recovered (10 for the Hth site mutation, and 8 for the Exd site mutation). To assess the importance of the Dlla and Dllb sites, position effects were minimized by using ϕC31-mediated transformation (Bischof et al. 2007) to target integration of mutant derivatives to the same site. Mutations in Dlla cause a dramatic reduction in expression, whereas mutations in Dllb have little or no effect (Fig. 4F). The double mutant Dlla Dllb is expressed to about the same level as the Dlla mutant. These observations indicate that the Dlla site is of key importance for activation by Dll. Significantly, Dlla is the site at which Antp competes with Dll for binding.
Although not central to this report, we find that D4 is also regulated by cut. Antennal expression of D4/lacZ is expanded proximally in cut- clones, and repressed within clones ectopically expressing Cut (Suppl. Fig. 1). These observations indicate that the proximal limit of D4/lacZ expression is set, at least in part, by Cut. Gel shift assays indicate that Cut binds to D4 at two sites simultaneously. These sites overlap the Dlla and Exd sites, and both are required for binding (Suppl. Fig. 1). Binding to these sites is likely mediated by different DNA binding domains within the Cut protein (see Nepveu 2001).
Antp represses D4 in the proximal leg
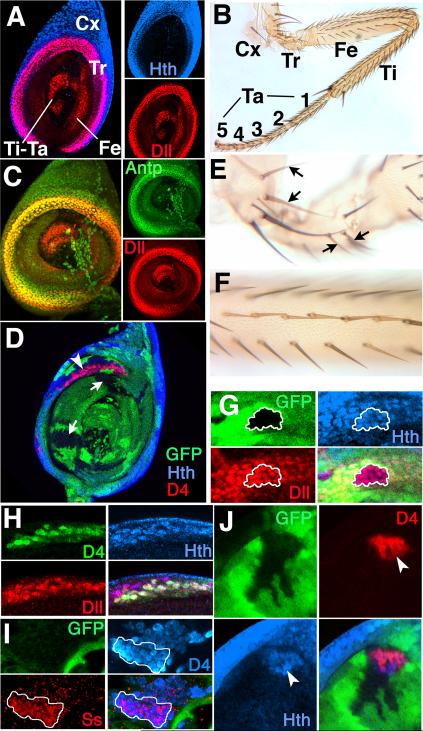
Our finding that Antp interacts directly with D4 was unexpected. What is the relevance of this finding to normal development? The answer turns out to be that the key, and perhaps sole, function of Antp during leg development is the repression of ss and other antennal target genes within a narrow proximal ring that coexpresses Dll, Hth, and Exd. This ring is shown in Fig. 5A. It is 5-7 cells wide, and is defined by Dll expression; Hth is expressed in the ring as well as more proximally. The function of the ring is not known with certainty. It appears in the early third instar, and overlaps the joint between the trochanter and the femur (Wu and Cohen 2000) (leg segments are shown in Fig. 5B). Although Antp is expressed throughout the leg primordium early in development (Casares and Mann 1998), during larval life its expression becomes limited to a broad proximal domain (Fig. 5C). Within this domain, Antp is most strongly expressed within the proximal ring.
Figure 5.
Antp represses D4/lacZ in the proximal Dll+Hth ring of the second leg imaginal disc. (A) A second leg disc stained for Hth and Dll. Hth is expressed in a broad proximal domain, whereas Dll is expressed in a 5-7 cell wide proximal ring whose distal border coincides with the distal limit of Hth. Dll is also expressed in the central (distal) region of the disc, which is only partly in the plane of focus. Cx=coxa, Tr=trochanter, Fe=femur, Ti=tibia, and Ta=tarsus. (B) An adult second leg. Abbreviations as in (A). (C) A second leg disc stained for Dll and Antp. Antp is expressed in a broad proximal region, and is upregulated within the Dll ring. (D) Antp- clones, marked by the loss of GFP, in a second leg disc. D4/lacZ is activated in Antp- clones where they overlap the Hth + Dll ring (arrowhead). Antp- clones that do not overlap the ring (arrows) show no activation of D4/lacZ, have interdigitated borders, and appear to develop normally. (E) Antp- clones in the coxa and trochanter marked by yellow bristles (arrows) produce normal cuticular structures. (F) An Antp- clone in the femur marked by yellow bristles produces normal structures. (G) An Antp- clone (outlined in white) in the proximal ring has no effect on expression of Hth or Dll. (H) All cells expressing D4/lacZ within an Antp- leg clone also express both Hth and Dll. Clones are not marked in this disc to allow direct comparison of Dll, Hth, and D4/lacZ. (I) A partially rounded up Antp- clone showing expression of Ss within part (white outline) of the D4/lacZ-expressing region. (J) An Antp- clone marked by the absence of GFP in a second leg disc showing activation of D4/lacZ and ectopic expression of Hth. Note that D4/lacZ and Hth are expressed in a rounded-up portion of the clone (arrowheads), which is presumably transformed to antenna, whereas the remainder of the clone is interdigitated. Although not visible in this focal plane, the region of the clone expressing D4/lacZ and Hth retains a connection to the proximal Hth + Dll ring.
In analyzing Antp- clones in second leg discs, we noted that there are two basic types: clones that are well integrated into the disc epithelium and whose borders are interdigitated with their wild-type neighbors, and clones that are rounded up and have smooth borders. Rounded-up clones appear to have reduced affinity for their neighboring cells, and their borders often coincide with novel folds in the disc. Interdigitated Antp- clones occur in all regions of the leg disc and appear to develop completely normally. In contrast, rounded-up clones almost always show some connection to the proximal ring, and express ss or other antennal markers, indicating they are transformed to antenna.
We first consider Antp- clones of the interdigitated type. Such clones can be induced at any time during larval development, and even very large interdigitated clones are well integrated into the disc (Fig. 5D). Clones of this type appear to develop completely normally, as most Antp- clones produce normal cuticular structures in adult second legs (Fig. 5E,F). However, when interdigitated Antp- clones overlap the proximal ring of Dll, Hth, and Exd expression, D4/lacZ becomes activated in Antp- cells of the ring (Fig. 5D). Importantly, expression of Dll and Hth is unaffected in such clones (Fig. 5G,H). A few cells at the proximal edge of the ring do not activate D4/lacZ expression. The reason is not known, but both teashirt and dachshund are differentially expressed within the ring (Wu and Cohen 2000), and may play a role in activating or repressing D4. Although D4/lacZ is activated in the proximal ring in Antp- clones of the interdigitated type, Ss itself is not expressed, indicating that such clones are not transformed to antenna (not shown).
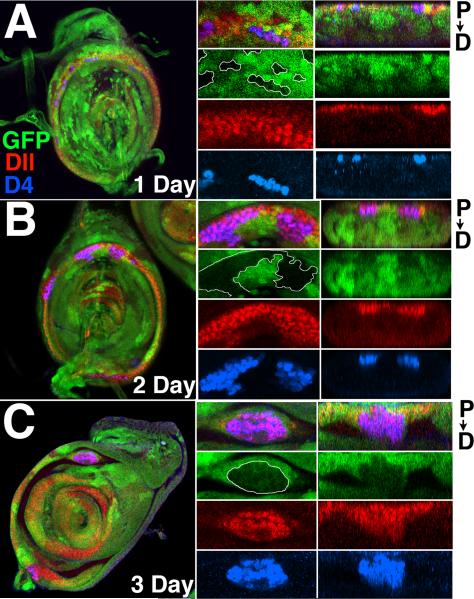
Rounded-up Antp- clones present a more complex picture. Such clones almost always express D4/lacZ, and usually extend distally from the ring of Dll, Hth and Exd coexpression. Rounded-up clones express Hth (Fig. 5J), Dll, and usually also Ss (Fig. 5I), indicating they are transformed to antenna. Often clones contain both rounded-up and interdigitated regions; in such cases the rounded-up portion is almost always associated with the ring (Fig. 5J). To determine the origin of rounded-up clones, we examined Antp- clones in late larval discs that were induced at progressively earlier times in development. D4/lacZ-expressing clones 0-24 hrs of age are almost exclusively of the interdigitated type, with D4/lacZ expression occurring only within the proximal ring (Fig. 6A). D4/lacZ-expressing clones 24-48 hrs old show some rounding up, causing distortion of the ring (Fig. 6B). By 48-72 hrs, rounding up of D4/lacZ-expressing clones is more pronounced (Fig. 6C). Moreover, most clones of this age extend distally from the proximal ring. This distal extension can become very pronounced, with some clones bridging the region between the ring and the distal expression domain of Dll (Fig. 7AB), which includes the tibial and tarsal portions of the disc. Occasionally, rounded up D4/lacZ expressing clones are found that are entirely distal and not connected to the proximal ring. The presence of intermediates in which distal extensions are connected to the proximal ring by a narrow isthmus (Fig. 7C) suggest that many or all of these strictly distal clones originate within the proximal ring. Of 106 D4/lacZ expressing clones scored from the 48-72 hr age group, 54 were of the interdigitated type, and 52 contained rounded-up regions. Of the rounded-up clones, only four lacked a connection to the proximal ring. At all times, clones not expressing D4/lacZ are of the interdigitated type and are well integrated into the disc.
Figure 6.
D4/lacZ activation in Antp- clones of increasing age in second leg discs. Antp- clones are marked by the loss of GFP (green), and all discs are stained for Dll (red) and D4/lacZ (blue). Left-hand panels show merged images of the entire disc. The central panels show an enlarged region, with the merged image at top. The right hand panels show cross sections at the same level as the central panels. Distal extension of clones from the ring is seen as downward extension in these cross sections. (A) 1 day old Antp- clones. Note in central panels that Antp- clones activate D4/lacZ in the distal part of the Dll ring, but not in the proximal portion. No distal extension of the D4/lacZ-expressing clones has taken place. (B) By day two, D4/lacZ-expressing clones are beginning to round up and distort the ring (central panels). Slight distal extension of these clones has occurred (right panels). (C) By day three, rounding up of D4/lacZ-expressing clones is advanced (middle panels), and significant distal extension is seen (right panels).
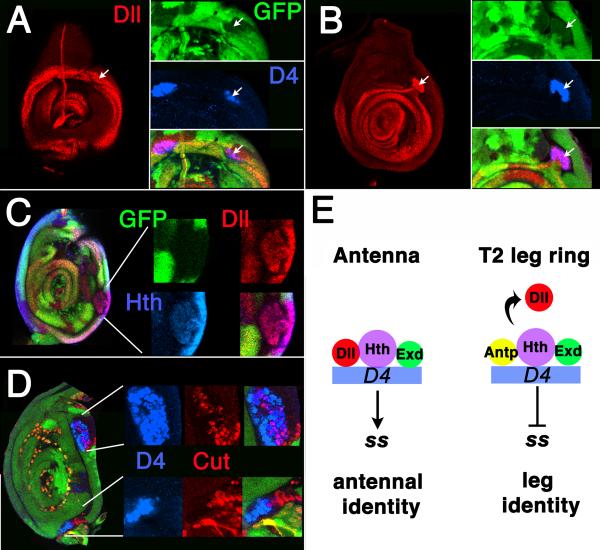
Figure 7.
In all panels, Antp- clones are marked by the loss of GFP (green). (A-B) A D4/lacZ-expressing Antp- clone in a second leg disc that extends from the proximal ring to the central domain of Dll expression. (A) Proximal focal plane, showing the Dll ring. Note activation of D4/lacZ in a clone overlapping the ring (arrows). (B) Distal focal plane, showing that the same transformed clone (arrows) connects to the distal domain of Dll expression. (C) A transformed Antp- clone in the second leg stained for Hth and Dll expression. Part of the clone has rounded up, but remains connected to the ring by a narrow isthmus. In some transformed clones, the connection is much narrower and thread-like. (D) Second leg disc containing Antp- clones stained for Cut and D4/lacZ expression. Note two rounded-up clones in which both Cut and D4/lacZ are expressed. Although Cut and D4/lacZ are coexpressed in many cells in the upper clone, Cut is expressed adjacent to a D4/lacZ expressing region in the bottom clone. (E) Model summarizing the control of D4 in the antenna and leg. Left: In the antenna, D4 is activated by binding of a Dll/Hth/Exd complex. Right: In the proximal ring of the leg, Antp displaces Dll and prevents activation of the enhancer.
Frequently, a subset of the cells in rounded-up clones expresses Cut, a marker for the A1 and A2 segments of the antenna (Fig. 7D). Cut-expressing and D4/lacZ expressing regions in such clones usually occupy distinct, although often overlapping, territories. Cut expression is usually not seen in Antp- clones of the interdigitated type, although sometimes Cut is weakly expressed in a few cells at the proximal edge of the ring in such clones. The emergence of Cut-expressing cells within transformed clones indicates that such clones can become organized internally to include distinct proximal (Cut-expressing) and distal (D4/lacZ-expressing) territories.
The overall picture that emerges is that Antp- clones in the second leg that lie proximal or distal to the ring of Dll, Hth, and Exd expression develop normally. However, antennal identity is triggered within clones that overlap this ring. Transformed clones then round up, become internally reorganized to include distinct proximal and distal territories, and appear to migrate or extend distally. The D4/lacZ reporter was of key importance in working out these events, as it allowed visualization of steps prior to the overt antennal transformation of Antp- clones.
Although Antp is expressed in a proximal ring in all three legs, Antp- clones show transformations to antenna only in the second leg (Struhl 1981, 1982a, Abbott and Kaufman 1986). A likely explanation is that antennal genes are repressed in the first and third legs by the Hox proteins Scr and Ubx, respectively, as well as by Antp (Struhl 1982a). Consistent with this possibility, we find that, like Antp, both Scr and Ubx repress D4/lacZ in the antenna when ectopically expressed on their own or in combination with Hth. In addition, both proteins bind D4 cooperatively with Hth and Exd (Suppl. Fig. 2).
Discussion
In this report we study the regulation of an enhancer from the antennal gene ss that drives expression specifically in the third antennal segment (A3). Our work provides the first look at how the homeodomain proteins Dll, Hth, and Exd function in the antenna to activate antennal target genes. We find that these proteins form a trimeric Dll/Hth/Exd complex on the enhancer, suggesting that Dll acts much like a Hox protein in antennal specification. Our work also reveals how the Hox protein Antp functions in the leg to repress antennal development. The conventional view has been that the primary function of Antp is to repress hth in the distal leg, which then prevents the activation of all downstream antennal genes. However, we find that Antp represses the ss A3 enhancer directly. This repression is essential within a proximal ring in the leg that coexpresses the antennal gene activators Dll, Hth, and Exd. We show that Antp competes with Dll for binding to the enhancer, and that this competition is part of a molecular switch that allows the ss A3 element to be activated in the antenna, but represses its activation in the leg (Fig. 7E). Our results suggest that repression of antenna-specific genes in the proximal ring is the sole function of Antp in the leg imaginal disc.
At 62 bp, the ss A3 enhancer (called D4) is one of the smallest enhancers to be identified in Drosophila, and yet it is quite strong; only a single copy is required to drive robust expression of lacZ reporters. The enhancer is also very specific, driving expression in A3 and nowhere else in imaginal discs. Dong et al. (2000) proposed that antennal identity in Drosophila is determined by the combined action of Dll, Hth, and Exd. Consistent with this proposal, we find that all three of these factors are required for D4 expression. Although these activators are coexpressed in both A2 and A3, D4/lacZ expression is restricted to A3 by Cut, which represses the enhancer in A2. Like ss itself (Duncan et al. 1998), D4/lacZ is also repressed by ectopically expressed Antp.
Surprisingly, Dll, Hth, Exd, Cut, and Antp all act directly upon D4. The activators Hth and Exd bind with strong cooperativity to directly adjacent sites. Their joint binding site matches the optimum site for in vitro binding of the mammalian homologs of Hth and Exd (Meis and Prep) (Chang et al. 1997), consistent with the robust activity of the enhancer in vivo. Mutation of either of these sites abolishes activity of the enhancer. The coactivator Dll binds three sites in D4; one of these sites (Dlla) is required for almost all activity of the enhancer. Dll shows strong cooperativity with Hth and Exd for binding to D4, indicating that Dll interacts physically with these proteins. This interaction requires DNA binding, as Dll protein containing a missense change that blocks DNA binding (a change of asn51 to ala in the homeodomain) shows no ability to associate with D4-bound Hth and Exd. A curious feature of the cooperativity seen in our binding studies is that although Hth and Exd increase the affinity of Dll for D4, Dll appears to have little effect on the affinity of Hth and Exd for the enhancer (see Fig. 3). Since Hth and Exd already bind cooperatively with one another, it may be that additional cooperative interactions with Dll have little effect. Alternatively, it may be that Hth and Exd interact with Dll only after binding DNA. If so, Hth and Exd would be expected to increase Dll binding to D4, but Dll would have little effect on the binding of Hth and Exd, as observed. Panganiban and Rubenstein (2002) have reported detecting interactions between Dll and Hth in the absence of DNA in immunoprecipitation experiments. However, we have been unable to repeat these observations (data not shown). Moreover, our finding that the asn51 mutant of Dll fails to associate with D4-bound Hth and Exd argues strongly against such interactions.
The repressor Cut also acts directly upon D4. Binding of Cut requires two sites, one overlapping Dlla and the other overlapping the joint Hth/Exd site. These binding sites suggest that D4 is controlled by Cut in much the same way that a structurally similar Abdominal-A (Abd-A) regulated enhancer from the rhomboid gene is controlled by the repressor Senseless (Sens) (Li-Kroeger et al. 2008). In the rhomboid enhancer, adjacent Hth and Exd sites are also present, and these create a binding site for Sens. Activity of the rhomboid enhancer is controlled by a competition between binding of the Sens repressor and binding of the activators Abd-A, Hth, and Exd. It seems likely that D4 is controlled similarly, with the repressor Cut competing for binding with the activators Dll, Hth, and Exd. It will be of interest to determine whether enhancers similar to D4 are used more widely to control Cut targets involved in its role as an external sense organ determinant.
A key finding in our work is that Antp represses D4 by direct interaction. We show that Antp binds a single site in D4, which overlaps or is identical to the Dlla binding site. Like Dll, Antp binds cooperatively with Hth and Exd. Using purified proteins, we show that binding of Dll and Antp to the Dlla site is mutually exclusive. This indicates that Antp represses the enhancer at least in part by competing with Dll for binding. Similar competition may occur at other enhancers; when Antp expression is driven artificially in the distal leg, variable deletions of the tarsal segments occur (Emerald and Cohen 2004). These defects might arise because Antp competes with Dll for binding to its target genes in the distal leg. In most other contexts examined, Antp is an activator of transcription (Capovilla et al. 2001; Winslow et al. 1989; Reuter and Scott 1990); why it fails to activate D4 is not clear. The similar behavior of Dll and Antp in binding to D4 supports the idea that Dll behaves like a Hox protein in activating D4.
Although our initial focus was on the antenna, the finding that Antp interacts directly with D4 led us to examine D4 regulation in the leg, where Antp is normally expressed. We find that in second leg imaginal discs, Antp is required only in a proximal ring of cells that coexpresses Dll and Hth. This ring appears in the early third instar, and is of uncertain function. Large Antp- clones in T2 leg discs that do not enter this ring appear to develop completely normally, regardless of whether they are located distal or proximal to the ring. However, clones that overlap the ring show activation of D4/lacZ within the ring cells. Importantly, such clones have no effect on the expression of Dll or Hth within the ring. By examining Antp- clones of increasing age the following sequence of events is inferred. First, D4/lacZ is activated in cells of the ring that are included within Antp- clones. Second, many such clones begin expressing the antennal markers Ss and Cut, indicating a transformation to antenna, and round up as if they have lost affinity for neighboring cells. Third, such clones appear to extend and move distally in the disc.
The events we describe for Antp- clones in the leg make sense of several previously enigmatic observations. Struhl (1981; 1982a) noted that many Antp- clones in the leg do not transform to antenna and appear to develop normally. Our finding that only clones that overlap the proximal ring undergo transformation accounts for this observation. Struhl also found that Antp- clones that do contain transformations usually show apparent nonautonomy in that not all cells in the clone are transformed to antenna. Our results account for this observation as well, since within an Antp- leg clone only those cells located in the proximal ring undergo transformation to antenna; cells located elsewhere in the clone retain normal leg identity. Most importantly, our observations provide an explanation for why ss is controlled directly by Antp. We find that Antp- clones have no effect on hth or Dll expression in the proximal ring. Therefore, Antp must function in the ring at the target gene level to repress antennal genes that would otherwise be activated by combined Hth and Dll (and Exd). Since several such targets are known (Dong et al. 2002), it seems likely that several, perhaps many, antennal genes in addition to ss are repressed directly by Antp.
The findings of McKay et al. (2009) challenge our inference that transformed Antp- clones extend or migrate distally in the leg. These authors show that distal migration of cells from the hth-expressing domain of the leg does not occur during normal development. However, coexpression of Dll and Hth in leg discs normally occurs only within the proximal ring, whereas such coexpression in the antenna extends far more distally, including all of A2 and A3. Therefore, Antp- cells from the proximal ring that transform to antenna likely assume a more distal identity as well as an altered segmental identity, perhaps allowing them to migrate more distally. Alternatively, it is possible that the transformed clones we interpret as having migrated distally were in fact generated early in leg development, when Hth expression overlaps Dll expression more distally in the leg primordium (McKay et al. 2009). We favor the first possibility because almost all transformed clones retain a clear, although sometimes tenuous, connection to the ring.
We confirm the finding of Casares and Mann (1998) that transformed Antp- clones in the leg often show ectopic hth expression in distal locations. If hth is not directly controlled by Antp in the leg, as we suggest, then why is hth ectopically expressed within such clones? A likely explanation is that downstream antennal genes that have become activated in such clones feed back to activate hth. This interpretation is strongly supported by the finding that ectopic expression of the antennal genes ss, dan, or danr in the distal leg causes ectopic activation of hth (Suzanne et al. 2003). Thus, the distal expression of hth seen in Antp- leg clones is likely a consequence rather than a cause of the transformation to antenna. Whether repression of hth in the antenna by ectopic Antp is also indirect is not clear. Dll is also expressed ectopically in transformed Antp- leg clones, suggesting that it is also subject to feedback activation by downstream antennal genes.
The function of the proximal Dll- and Hth-expressing ring in the proximal leg is not well understood. The ring is highly conserved among the insects (Angelini and Kaufman 2005), and may serve as a boundary between the proximal and distal portions of the legs (Wu and Cohen 1999; McKay et al. 2009). In the context of our work, a striking feature of the ring is that it contains a microcosm of gene expression domains corresponding to the three major antennal segments. Thus, proceeding from proximal to distal through the ring, cells express hth alone, hth + Dll, and hth + Dll + strong dachshund (Wu and Cohen 2000). These expression combinations are characteristic of the A1, A2, and A3 antennal segments, respectively. Looked at in this way, the ring would appear to resemble a repressed antennal primordium within the leg.
It has been known for almost thirty years that Antp is required in the leg to repress antennal identity. However, an understanding of how this repression occurs has been lacking. Our results indicate that Antp functions within the proximal ring to directly repress antennal genes that would otherwise be activated by combined expression of Dll, Hth, and Exd. This appears to be the only function of Antp in the leg, at least during the third instar larval stage. Our results are entirely consistent with the ideas of Struhl (1981; 1982a), who argued that second leg is the “ground state” ventral appendage (the limb type that develops in the absence of identity specification) and that the role of Antp in the leg is to preserve this ground state by repressing the activation of “head-determining” genes.
Experimental Procedures
Antibody Staining
Antibody stainings were performed as described previously (Kankel et al. 2004). Primary antibodies used were mouse anti Dll (Duncan et al. 1998), rabbit anti Dll (gift of Grace Boekhoff-Falk), rabbit anti Hth (gift of A. Salzberg), mouse anti Ubx and mouse anti Exd (gifts of R. White), guinea pig anti Ss (gift of Michael Kim), mouse anti Cut, mouse anti Scr, and mouse anti Antp (all from the Developmental Studies Hybridoma Bank), mouse anti β-galactosidase (Promega), and rabbit anti β-galactosidase (Cappel). Secondary antibodies used were Cy3 donkey anti rabbit, Cy3 donkey anti mouse, Cy3 donkey anti guinea pig, Cy5 donkey anti rabbit, Cy5 donkey anti mouse (Jackson), and FITC goat anti rabbit (Cappel). Images were captured on a Nikon A1 scanning confocal microscope.
Gel shift assays
Unless otherwise noted, all chemicals were from Sigma-Aldrich. The sequences of the oligonucleotides used as probes are in Figure 5. Oligonucleotides were labeled with α 32P dCTP (Perkin Elmer) using the Klenow fragment of DNA pol I (New England Biolabs). 10 to 50 ng of annealed oligonucleotide was used per reaction. Unincorporated label was removed with P6DG spin columns (Biorad) and amounts were normalized using DE81 filters (Whatman).
All proteins were produced by in vitro translation using the TnT T7 Rabbit Reticulocyte Lysate kit (Promega). 1 μg of circular plasmid DNA was used per reaction and 5% of the translation was incubated with 35S methionine to assay translation efficiency. The unlabelled translated protein was used without further purification. 1 to 10 μl of in vitro translation reaction product were used per reaction. Total protein was kept constant among samples in an experiment by addition of control luciferase translations. Luciferase translations were also used in control lanes to assess non-specific binding of proteins in the lysate. Poly (dI.dC) was used to reduce nonspecific binding. For super-shift experiments, 1 μl of antibody (1:10 dilution in PBS) was added for the final 5 minutes of incubation prior to gel loading.
The plasmids used for in vitro translation were as follows: Dll, Hth, and Exd constructs contain the full-length coding regions of the respective genes cloned into pT7βplink (Dalton and Treisman 1992), a generous gift of G. Boekhoff-Falk. The Dll coding sequence used includes an additional 20 codons relative to the standard sequence due to alternate splicing between exons 2 and 3. Antp, Scr and Ubx constructs contain the full length coding regions cloned into pTnT (Promega). The Cut construct includes nucleotides 2632-5434 (numbering as in Blochlinger et al. 1988) of the cut cDNA, which includes the coding sequences for all three Cut domains and the homeodomain, cloned into pTnT.
For competition assays, in vitro translation products were purified from the lysate and concentrated using oligonucleotide selection as described by Ozyhar et al. 1992. Based on Coomassie staining of pre- and post-purification lysate, greater than 90% of nonspecific lysate protein was removed by this protocol. Copper phenanthroline footprinting of shifted bands excised from gels was as described by Sigman et al. (1991).
Generation of lacZ reporter lines
Deletion derivatives of the ss522 sequence were generated by recombinant PCR, verified by sequencing, and subcloned into either pCaSpeR-hs43-βgal (Thummel and Pirrotta, 1992) or placZattB (Bischof et al. 2007). pCaSper-hs43-βgal constructs were transformed into y w67c23 flies by standard methods, and a minimum of 5 separate insertions per construct assayed. placZattB constructs were transformed as described by Bischof et al. (2007). For placZattB, all integrations were at a site at 22A. X-Gal staining was as described by Emmons et al. (2007).
Mitotic recombination clones: A D4/lacZ reporter line containing a dimer of the D4 sequence inserted at the 22A site (line 81d42), was used in almost all experiments. In a few early experiments, a P-element lacZ reporter containing a multimer of D4 was used and gave similar results. Clones were generated by the FLP-FRT method using the following chromosomes: exd1 FRT18E, cut145 FRT18E, FRT82B hth64-1, FRT82B Antp25, and FRT42D DllSA1. In all cases, mitotic recombination clones were identified in discs by the loss of the Ubi-GFP marker. hs-FLP122 and hs-FLP38 were used as sources of recombinase. Crosses were made in plastic vials, and cultures were immersed for 30 minutes (hs-FLP122) or 1 hr (hs-FLP38) in a water bath at 37° to induce recombinase expression.
Ectopic expression clones: Males carrying appropriate UAS constructs were crossed to y w hs-FLP12/y w hs-FLP12; D4lacZ 81d42/D4lacZ 81d42; Act>y+>Gal4 UAS-GFP/TM6B, Tb females or to y w hs-FLP12/y w hs-FLP12; Act>y+>Gal4 UAS-GFP/TM6B, Tb females. Clones were induced by immersion at 37° for 8 minutes. The UAS lines used were UAS-Cut (provided by Helen McNeil), UAS-Hth (line 12; provided by Henry Sun), UAS-Antp (provided by T. Kaufman), and UAS-Scr, UASUbx, and UAS-Dll (all from the Bloomington Stock Center).
Supplementary Material
Acknowledgements
We thank Grace Boekhoff-Falk, Steve Cohen, Tom Kaufman, Michael Kim, Richard Mann, Tony Percival-Smith, Adi Salzberg, and Rob White for providing stocks and reagents. We are particularly grateful to Yehuda Ben-Shahar, Doug Chalker, and Jim Skeath for discussions and help with the manuscript, and to an anonymous reviewer for drawing our attention to the asymmetric nature of the cooperativity of Dll with Hth and Exd. Our work was supported by a grant from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott MK, Kaufman TC. The relationship between the functional complexity and the molecular organization of the Antennapedia locus of Drosophila melanogaster. Genetics. 1986;114:919–942. doi: 10.1093/genetics/114.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Shaar M, Mann RS. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development. 1998;125:3821–3830. doi: 10.1242/dev.125.19.3821. [DOI] [PubMed] [Google Scholar]
- Ades SE, Sauer RT. Specificity of minor-groove and major-groove interactions in a homeodomain-DNA complex. Biochemistry. 1995;34:14601–14608. doi: 10.1021/bi00044a040. [DOI] [PubMed] [Google Scholar]
- Angelini DR, Kaufman TC. Insect appendages and comparative ontogenetics. Dev Biol. 2005;286:57–77. doi: 10.1016/j.ydbio.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K, Bodmer R, Jack J, Jan LY, Jan YN. Primary structure and expression of a product from cut, a locus involved in specifying sensory organ identity in Drosophila. Nature. 1988;333:629–635. doi: 10.1038/333629a0. [DOI] [PubMed] [Google Scholar]
- Capovilla M, Kambris Z, Botas J. Direct regulation of the muscle-identity gene apterous by a Hox protein in the somatic mesoderm. Development. 2001;128:1221–1230. doi: 10.1242/dev.128.8.1221. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. The ground state of the ventral appendage in Drosophila. Science. 2001;293:1477–1480. doi: 10.1126/science.1062542. [DOI] [PubMed] [Google Scholar]
- Chang C-P, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric PBX proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Brönner G, Küttner F, Jürgens G, Jäckle H. Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature. 1989;338:432–434. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- Cohen SM, Jürgens G. Proximal-distal pattern formation in Drosophila: cell autonomous requirement for Distal-less gene activity in limb development. EMBO J. 1989;8:2045–2055. doi: 10.1002/j.1460-2075.1989.tb03613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- Dong PDS, Chu J, Panganiban G. Coexpression of the homeobox genes Distalless and homothorax determines Drosophila antennal identity. Development. 2000;127:209–216. doi: 10.1242/dev.127.2.209. [DOI] [PubMed] [Google Scholar]
- Dong PDS, Dicks JS, Panganiban G. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development. 2002;129:1967–1974. doi: 10.1242/dev.129.8.1967. [DOI] [PubMed] [Google Scholar]
- Duncan DM, Burgess EA, Duncan I. Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 1998;12:1290–1303. doi: 10.1101/gad.12.9.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerald BS, Cohen SM. Spatial and temporal regulation of the homeotic selector gene Antennapedia is required for the establishment of leg identity in Drosophila. Dev Biol. 2004;267:462–472. doi: 10.1016/j.ydbio.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Emmons RB, Duncan D, Duncan I. Regulation of the Drosophila distal antennal determinant spineless. Dev Biol. 2007;302:412–426. doi: 10.1016/j.ydbio.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischer LE, Hagen FS, Garber RL. An inversion that disrupts the Antennapedia gene causes abnormal structure and localization of RNAs. Cell. 1986;47:1017–1023. doi: 10.1016/0092-8674(86)90816-0. [DOI] [PubMed] [Google Scholar]
- González-Crespo S, Morata G. Control of Drosophila adult pattern by extradenticle. Development. 1995;121:2117–2125. doi: 10.1242/dev.121.7.2117. [DOI] [PubMed] [Google Scholar]
- Kankel MW, Duncan DM, Duncan I. A screen for genes that interact with the Drosophila pair-rule segmentation gene fushi tarazu. Genetics. 2004;168:161–180. doi: 10.1534/genetics.104.027250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant E, Pai C-Y, Sharf R, Halachmi N, Sun YH, Salzberg A. dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- Li-Kroeger D, Witt LM, Grimes HL, Cook TA, Gebelein B. Hox and Senseless antagonism functions as a molecular switch to regulate EGF secretion in the Drosophila PNS. Dev Cell. 2008;15:298–308. doi: 10.1016/j.devcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. Hox specificity: Unique roles for cofactors and collaborators. Cur. Top. Dev. Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ, Estella C, Mann RS. The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development. 2009;136:61–71. doi: 10.1242/dev.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factors in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Ozyhar A, Gries M, Kiltz HH, Pongs O. Magnetic DNA affinity purification of ecdysteroid receptor. J Steroid Biochem Mol Biol. 1992;43:629–634. doi: 10.1016/0960-0760(92)90287-s. [DOI] [PubMed] [Google Scholar]
- Pai C-Y, Kuo T-S, Jaw TJ, Kurant E, Chen C-T, Bessarab DA, Salzberg A, Sun YH. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, Extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panganiban G, Rubenstein JLR. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- Reuter R, Scott MP. Expression and function of the homeotic genes Antennapedia and Sex combs reduced in the embryonic midgut of Drosophila. Development. 1990;109:289–303. doi: 10.1242/dev.109.2.289. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of Extradenticle requires homothorax, which encodes an Extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Sigman DS, Kuwabara MD, Chen CH, Bruice TW. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 1991;208:414–433. doi: 10.1016/0076-6879(91)08022-a. [DOI] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. Website: http://genome.ucsc.edu/. Release date April 2006 (BDGP R5/dm3) [DOI] [PMC free article] [PubMed]
- Struhl G. A homeotic mutation transforming leg to antenna in Drosophila. Nature. 1981;292:635–638. doi: 10.1038/292635a0. [DOI] [PubMed] [Google Scholar]
- Struhl G. Genes controlling segmental specification in the Drosophila thorax. Proc Natl Acad Sci USA. 1982a;79:7380–7384. doi: 10.1073/pnas.79.23.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. spineless-aristapedia: a homeotic gene that does not control the development of specific compartments in Drosophila. Genetics. 1982b;102:737–749. doi: 10.1093/genetics/102.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel CE, Whittle JRS. Brista: a gene involved in the specification and differentiation of distal cephalic structures in Drosophila melanogaster. Wilhelm Roux Arch Dev Biol. 1987;196:124–132. doi: 10.1007/BF00402034. [DOI] [PubMed] [Google Scholar]
- Suzanne M, Estella C, Calleja M, Sánchez-Herrero E. The hernandez and fernandez genes of Drosophila specify eye and antenna. Dev Biol. 2003;260:465–483. doi: 10.1016/s0012-1606(03)00249-5. [DOI] [PubMed] [Google Scholar]
- Thummel C, Pirrotta V. New pCaSpeR P-element vectors. Drosoph Inf Serv. 1992;71:150. [Google Scholar]
- Winslow GM, Hayashi S, Krasnow M, Hogness DS, Scott MP. Transcriptional activation by the Antennapedia and fushi tarazu proteins in cultured Drosophila cells. Cell. 1989;57:1017–1030. doi: 10.1016/0092-8674(89)90340-1. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Proximodistal axis formation in the Drosophila leg: subdivision into proximal and distal domains by Homothorax and Distal-less. Development. 1999;126:109–117. doi: 10.1242/dev.126.1.109. [DOI] [PubMed] [Google Scholar]
- Wu J, Cohen SM. Proximal distal axis formation in the Drosophila leg: distinct functions of Teashirt and Homothorax in the proximal leg. Mech Dev. 2000;94:47–56. doi: 10.1016/s0925-4773(00)00311-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.